Investigation of the Stereochemical-Dependent DNA and RNA Binding of Arginine-Based Nucleopeptides
Abstract
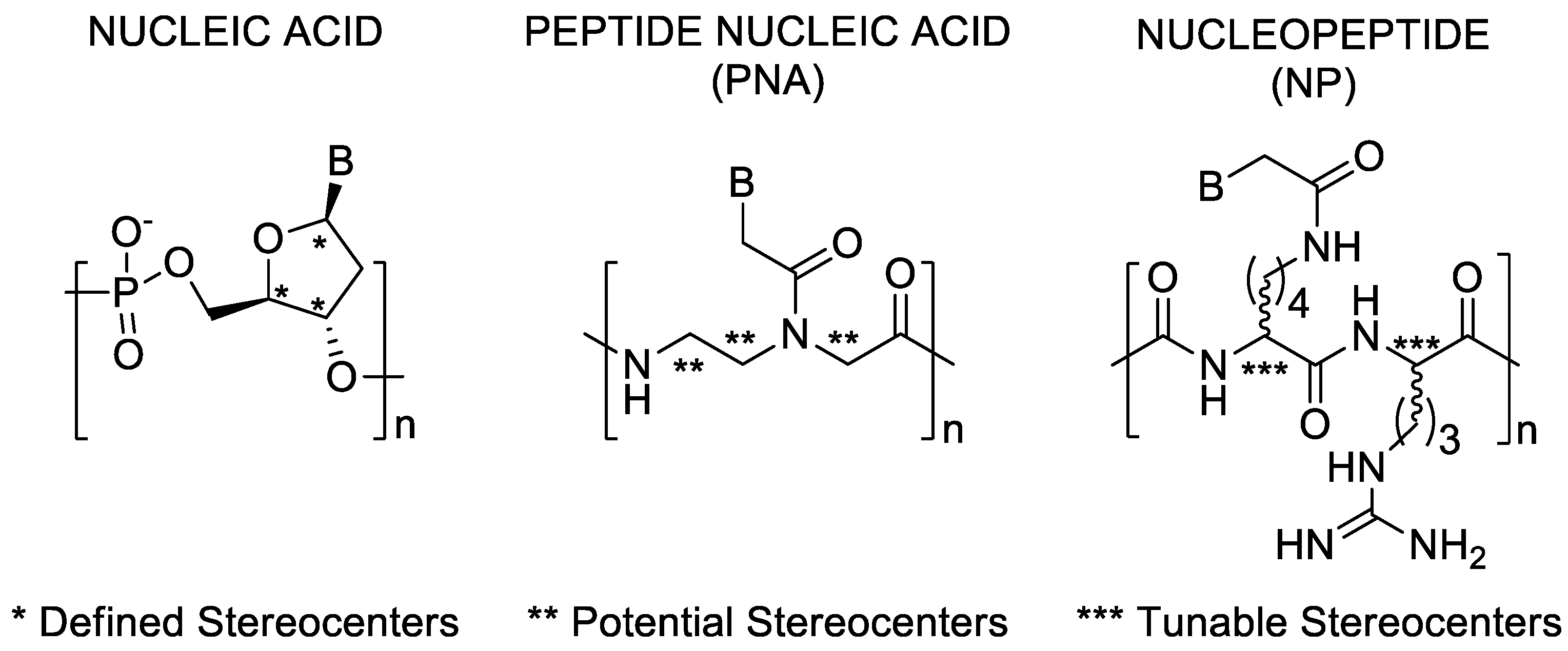
1. Introduction
2. Materials and Methods
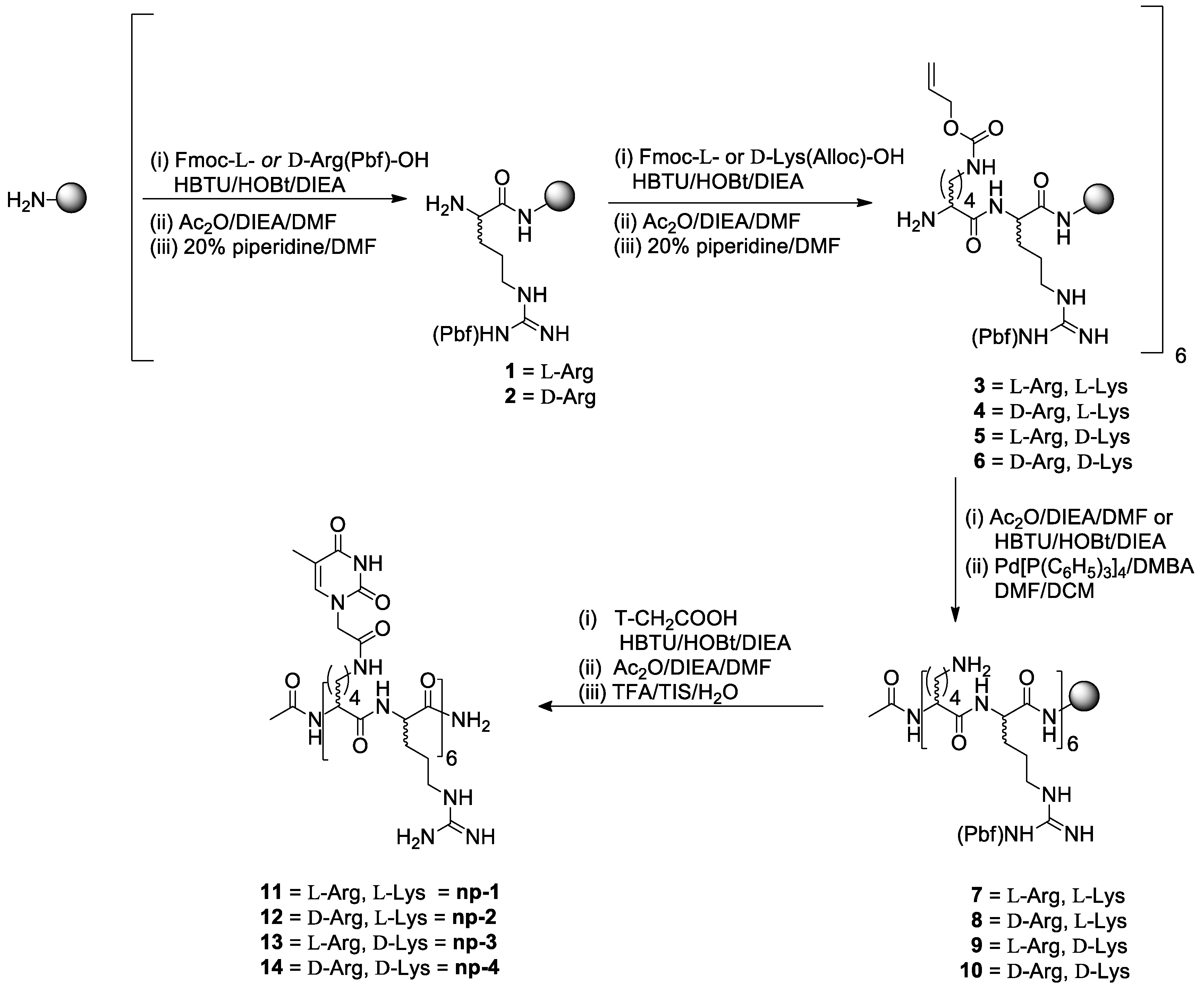
2.1. Synthesis of Nucleopeptides np-1/4
2.2. Cleavage, HPLC Purification and Characterization of Nucleopeptides np-1/4
2.3. UV and CD procedures
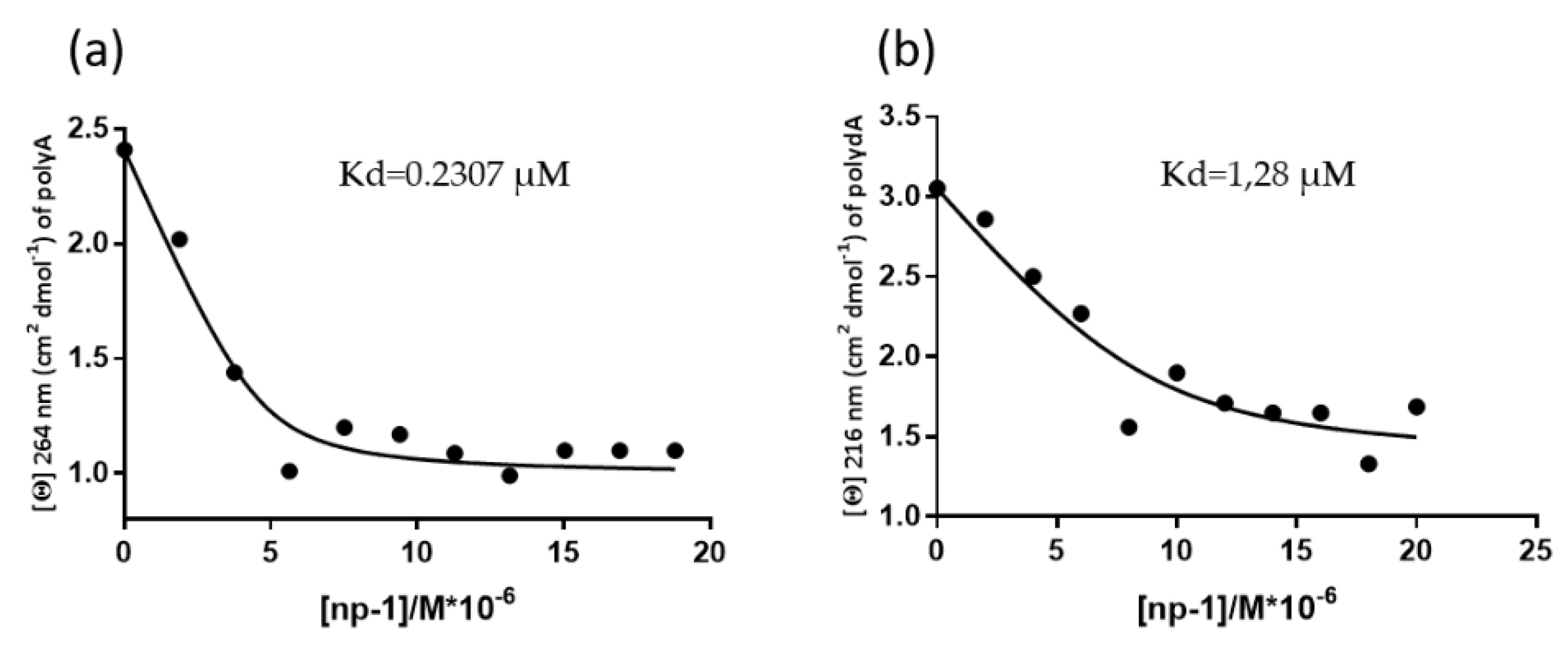
2.4. CD Measurements of Apparent Kd Determination of np-1/polyA and np1/polydA
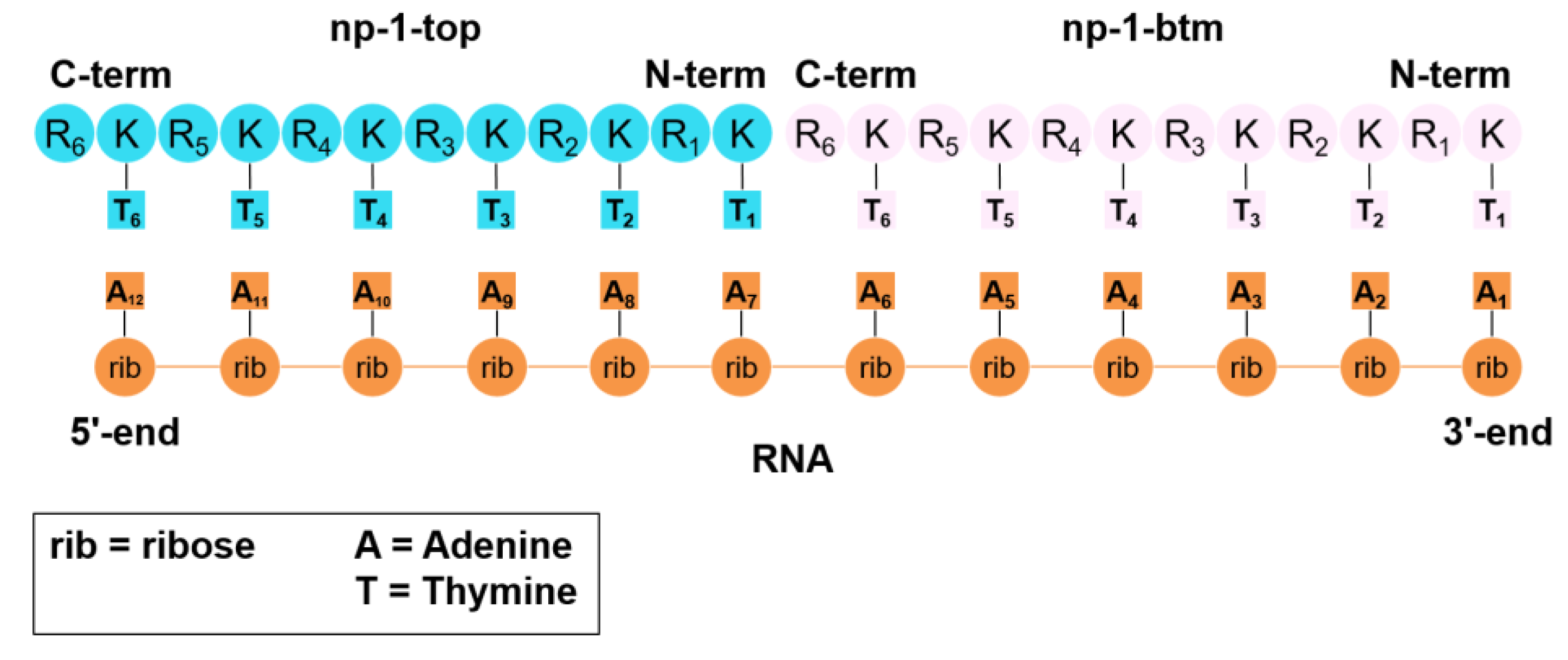
2.5. Building of np-1/RNA Complex
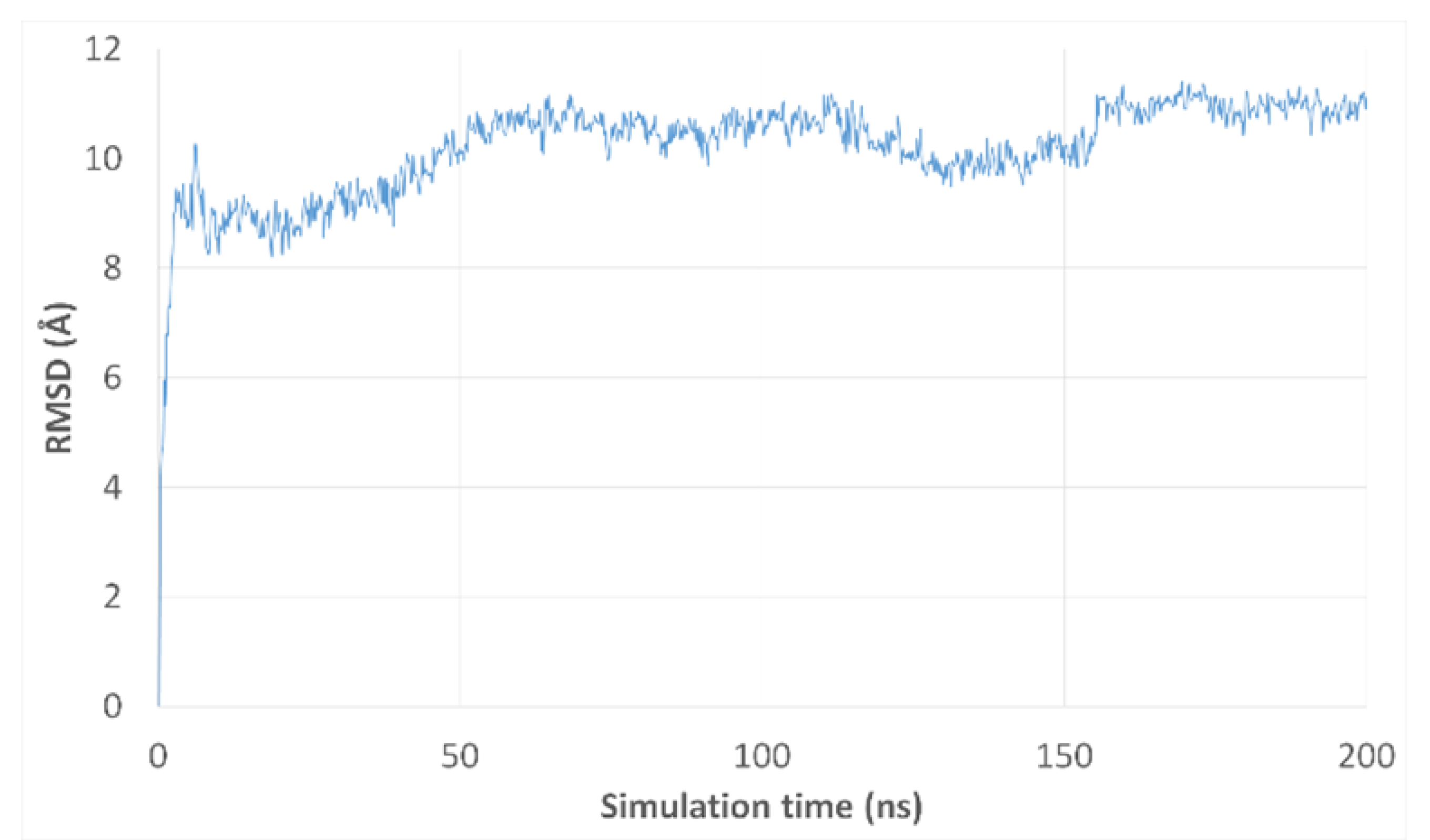
2.6. Molecular Dynamics Simulation
2.7. RMSD Clustering Analysis
3. Results and Discussion
3.1. Synthesis
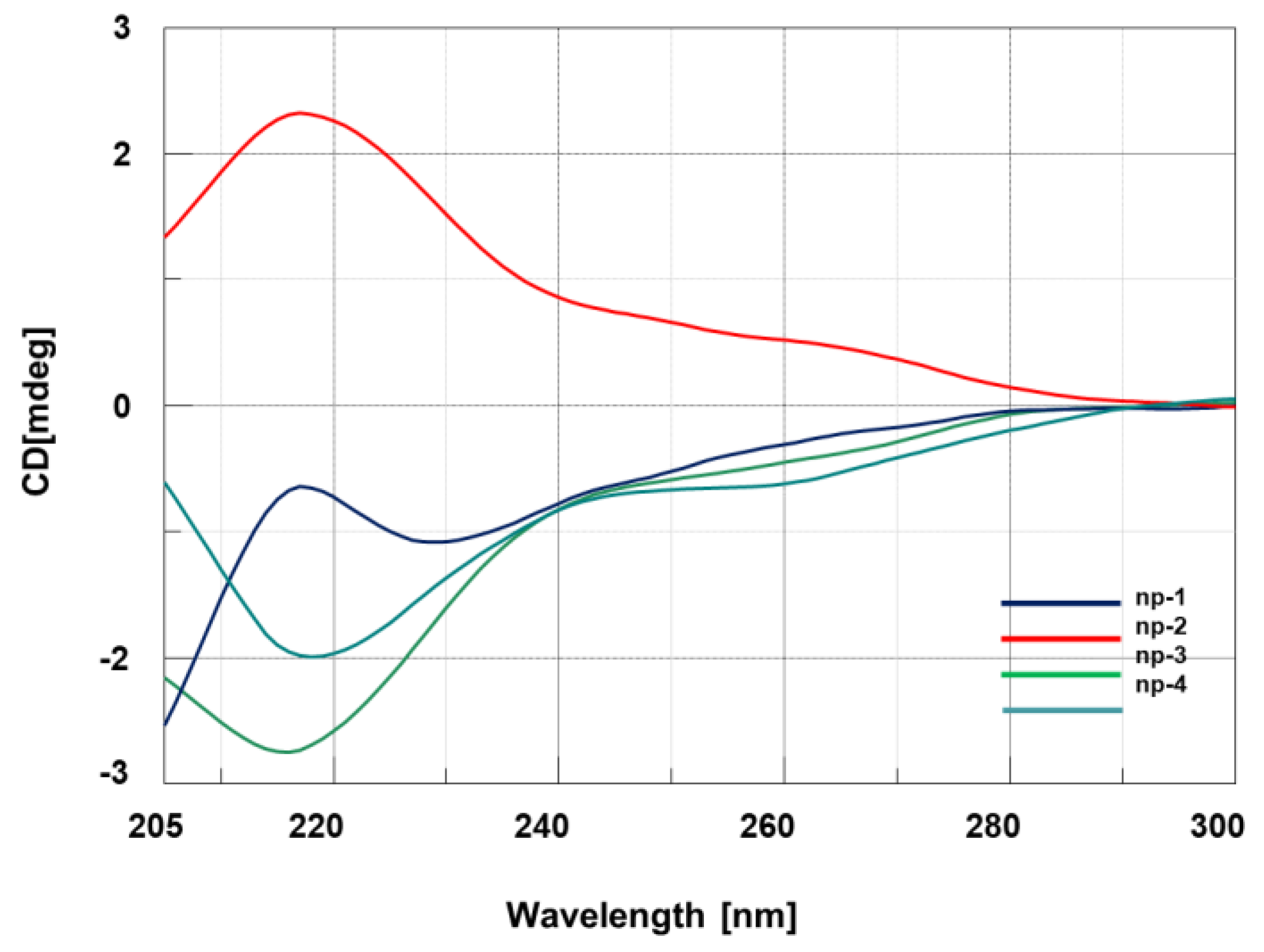
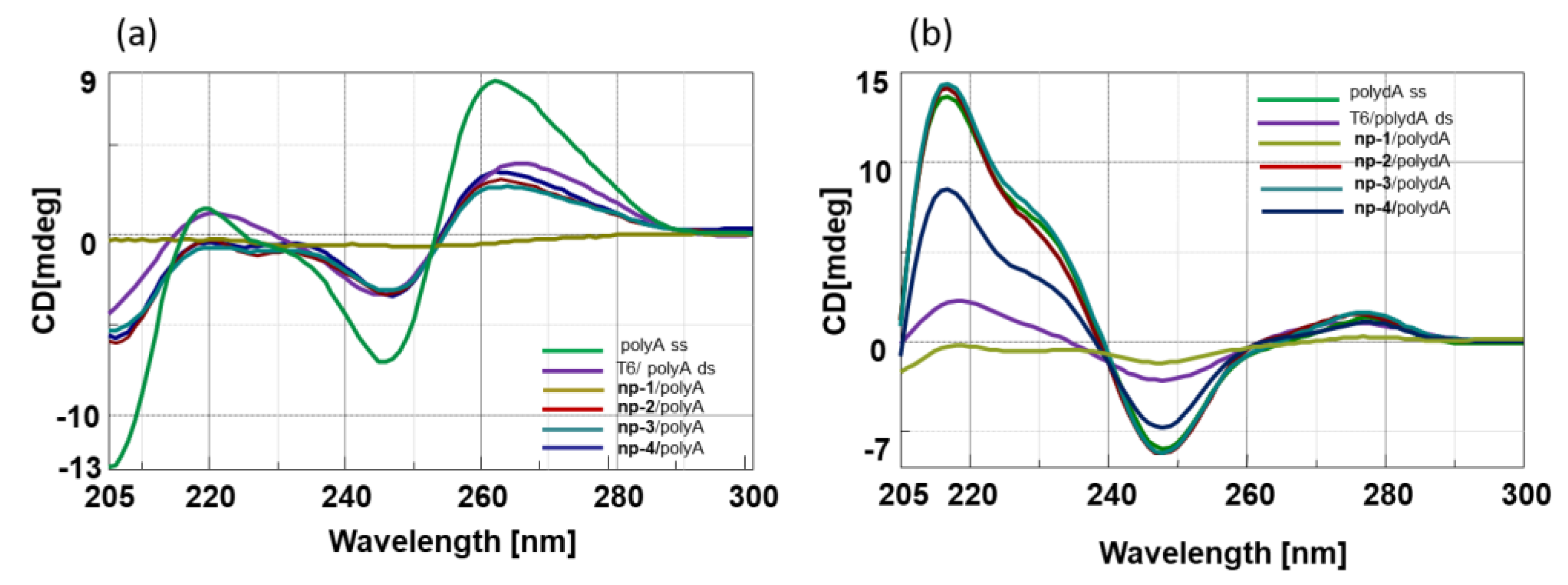
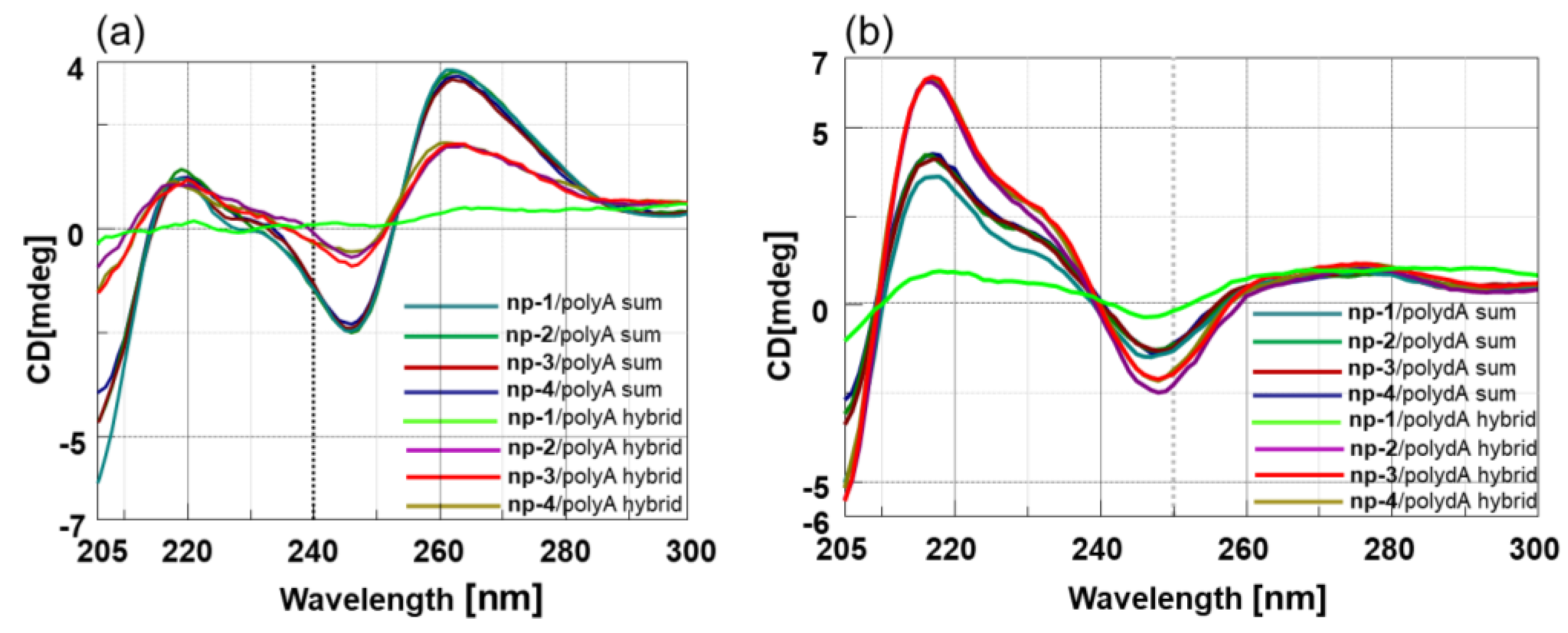
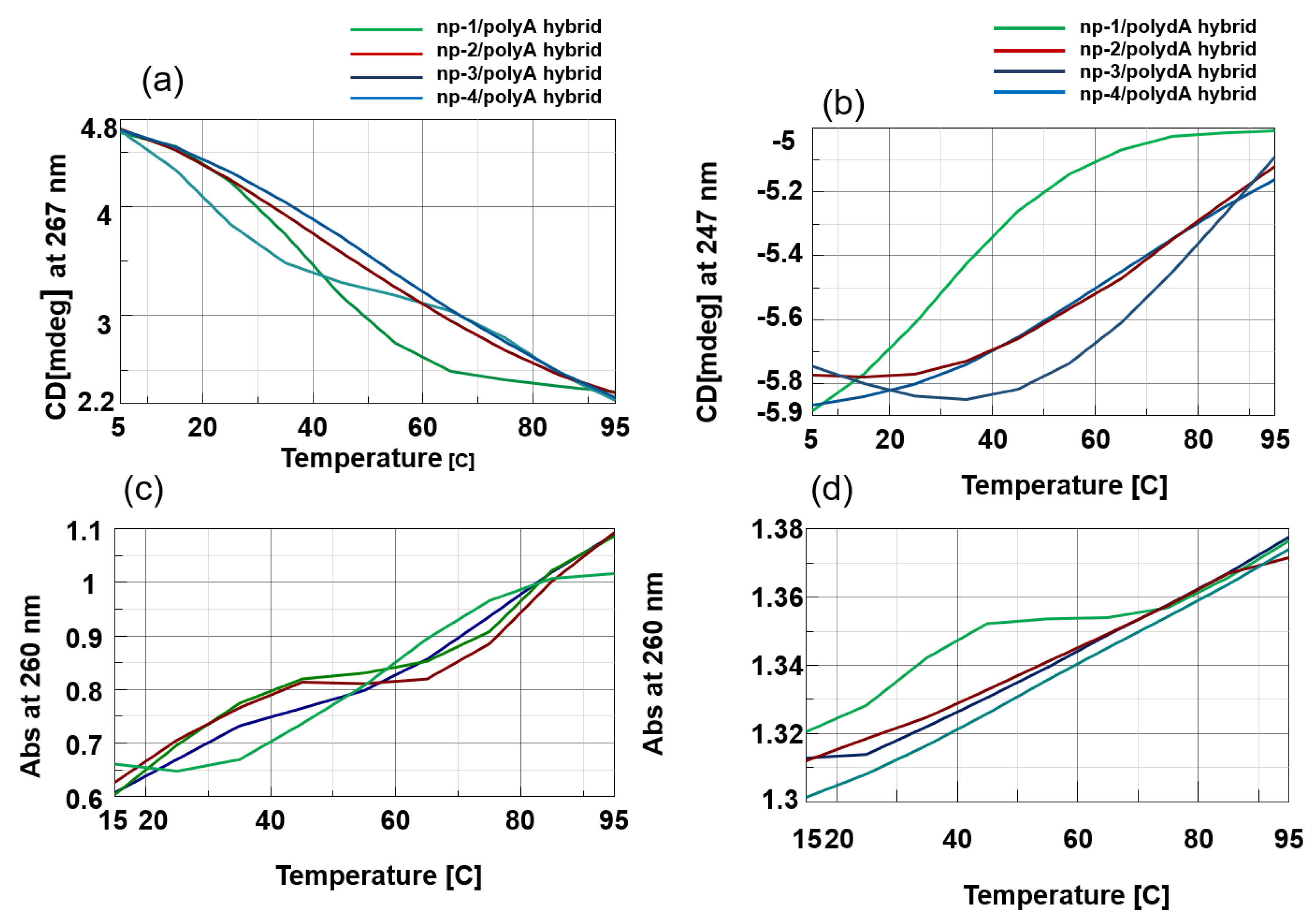
3.2. CD Spectroscopic Studies
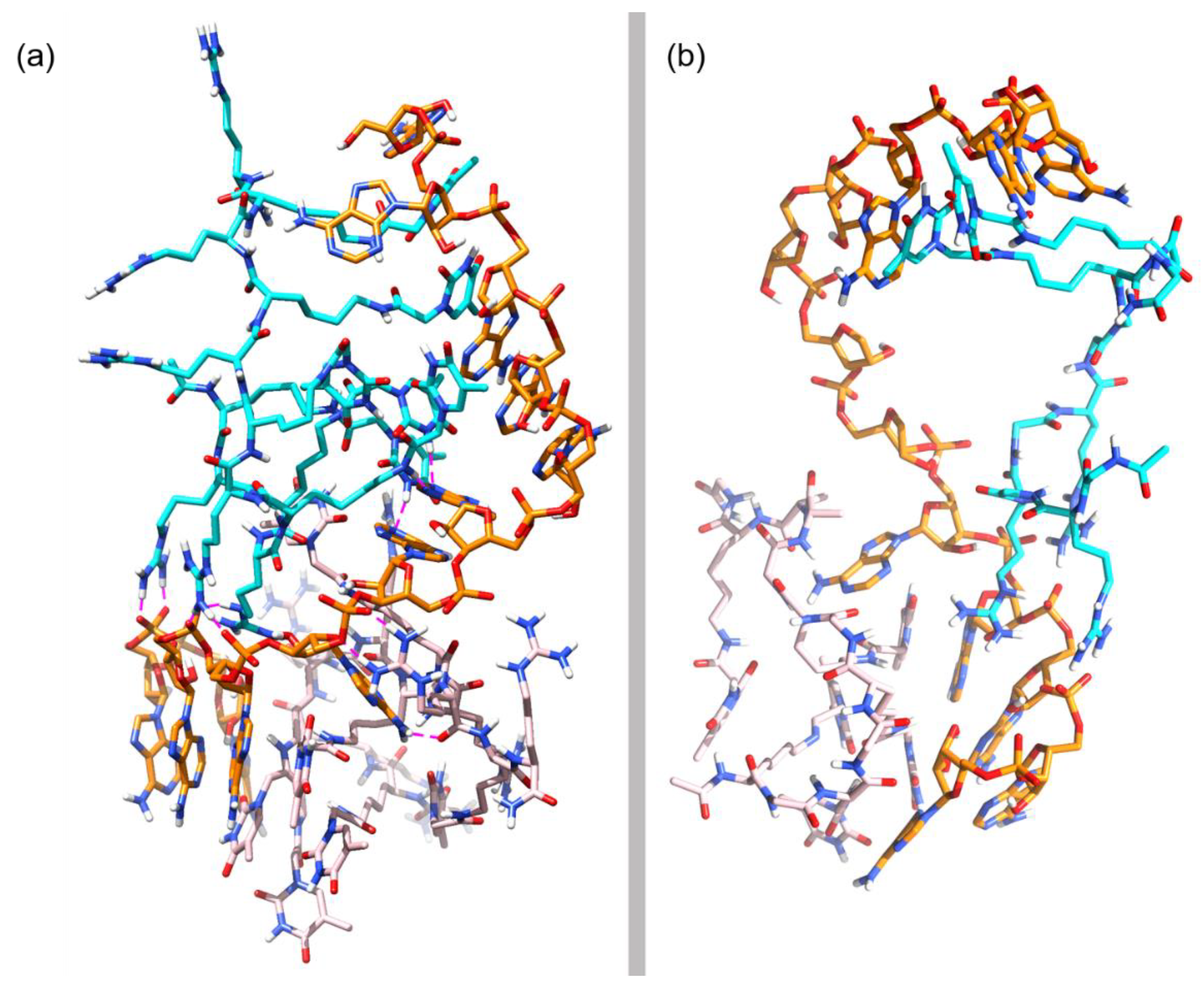
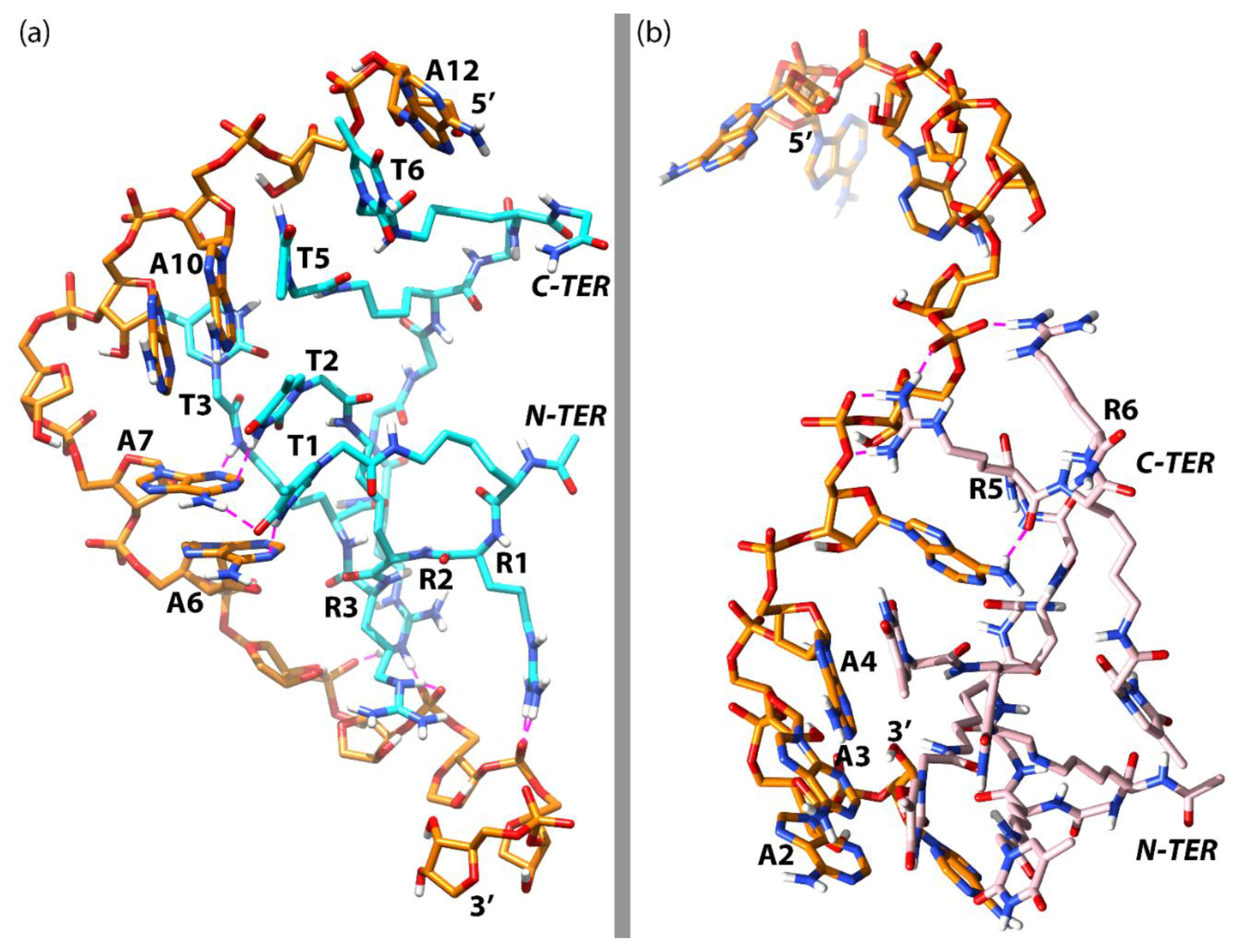
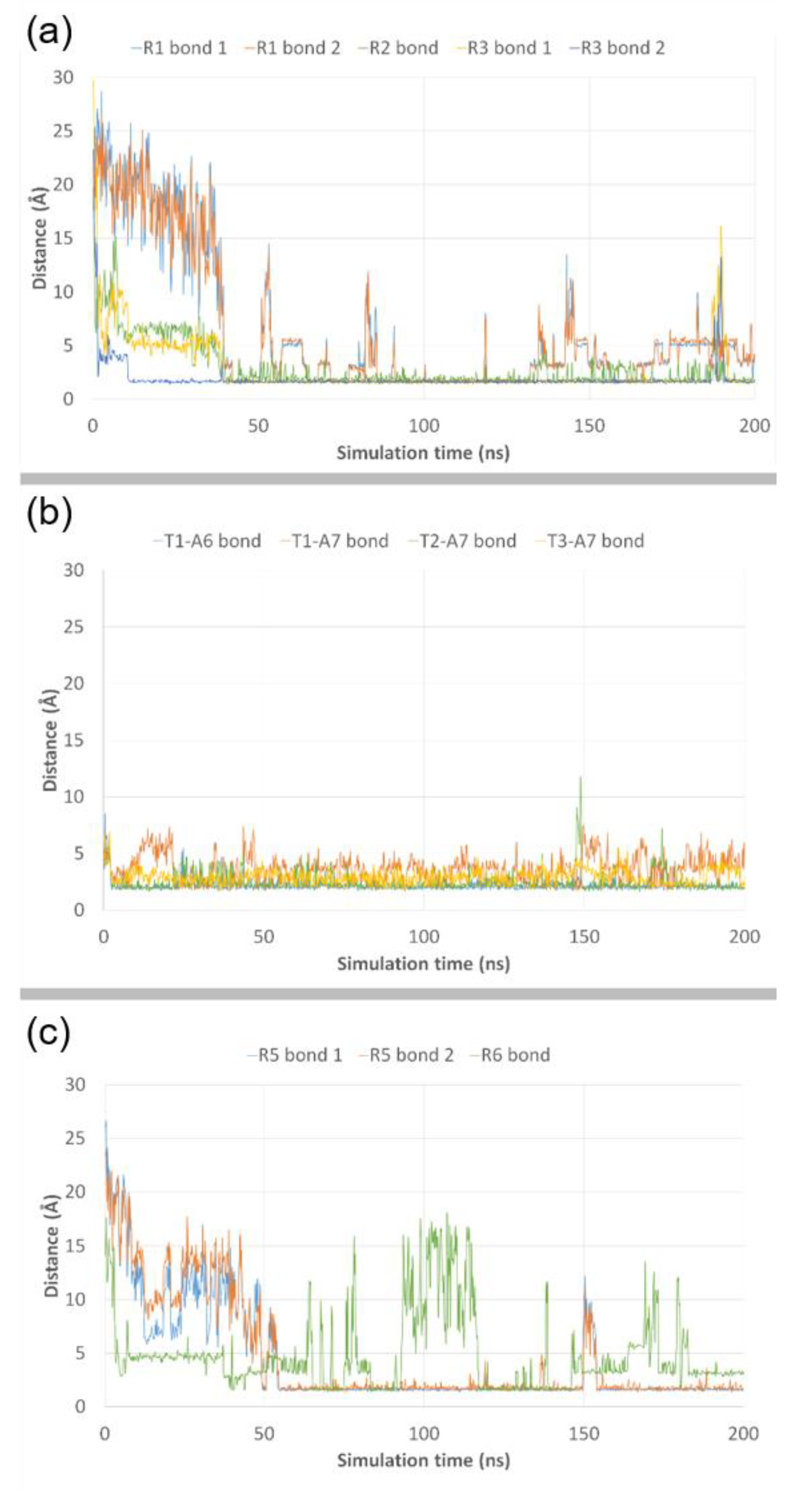
3.3. Molecular Dynamics Simulations
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Caprara, M.G.; Nilsen, T.W. RNA: Versatility in form and function. Nat. Struct. Biol. 2000, 7, 831–833. [Google Scholar] [CrossRef]
- Hudson, W.H.; Ortlund, E.A. The structure, function and evolution of proteins that bind DNA and RNA. Nat. Rev. Mol. Cell Biol. 2014, 15, 749–760. [Google Scholar] [CrossRef]
- Brennan, R.G.; Matthews, B.W. The helix-turn-helix DNA binding motif. J. Biol. Chem. 1989, 264, 1903–1906. [Google Scholar]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef]
