Abstract
DNA double-strand breaks (DSBs) jeopardize genome integrity and can—when repaired unfaithfully—give rise to structural rearrangements associated with cancer. Exogenous agents such as ionizing radiation or chemotherapy can invoke DSBs, but a vast amount of breakage arises during vital endogenous DNA transactions, such as replication and transcription. Additionally, chromatin looping involved in 3D genome organization and gene regulation is increasingly recognized as a possible contributor to DSB events. In this review, we first discuss insights into the mechanisms of endogenous DSB formation, showcasing the trade-off between essential DNA transactions and the intrinsic challenges that these processes impose on genomic integrity. In the second part, we highlight emerging methods for genome-wide profiling of DSBs, and discuss future directions of research that will help advance our understanding of genome-wide DSB formation and repair.
1. Introduction
Out of the thousands of DNA insults that hit our cells every day [1], double-strand breaks (DSBs) are among the most deleterious lesions. DSBs can trigger cell death or give rise to structural genomic rearrangements associated with carcinogenesis and other diseases [2,3,4]. The frequency of DSBs is estimated to be 10–50 events per cell per day [5,6]. While exogenous genotoxic insults contribute to this [7,8], in healthy individuals the majority of DSBs is thought to originate from within the nucleus, where DSBs form during fundamental processes such as DNA replication, meiosis, antibody diversification, gene transcription, and—although indirectly—cellular metabolism [1,9,10,11,12,13]. Fortunately, most DSB events are quickly sensed and—orchestrated by the cell’s DNA damage response (DDR) [1,7]—faithfully resolved by two partially redundant repair pathways: homologous recombination (HR) and classical non-homologous end-joining (C-NHEJ) [1,14]. While HR mainly operates in the G2 and S phases of the cell cycle, C-NHEJ is active throughout the cell cycle and is considered the predominant DSB repair pathway in cycling cells. Although C-NHEJ is more error-prone than HR, mistakes of this classical NHEJ repair route are generally minimal [5,15,16]. However, errors become more frequent when alternative end joining pathways (a-EJ) such as microhomology-mediated end joining (MMEJ) engage [17,18,19].
In Part I of this review, we describe how DSBs can form during essential DNA transactions—especially during DNA replication, transcription, and 3D genome folding—after which we touch upon the undesired outcomes of DSB repair and their possible consequences. In Part II we attempt to provide a comprehensive overview of the diverse methods that have emerged to profile genome-wide DSB landscapes and we hypothesize how future efforts can help to answer some of the open questions that exist in the field.
Part I—Endogenous Causes of DSBs
2. DNA Replication as a Source of Endogenous DSBs
During the S phase of the cell cycle, thousands of replication forks work in a highly coordinated manner to precisely replicate the entire genome exactly once. In the absence of perturbations, a replication fork proceeds undisturbed until it approaches a neighboring fork traveling in the opposite direction. At this point, the two forks fuse, the replication machineries disassemble, and the catenated sister chromatids separate [20,21,22,23]. However, numerous factors can hinder fork progression or interfere with proper execution of the replication program, altogether referred to as replication stress.
2.1. Exhaustion of Replication Resources and Disturbed Replication Programs
Replication forks are forced to slow or stall in a global manner when resources required for faithful completion of replication run out [24]. Essential components such as dNTPs, replication protein A, or chromatin constituents—required for repacking of the replicated DNA and packaging of the newly produced copy—can, for instance, be depleted as a result of perturbed replication timing [24]. Proper licensing of replication origins is crucial for replication precision, and deregulated licensing—which induces systemic replication stress—is believed to contribute to carcinogenesis [25,26]. Activated oncogenes can perturb replication timing by increasing origin firing—which concomitantly depletes replication factors—or by invoking origin re-firing. The latter leads to re-replication, which is particularly detrimental as it not only increases the consumption of resources, but also directly transforms unligated Okazaki fragments into single-ended DSBs [27,28,29,30]. Although oncogene-triggered perturbations of replication timing can lead to genome instability and, as such, play a key role during early cancer development [31,32], oncogene activation can also provoke premature cellular senescence in a process called oncogene-induced senescence, which has been uncovered as an important tumor-suppressing strategy in premalignant cells [33,34].