- Vinson, C.R.; Sigler, P.B.; McKnight, S.L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science 1989, 246, 911–916. [Google Scholar] [CrossRef]
- Pavletich, N.P.; Pabo, C.O. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 A. Science 1991, 252, 809–817. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J.J. Cell-type-specific: Aptamer-functionalized agents for targeted disease therapy. Mol. Ther. Nucleic Acids 2014, 3, e169. [Google Scholar] [CrossRef]
- Vaught, J.D.; Bock, C.; Carter, J.; Fitzwater, T.; Otis, M.; Schneider, D.; Rolando, J.; Waugh, S.; Wilcox, S.K.; Eaton, B.E. Expanding the chemistry of DNA for in vitro selection. J. Am. Chem. Soc. 2010, 132, 4141–4151. [Google Scholar] [CrossRef]
- Gupta, S.; Hirota, M.; Waugh, S.M.; Murakami, I.; Suzuki, T.; Muraguchi, M.; Shibamori, M.; Ishikawa, Y.; Jarvis, T.C.; Carter, J.D.; et al. Chemically Modified DNA Aptamers Bind Interleukin-6 with High Affinity and Inhibit Signaling by Blocking Its Interaction with Interleukin-6 Receptor. J. Biol. Chem. 2014, 289, 8706–8719. [Google Scholar] [CrossRef]
- Matsumura, S.; Takahashi, T.; Ueno, A.; Mihara, H. Complementary nucleobase interaction enhances peptide-peptide recognition and self-replicating catalysis. Chem.-A Eur. J. 2003, 9, 4829–4837. [Google Scholar] [CrossRef]
- Matsumura, S.; Ueno, A.; Mihara, H. Peptides with nucleobase moieties as a stabilizing factor for a two-stranded R-helix. Chem. Commun. 2000, 1615–1616. [Google Scholar] [CrossRef]
- Takahashi, T.; Hamasaki, K.; Kumagai, I.; Ueno, A.; Mihara, H. Design of a nucleobase-conjugated peptide that recognizes HIV-1 RRE IIB RNA with high affinity and specificity. Chem. Commun. 2000, 349–350. [Google Scholar] [CrossRef]
- Takahashi, T.; Hamasaki, K.; Ueno, A.; Mihara, H. Construction of peptides with nucleobase amino acids: Design and synthesis of the nucleobase-conjugated peptides derived from HIV-1 Rev and their binding properties to HIV-1 RRE RNA. Bioorg. Med. Chem. 2001, 9, 991–1000. [Google Scholar] [CrossRef]
- Takahashi, T.; Ueno, A.; Mihara, H. Nucleobase amino acids incorporated into HIV-1 nucleocapsid protein increased the binding affinity and specificity to a hairpin RNA. ChemBioChem. 2002, 3, 543–549. [Google Scholar] [CrossRef]
- Takahashi, T.; Yana, D.; Mihara, H. Utilization of l-a-Nucleobase Amino Acids (NBAs) as Protein Engineering Tools: Construction of NBA-Modified HIV-1 Protease Analogues and Enhancement of Dimerization Induced by Nucleobase Interaction. ChemBioChem. 2006, 7, 729–732. [Google Scholar] [CrossRef]
- Bai, X.; Talukder, P.; Daskalova, S.M.; Roy, B.; Chen, S.; Li, Z.; Dedkova, L.M.; Hecht, S.M. Enhanced Binding Affinity for an i-Motif DNA Substrate Exhibited by a Protein Containing Nucleobase Amino Acids. J. Am. Chem. Soc. 2017, 139, 4611–4614. [Google Scholar] [CrossRef]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef]
- Egholm, M.; Buchardt, O.; Christensen, L.; Behrens, C.; Freier, S.M.; Driver, D.; Berg, R.H.; Kim, S.K.; Norden, B.; Nielsen, P.E. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature 1993, 365, 566–568. [Google Scholar] [CrossRef]