2.2. Conflicts with Pre-Existing DNA Damage and Secondary Structures
Individual replication forks can be brought to stall when an obstacle on the DNA prevents helicase or polymerase activity of the replisome (Figure 1A). If stalling is not properly resolved, the fork can collapse and endanger genome integrity [35,36,37], as we will discuss below in Section 2.4. Pre-existing DNA lesions—including base alterations and strand damage such as inter-strand crosslinks—can cause replication fork stalling [36]. Furthermore, secondary non-B DNA structures that can form at repetitive sequences upon transient strand separation also block fork progression, rendering certain regions of the genome particularly difficult to replicate. Examples of such regions include repetitive telomeric sequences, which can form G-quadruplexes held together by strong Hoogsteen hydrogen bonds [38,39,40,41], centromeres, which are rich in protein-bound heterochromatic AT-rich alpha satellite repeats [42], and tightly compacted, repeat-rich, heterochromatin [24]. In general, obstacles that impede fork progression are believed to have worse consequences when they are encountered on the leading strand, as discontinuous DNA synthesis on the lagging strand may help to bypass obstructions [43,44]. In the next section, we look at another major impediment for replication fork progression: the transcription bubble.
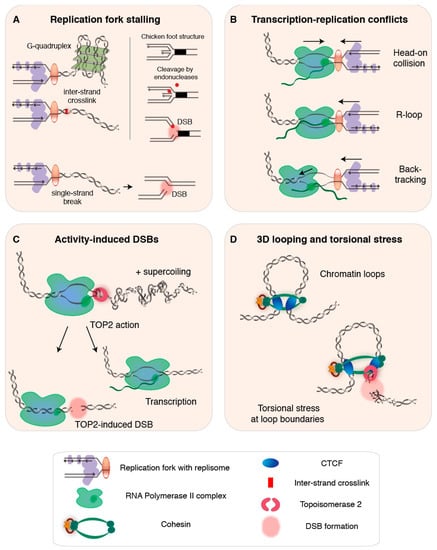
Figure 1.
Endogenous DNA double-strand breaks (DSBs) during DNA transactions. (A) Left panel: during DNA replication, pre-existing DNA lesions such as G-quadruplexes and inter-strand crosslinks invoke replication stress and cause replication fork stalling. Right panel: an example of how intermediate structures during fork remodeling can lead to formation of a single-ended DSB. Bottom: replication through a single-strand break or nick can result in a single-ended DSB [36,39,41]. (B) Transcription complexes obstruct replication fork progression, particularly when transcription-replication encounters are head-on (top) [68], when the nascent RNA has formed an R-loop that stabilizes the RNAPII association with the DNA (middle) [62], and when the RNAPII complex is paused and displays backtracking (bottom, arrow indicates backward sliding of the RNAPII) [59]. (C) Transcription-related activity-induced DSBs emerge at sites of topoisomerase 2 (TOP2) action [56,71], which is required to release positive (+) supercoiling building up ahead of the RNAPII complex. TOP2-mediated DSBs enable transcription but can also lead to non-resolved DSBs when repair is escaped or fails [72]. (D) Genomic regions involved in 3D genome looping experience torsional stress that requires TOP2 activity to be resolved [73]. As a result, chromatin loop boundaries or anchors may accumulate TOP2-dependent DSBs [74].
2.3. Transcription-Replication Conflicts, R-Loops, and Backtracking
Transcription bubbles present a natural obstacle for the replication fork [45,46,47] (Figure 1B), and consequences of so-called transcription-replication conflicts (TRCs) are believed to be particularly detrimental when the two machineries encounter each other head-on [46,48,49]. Although DNA replication and transcription are spatiotemporally separated—they globally anti-correlate, occur in distinct cell cycle phases and nuclear sub-compartments, and are presumably subjected to co-orientation bias, which suppresses head-on encounters [21,50,51,52,53,54]—both transactions use the same DNA template, and collisions are inevitable, especially for long genes that require more than one interphase to be transcribed [55]. While TRCs may be rare in healthy human cells, their frequency increases when replication timing is perturbed and in situations in which the transcription machinery lingers [27,56].