- Wittung, P.; Nielsen, P.E.; Buchardt, O.; Egholm, M.; Norden, B. DNA-like double helix formed by peptide nucleic acid. Nature 1994, 368, 561–563. [Google Scholar] [CrossRef]
- Nielsen, P.E. Peptide nucleic acid: A versatile tool in genetic diagnostics and molecular biology. Curr. Opin. Biotechnol. 2001, 12, 16–20. [Google Scholar] [CrossRef]
- Nielsen, P.E. Peptide Nucleic Acid. A Molecule with Two Identities. Acc. Chem. Res. 1999, 32, 624–630. [Google Scholar] [CrossRef]
- Ganesh, K.N.; Nielsen, P.E. Peptide Nucleic Acids Analogs and Derivatives. Curr Org. Chem. 2000, 4, 931–943. [Google Scholar] [CrossRef]
- Kumar, V.A.; Ganesh, K.N. Conformationally Constrained PNA Analogues: Structural Evolution toward DNA/RNA Binding Selectivity. Acc. Chem. Res. 2005, 38, 404–412. [Google Scholar] [CrossRef]
- Kumar, V.A.; Ganesh, K.N. Structure-Editing of Nucleic Acids for Selective Targeting of RNA. Curr. Top. Med. Chem. 2007, 7, 715–726. [Google Scholar] [CrossRef]
- Sforza, S.; Corradini, R.; Ghirardi, S.; Dossena, A.; Marchelli, R. DNA Binding of A D-Lysine-Based Chiral PNA: Direction Control and Mismatch Recognition. Eur. J. Org. Chem. 2000, 2905–2913. [Google Scholar] [CrossRef]
- Tedeschi, T.; Sforza, S.; Dossena, A.; Corradini, R.; Marchelli, R. Lysine-based peptide nucleic acids (PNAs) with strong chiral constraint: Control of helix handedness and DNA binding by chirality. Chirality 2005, 17, 196–204. [Google Scholar] [CrossRef]
- Bose, T.; Banerjee, A.; Nahar, S.; Maiti, S.; Kumar, V.A. β,γ-Bis-substituted PNA with configurational and conformational switch: Preferred binding to cDNA/RNA and cell-uptake studies. Chem. Commun. 2015, 51, 7693–7696. [Google Scholar] [CrossRef]
- Futaki, S.; Nakase, I. Cell-Surface Interactions on Arginine-Rich Cell-Penetrating Peptides Allow for Multiplex Modes of Internalization. Acc. Chem. Res. 2017, 50, 2449–2456. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Wang, M.; Du, L.; Fisher, G.W.; Waggoner, A.; Ly, D.H. Novel Binding and Efficient Cellular Uptake of Guanidine-Based Peptide Nucleic Acids (GPNA). J. Am. Chem. Soc. 2003, 125, 6878–6879. [Google Scholar] [CrossRef]
- Dragulescu-Andrasi, A.; Zhou, P.; He, G.; Ly, D.H. Cell-permeable GPNA with appropriate backbone stereochemistry and spacing binds sequence-specifically to RNA. Chem. Commun. 2005, 244–246. [Google Scholar] [CrossRef]
- Diederichsen, U. Pairing Properties of Alanyl Peptide Nucleic Acids Containing an Amino Acid Backbone with Alternating Configuration. Angew. Chem. Int. Ed. Engl. 1996, 35, 445–448. [Google Scholar] [CrossRef]
- Diederichsen, U.; Schmitt, H.W. β-Homoalanyl PNAs: Synthesis and Indication of Higher Ordered Structures. Angew. Chem. Int. Ed. 1998, 37, 302–305. [Google Scholar] [CrossRef]
- Hoffmann, M.F.H.; Bruckner, A.M.; Hupp, T.; Engels, B.; Diederichsen, U. Specific Purine-Purine Base Pairing in Linear Alanyl-Peptide Nucleic Acids. Helv. Chim. Acta 2000, 83, 2580–2593. [Google Scholar] [CrossRef]
- Chakraborty, P.; Diederichsen, U. Three-Dimensional Organization of Helices: Design Principles for Nucleobase-Functionalized β-Peptides. Chem. Eur J. 2005, 11, 3207–3216. [Google Scholar] [CrossRef]