Genome-wide transcription rates are inherently heterogeneous [57,58], and are influenced by regulatory events such as RNA polymerase II (RNAPII) pausing and/or backtracking [59,60]. Two transcriptional intermediates that have been implicated in TRCs are DNA/RNA hybrids known as R-loops and backtracked RNAPII complexes (Figure 1B). R-loops can form co-transcriptionally when the nascent RNA behind RNAPII anneals back to its DNA template, creating a stable triple-stranded hybrid structure [61,62]. R-loops have been identified in genic regions—at promoters, sites of RNAPII pausing, and transcription terminal regions of genes with short intergenic distances—and, more generally, in regions rich in guanines, expanded trinucleotide repeats, supercoils, and DNA nicks [53,63,64]. Recent work has revealed that R-loops can result from head-on TRCs, whereas co-directional TRCs act as R-loop erasers [65]. Although R-loops can exert regulatory roles [63,66,67], they can also obstruct replication and transcription, and are frequently associated with genomic instability and mutagenic potential, especially when longer stretches of DNA are involved [62,68,69,70].
A backtracked transcription complex—composed of a dangling 3’ end of the nascent RNA and an inactive RNAPII that might slide backwards—presents another direct impediment to replication fork progression [59]. Backtracking is associated with promoter-proximal RNAPII pausing—which is considered essential for controlling the accuracy and rate of transcriptional elongation [60,75]—as well as with transcriptional pausing relevant to transcription termination, co-transcriptional RNA folding and processing, and handling of pre-existing obstacles such as DNA adducts and R-loops [59,76]. When not swiftly released or removed, backtracked transcription complexes increase the chance that a replisome switches from the DNA strand onto RNA, which generates a DNA single-strand break (SSB) that can give rise to a DSB during subsequent replication [59]. Although both R-loops and backtracking are frequently related to genomic instability, revealing their direct contribution is complicated by the many co-occurring sources of instability [62].
2.4. The Cellular Response to Replication Stress and the Impact of Replication Timing
In general, sensing of replication stress leads to activation of the replication checkpoint, temporary cell cycle arrest, and orchestration of the DDR. Diverse DNA repair pathways can then engage to attempt stabilization of the stalled replication fork—aimed at preventing irreversible fork collapse—and promote removal or repair of blocking impediments or lesions. Ultimately, to restart replication the stabilized fork can be reprimed or, if this fails, replication can be rescued by firing nearby dormant replication origins [24,26,35,37,77,78,79,80,81,82]. Importantly, sustained DDR activation, for example due to failed repair of a lesion, can also lead to apoptosis or senescence [33,83].
2.4.1. Fork Stabilization and Restart
Stalled replication forks are predominantly stabilized by components of the HR repair pathway, which also protect nascent DNA at the stalled fork and support repair of DSBs induced by replication stress [24]. Furthermore, the NHEJ pathways C-NHEJ and MMEJ have been identified at stalled replication forks, although they are believed to be mainly implicated in repair of single-ended DSBs at collapsed forks [78,84,85]. Transcription-coupled nucleotide excision repair is thought to assist removal of obstacles that cause replication stress such as R-loops, bulky lesions, and arrested transcription complexes [86,87,88], although rather than stalling, the replisome may also bypass encountered lesions and other impediments with the help of specialized but error-prone DNA polymerases [89,90].
After removal of the source of replication stress, restart of the replication fork is believed to occur with the help of HR factors and via repriming mediated by local ssDNA, although the precise underlying fork remodeling and reversal mechanisms remain to be elucidated further (for the main proposed models we refer elsewhere [24,91,92]). While HR is among the most faithful approaches to repair, it can prime error-prone replication restart [78,93]. In Section 5, we elaborate on the consequences of this and discuss more notorious repair processes linked to rearrangements introduced during fork restart.
2.4.2. DSB Formation from Stalled Replication Forks
When fork stabilization and restart fail the persistently stalled replication fork can collapse, which terminally inactivates the replisome and can result in the formation of a single-ended DSB [24,27,78]. Furthermore, single-ended DSBs can arise at stalled forks that undergo fork reversal or remodeling, for example due to endonuclease-mediated cleavage of chicken foot structures (Figure 1A) or other remodeling intermediates that have been hypothesized to allow DSB-mediated fork restart [77,92,94,95,96]. Single-ended DSBs can also form as the result of replication through nicked DNA [97] (Figure 1A), in case of extensive origin refiring—with or without actual fork collisions (comprehensively reviewed in [30]), or from passively broken ssDNA near stalled forks [24,30,35,98,99].