- Miyanishi, H.; Takahashi, T.; Mihara, H. De Novo Design of Peptides with l-α-Nucleobase Amino Acids and Their Binding Properties to the P22 boxB RNA and Its Mutants. Bioconjugate Chem. 2004, 15, 694–698. [Google Scholar] [CrossRef]
- Watanabe, S.; Tomizaki, K.; Takahashi, T.; Usui, K.; Kajikawa, K.; Mihara, H. Interactions between peptides containing nucleobase amino acids and T7 phages displaying S. cerevisiae proteins. Peptide Sci. 2007, 88, 131–140. [Google Scholar] [CrossRef]
- Mercurio, M.E.; Tomassi, S.; Gaglione, M.; Russo, R.; Chambery, A.; Lama, S.; Stiuso, P.; Cosconati, S.; Novellino, E.; Di Maro, S.; et al. Switchable Protecting Strategy for Solid Phase Synthesis of DNA and RNA Interacting Nucleopeptides. J. Org. Chem. 2016, 81, 11612–11625. [Google Scholar] [CrossRef]
- Tomassi, S.; Ieranò, C.; Mercurio, M.E.; Nigro, E.; Daniele, A.; Russo, R.; Chambery, A.; Baglivo, I.; Pedone, P.V.; Rea, G.; et al. Cationic nucleopeptides as novel non-covalent carriers for the delivery of peptide nucleic acid (PNA) and RNA oligomers. Bioorg. Med. Chem. 2018, 26, 2539–2550. [Google Scholar] [CrossRef]
- Roviello, G.N.; Gaetano, S.D.; Capasso, D.; Franco, S.; Crescenzo, C.; Bucci, E.M.; Pedone, C. RNA-Binding and Viral Reverse Transcriptase Inhibitory Activity of a Novel Cationic Diamino Acid-Based Peptide. J. Med. Chem. 2011, 54, 2095–2101. [Google Scholar] [CrossRef]
- Wang, H.; Feng, Z.; Qin, Y.; Wang, J.; Xu, B. Nucleopeptide Assemblies Selectively Sequester ATP in Cancer Cells to Increase the Efficacy of Doxorubicin. Angew. Chem. Int. Ed. 2018, 130, 5025–5029. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the ACM/IEEE Conference on Supercomputing (SC06), Tampa, FL, USA, 11–17 November 2006. [Google Scholar]
- Schrödinger Release 2018-3: Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2018. Available online: http://www.schrodinger.com/ (accessed on 17 April 2019).
- Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, USA, 2018. Available online: http://www.schrodinger.com/ (accessed on 17 April 2019).
- Martyna, G.J.; Klein, M.L.; Tuckerman, M. Nosé–Hoover chains: The canonical ensemble via continuous dynamics. J. Chem. Phys. 1992, 97, 2635–2643. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Kunz, H.; Unverzagt, C. The Allyloxycarbonyl (Aloc) Moiety-Conversion of an Unsuitable into a Valuable Amino Protecting Group for Peptide Synthesis. Angew. Chem. Int. Ed. Engl. 1984, 23, 436–437. [Google Scholar] [CrossRef]
- Di Maro, S.; Zizza, P.; Salvati, E.; De Luca, V.; Capasso, C.; Fotticchia, I.; Pagano, B.; Marinelli, L.; Gilson, E.; Novellino, E.; et al. Shading the TRF2 Recruiting Function: A New Horizon in Drug Development. J. Am. Chem. Soc. 2014, 136, 16708–16711. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, F.S.; Tomassi, S.; Di Maro, S.; Reichart, F.; Notni, J.; Dangi, A.; Marelli, U.K.; Brancaccio, D.; Merlino, F.; Wester, H.J.; et al. From a Helix to a Small Cycle: Metadynamics-Inspired αvβ6 Integrin Selective Ligands. Angew. Chem. Int. Ed. Engl. 2018, 57, 14645–14649. [Google Scholar] [CrossRef] [PubMed]
| ID | Yield (%) | Purity | tR (min) | Mass Calcd | Mass Found [M+H]+ |
|---|---|---|---|---|---|
| np-1 | 76% | ≥95% | 11.90 | 2761.45 | 2762.30 |
| np-2 | 64% | ≥95% | 13.60 | 2761.45 | 2762.65 |
| np-3 | 68% | ≥95% | 12.70 | 2761.45 | 2762.62 |