2.4.3. Common Fragile Sites
DSBs associated with replication stress occur at higher frequency in certain regions of the genome such as common fragile sites (CFSs). CFSs are defined as chromosomal cytobands that appear broken in metaphase spreads following induced replication stress [100,101,102]. CFSs are largely cell type-specific, overlap with recurrent copy number alterations (CNAs) seen in cancer—especially with large deletions—and contain genes with presumed tumor-suppressor activity [102], or belonging to larger functional hubs associated with cancer-related loss of homeostasis [103]. CFS fragility has been attributed to various features that can convey replication stress: repetitive sequence content, premature mitosis, late replication timing or paucity of replication origins, and delayed replication of long active genes [101,102,104,105].
2.4.4. Replication Timing and Fragility
In contrast to CFSs, early replicating fragile sites (ERFSs) represent a different class of fragile genomic regions that replicate early, reside in gene-rich accessible chromatin, and overlap with sites of recurrent translocations and rearrangements observed in cancer [106]. In a large study of somatic CNAs across human cancers, breakpoints of large duplications were found to more often reside in early replicating genomic domains, while deletions and overall CNA levels were enriched in late-replicating chromatin [107]. Similarly, translocation breakpoints in neuroblastoma cells mapped more frequently to early-replicating regions, where the overall breakpoint frequency was estimated to be more than three times higher compared to the rest of the genome [108]. In contrast, point mutation rates are generally higher in late-replicating genomic regions across eukaryotic species [109,110].
Although still poorly understood, the connection between replication timing and both fragility and the outcome of erratic repair might in part relate to inherent differences between early and late replication. Regions with early versus late replication timing exhibit roughly mirroring patterns of DNA accessibility, gene activity, and nuclear positioning—in line with A/B compartmentalization. Simultaneously replicating regions may thus reside in closer spatial proximity [111,112]. The more permissive state of chromatin in early-replicating regions likely allows efficient replication, whereas late-replicating—and oftentimes more compact—chromatin requires more origins and thus shorter inter-origin distances [113,114]. The presence of many concurrently active forks in a tighter space in late-replicating regions is thought to present more recombination partners and increase the probability of deletions [110]. In contrast, early-replicating regions are at higher risk of re-replication because a longer period is spent in an already replicated state. As a result, the chances of short-range duplications and rearrangements within the borders of topologically associating domains (TADs)—with intra-TAD rearrangements ranging from 10 to 300 Kb—may be increased [110,115]. Larger and inter-TAD rearrangements (>500 Kb) often seen in cancers [116] are believed to be favored by long-range chromatin contacts between spatially juxtaposed domains with similar characteristics in terms of replication timing and accessibility [107,110,117,118,119].
Although we already briefly touched upon how transcription bubbles contribute to replication stress, accumulating evidence also connects the act of transcription activation itself to elevated levels of DSBs and genomic alterations. In the following section we will therefore focus further on the relationship between transcription and DSB formation.
3. Transcription as a Source of Endogenous DSBs
Besides its share in hampering replication fork progression and generating roadblocks in the shape of R-loops and backtracked transcription complexes, transcription is in itself, in a replication-independent manner, considered to be a source of DSBs [53,56,71,120,121]. As during replication, various types of transcription stress can affect progression of the transcription machinery, and the transient strand separation during transcription is thought to render the non-transcribed strand particularly vulnerable. Although we specifically focus on transcription as a cause of DSBs here, it should be noted that the relationship between transcription and genome instability and mutation is not limited to DSB events, but also encompasses other types of DNA damage. For an extensive overview of transcription-related DNA damage and the connection between transcription and DDR, we refer the reader to an excellent review [121].
3.1. DSBs Accumulate around Activated Genes
The genome-wide frequency of DSBs is not only elevated in CFSs, ERFSs, and long, late-replicating genes, but also in accessible chromatin and, in particular, near transcriptionally active genes, with a significant enrichment around the transcription start site (TSS) [122,123,124,125,126]. Transcription-induced DSBs have, for example, been identified near genes responding to signaling invoked by sex hormones and transcription factors in human cancer cells [120], in activated promoters of stimulated neuronal stem/progenitor cells (NSPCs) [127], and DSB levels in mouse brain have been shown to increase upon physiological neuronal activity [128]. In turn, translocation breakpoint clusters, indicative of frequent DSB formation, were found near long, active genes in NSPCs and in activated B cells [124,129].