| np-4 | 71% | ≥95% | 12.50 | 2761.45 | 2762.72 |
| Entry | Sequence | polyA (Tm) | polydA (Tm) |
|---|---|---|---|
| T6 | 3’-TTTTTT-5’ | 35 | 25 |
| np-1 | Ac-K(T)-R-K(T)-R-K(T)-R-K(T)-R-K(T)-R-K(T)-RCONH2 | 45 | 35 |
| np-2 | Ac-K(T)-r-K(T)-r-K(T)-r-K(T)-r-K(T)-r-K(T)-rCONH2 | Not Detected | Not Detected |
| np-3 | Ac-k(T)-R-k(T)-R-k(T)-R-k(T)-R-k(T)-R-k(T)-RCONH2 | Not Detected | Not Detected |
| np-4 | Ac-k(T)-r-k(T)-r-k(T)-r-k(T)-r-k(T)-r-k(T)-rCONH2 | Not Detected | Not Detected |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomassi, S.; Montalban, F.F.; Russo, R.; Novellino, E.; Messere, A.; Di Maro, S. Investigation of the Stereochemical-Dependent DNA and RNA Binding of Arginine-Based Nucleopeptides. Symmetry 2019, 11, 567. https://doi.org/10.3390/sym11040567
Tomassi S, Montalban FF, Russo R, Novellino E, Messere A, Di Maro S. Investigation of the Stereochemical-Dependent DNA and RNA Binding of Arginine-Based Nucleopeptides. Symmetry. 2019; 11(4):567. https://doi.org/10.3390/sym11040567
Chicago/Turabian StyleTomassi, Stefano, Francisco Franco Montalban, Rosita Russo, Ettore Novellino, Anna Messere, and Salvatore Di Maro. 2019. "Investigation of the Stereochemical-Dependent DNA and RNA Binding of Arginine-Based Nucleopeptides" Symmetry 11, no. 4: 567. https://doi.org/10.3390/sym11040567
APA StyleTomassi, S., Montalban, F. F., Russo, R., Novellino, E., Messere, A., & Di Maro, S. (2019). Investigation of the Stereochemical-Dependent DNA and RNA Binding of Arginine-Based Nucleopeptides. Symmetry, 11(4), 567. https://doi.org/10.3390/sym11040567