3.2. Transcription Activation through DNA Damage
In contrast to previous work that related DSBs and, more generally, DNA damage to the suppression of transcription [130,131], the observed association between fragility and transcription activation suggests that DSBs, in certain situations, may be positively involved in transcription [121,132,133]. Similar concepts have been described before for SSBs generated by type I topoisomerase (TOP1), which emerged at sex hormone-responsive transcribed regulatory elements upon hormone stimulation of prostate cancer cells. In this context, depletion of TOP1 led to reduced transcription of these elements [134]. Furthermore, TOP1 has been implicated in the suppression of long R-loops, by resolving negative supercoiling [62].
Examples of transcriptional activation through DSBs—formed as a consequence of DDR pathways—are plentiful [133]. For example, in estrogen-responsive breast cancer cells, DSBs form at estrogen-responding genes through base excision repair (BER), aimed at repairing cytosines deaminated by the action of the APOBEC3B enzyme, which is frequently deregulated in cancer cells [11,121]. In line with the observation that APOBEC3B knockdown led to reduced transcription of the responsive genes, APOBEC3B-induced DSBs are thought to be essential for the recruitment of RNAPII and histone modifications that promote transcription [121]. Furthermore, upon pathogen infection of intestinal cells, endonucleases involved in nucleotide-excision repair (NER) have been identified to cause genome fragmentation. These infection-induced DSBs were shown to be required for activation of the key native immune response, and counteracted infection-associated apoptosis of the infected cells [135,136].
3.3. Transcription Activation Assisted by TOP2-Induced DSBs
Transcription-induced DSBs near activated genes in physiological contexts, such as stimulated NSPCs, have been attributed to the action of type II topoisomerase isoforms α (TOP2A) and β (TOP2B), and TOP2-induced DSBs have been suggested to be needed for the transcription of some or all genes [120,127,137,138,139,140] (Figure 1C). This is in line with previous work that found TOP2B-induced DSBs in gene promoters to be required for transcription activation upon hormone stimulation [121,137], and similar observations have since then been described in response to diverse stimuli, including heat shock, serum induction, and various hormonal and neuronal stimuli [11,133].
TOP2 forms transient DSBs that help resolve positive DNA supercoiling, which builds up ahead of transcription and replication forks, halting their progression or leading to strand breaks [72,141,142,143,144]. Although the precise mechanism underlying DSB-induced transcription activation remains elusive, TOP2-mediated release of the topological stress brought about by supercoils may be sufficient to stimulate RNAPII processivity [121]. Other postulated models associate both TOP2 and TOP1 activity at transcribed elements to the release of promoter-proximal pausing of RNAPII, allowing transcriptional elongation, or suggest that chromatin alterations upon DSB formation may induce a more transcription-compatible state [121].
3.4. TOP2 Poisons Are Associated with Therapy-Related Acute Myeloid Leukemias
Despite the vital function of TOP2 in removing supercoils, its activity is a double-edged sword. On the one hand, TOP2-induced DSBs are thought to be rapidly religated, thereby securing genome integrity [72,145,146]. On the other, the TOP2 religation cycle can occasionally fail, which leaves TOP2 covalently trapped onto cleaved DNA. When the trapped adduct is not removed and repaired via the action of, among others, BRCA1 [147], it can lead to the emergence of a persistent DSB. The danger of this is demonstrated by the TOP2 poison etoposide, a widely used anti-cancer drug that prevents the release of TOP2 cleavage complexes, and thereby generates protein-linked DSBs that block transcription and replication, and eventually kill cancer cells [148]. Despite its successful use in chemotherapy, etoposide has also been associated with the emergence of therapy-related secondary acute myeloid leukemias driven by recurrent genomic translocations between regions coinciding with etoposide-induced DSBs [74,149,150,151]. Of note, etoposide is not the only chemotherapeutic agent that is recognized to increase the chance of secondary malignancies: other TOP2 inhibitors, alkylating agents, and anthracyclines are also associated with an increased risk of developing acute myeloid leukemia, whereas cyclophosphamide treatment increases the risk of bladder cancer [152]. Furthermore, radiotherapy-related secondary cancers can emerge throughout the body depending on the type of irradiation [152,153].
In the previous two sections we have discussed relevant work that shows how strongly transcription and formation of endogenous DSBs are interconnected—with and without the interplay with DNA replication. Furthermore, we have seen how transcription activation frequently involves the formation of a transient DSB by TOP2. In the next section we will focus on another functional role of TOP2: resolving topological issues that arise during 3D genome folding.
4. 3D Genome Architecture and DSBs
Topological or torsional stress brought about by DNA overwinding not only affects replication or transcription, in which traversing of the linear genome must be made possible, but it can also impact processes involved in organizing the higher-order three-dimensional folding of the genome. Furthermore, supercoils have been hypothesized to transit through chromatin structure and as such, influence larger stretches of chromatin [144]. In this section, we highlight recent work that has suggested elevated DSB susceptibility at genomic regions involved in 3D genome organization.
4.1. TOP2-Induced DSBs at Chromatin Loop Anchors
In addition to its binding to gene promoter regions, TOP2B has been identified at cis-regulatory genomic elements bound by CCCTC-binding factor (CTCF) or by both CTCF and cohesin [73,154]. These TOP2B/CTCF/cohesin-bound elements overlap regions uncovered to be relevant for 3D genome organization, such as the borders or anchors of transcriptionally active supercoiling domains [143] and TADS [155,156], as well as the elements involved in regulatory chromatin loops [157]. At these sites, genomic knots and tangles—which require the remodeling activity of TOP2 and perhaps other enzymes—are thought to result from processes proposed to govern chromatin loop dynamics, such as loop extrusion [74,158,159] (Figure 1D).
Accordingly, chromatin loop borders and loop anchor points are increasingly associated with enhanced levels of DSBs and genomic breakpoints underlying structural variants (SV), and as a result, genome instability and cancer [159,160,161]. While the levels of etoposide-induced DSBs correlate with gene expression levels [126,162], TOP2B-induced DSBs have been reported to be largely transcription-, replication-, and cell type-independent [74], and instead related to the action of TOP2B at the border of chromatin loops [73,74]. However, with chromatin loops forming throughout the genome—often coinciding with active genes [163]—an intertwined relationship between transcription, torsional stress, and TOP2-related DSBs cannot be excluded easily. Indeed, a recent study demonstrated that the frequency of TOP2-induced DSBs enriched at CTCF/cohesin bound loop anchors correlates with expression levels and directionality of coinciding highly transcribed genes—such as those frequently involved in oncogenic translocations in leukemias [164]. Furthermore, inhibition of transcription elongation led to a decrease in DSB levels across transcribed regions and reduced formation of gene fusions, altogether suggesting that both transcription and 3D chromatin folding contribute to TOP2-related genomic instability [164].
4.2. Intertwined Actions Predispose Regulatory Regions to Fragility
The topological stress that results from chromatin loop extrusion and other chromatin-related remodeling activities can, besides directly impacting strand integrity, in turn also affect the transcription machinery. As torsional stress can contribute to transcription regulation [142,165], supercoiling and knots emerging during chromatin looping could be involved in, for instance, safeguarding timely termination of transcription, transcription direction, or seclusion of regulatory effects [166]. Furthermore, as mentioned above, the increased susceptibility to indirect breakage at both overwound and underwound DNA [144] can, in principle, affect any region involved in looping, transcription, regulation, or genome condensation. Together with the previous sections, this notion illustrates the complexity that we face when attempting to unravel the mechanisms that underlie the formation of DSBs at a given genomic location, and emphasizes the concept that genomic regions with many concurrent regulatory transactions, such as highly active genes and TAD boundaries in active chromatin, have an inherently higher risk of suffering attacks on strand integrity under physiological as well as diseased conditions.
4.3. Special Cases of Genome Rewiring Require Programmed DSBs
In the sections above, we have covered DNA replication, transcription, and 3D chromatin folding as important endogenous contributors to genome-wide DSB landscapes. However, this overview of endogenous causes is far from complete, as we have deliberately omitted several specialized processes that induce programmed DSBs in specific gene classes or during specific moments in development. We refer elsewhere for excellent literature on programmed physiological DSBs [133], which form in a highly regulated manner during meiotic recombination and the lymphocyte-specific processes V(D)J recombination—which shuffles gene segments to contribute to antigen receptor diversity—and class switch recombination (CSR)—which occurs in mature B lymphocytes and involves a second round of exon substitution to diversify effector antibodies [133,167,168,169]. DSBs essential for V(D)J recombination and CSR are brought about by recombination-activating gene (RAG) endonucleases and activation-induced cytidine deaminase (AID), respectively. Despite the importance of recombination for proper functioning and adaptation of the immune system, the dependency on programmed DSBs poses oncogenic risks [168,170]. For an overview of the protective mechanisms in place to suppress adverse outcomes of DSBs during lymphocyte maturation, we refer to a recent review [171].
Although the DDR can in principle repair programmed and accidental DSBs quickly and faithfully, and rescue stalled replication forks to prevent their collapse, DSBs are drivers of most of the structural genomic rearrangements observed in human (cancer) genomes [172]. In the following section, we will summarize some of the main insights into unsuccessful and/or unfaithful DSB repair, and how this can have detrimental effects on genome integrity and stability.
5. Adverse Outcomes of DSB Repair
Imbalances in the regulatory circuitries of the nucleus can cause DNA replication, cell cycle checkpoints, and DDR to go haywire and invoke an avalanche of increasing replication stress, DNA damage, and genomic instability [32,173]. During early carcinogenesis, increasing levels of DSBs—due to enhanced replication stress and transcriptional rewiring—can exhaust the DDR and eventually compromise faithful DSB repair. When C-NHEJ and HR are too slow or simply fail to repair DSBs—for example, due to scarcity or mutational inactivation of one or more of the required factors—alternative mechanisms can engage, of which most are considered to be typically error-prone and hence implicated in the genesis of structural genomic rearrangements [174,175,176]. When DSB repair becomes increasingly erratic during cancer progression, the formation of more oncogenic fusions and CNAs is promoted, which can in turn fuel intra-tumor evolution and heterogeneity. This process is of high clinical relevance, since it can lead to therapy resistance and/or the development of distant metastases [32,173,177].
In the following subsections, we first focus on various processes known to be involved in the transformation of a DSB or a stalled replication fork into potentially harmful non-natural junctions. Then, we describe how profiling of the resulting repair signatures—encountered with variable frequencies in cancer genomes—have become a tool to better understand cancer etiology, and predict disease and therapy outcome.
5.1. Mechanisms Underlying Structural Genomic Alterations
Copy number variation (CNV) is widespread in the human genome and underlies natural variation and evolution, but also cancer and developmental and neurological disorders [175,178]. CNVs—large segmental duplications or deletions—require the formation of junctions between sequences that are not normally juxtaposed in the reference genome [175]. The chaotic reassembly of DSB ends or entire genomic fragments—which can give rise to CNVs but also to highly complex genomic rearrangements seen in cancer genomes—has been attributed to complex processes that entail one or multiple rounds of DSB formation and low-fidelity repair, possibly combined with erratic template switching—a strand switch within the same or between distinct replication forks—or replication restart, for example. Below we briefly discuss some of the major processes thought to be involved in generating structural alterations, but for detailed and more complete reviews of proposed mechanisms underlying structural change in the genome we refer the reader elsewhere [174,175].
Although HR is considered to be more faithful than C-NHEJ because it utilizes homologous sequences for repair, homology-directed repair is intrinsically mutagenic. In contrast to healthy cells, where HR is strictly controlled [175] and minimized to S-G2 phase and specific recombinogenic processes such as meiotic crossover—which uses allelic HR (AHR)—and V(D)J recombination, HR can jeopardize genomic integrity in oncogenic situations [93,175]. HR repair can cause the formation of genomic rearrangements via abortive intermediates, and at stalled replication forks it can give rise to recurrent CNVs, via non-allelic HR (NAHR), or to non-recurrent CNVs, via error-prone homology-directed break-induced replication (BIR) or single-strand annealing (SSA) [78,93,174,175]. Non-recurrent CNVs can also be formed by NHEJ and other replication-based repair mechanisms, including microhomology-mediated BIR (MMBIR), fork stalling and template switching (FoSTeS), or serial replication slippage (SRS) [174,175]. It has been suggested that multiple rounds of FoSTeS and MMBIR can, for example, underlie the formation of complex rearrangements, while highly complex rearrangements classified as chromothripsis have been hypothesized to emerge during a single catastrophic breakage event, after which the generated chromosomal fragments are erroneously stitched back together, most likely via NHEJ and MMEJ [24,172,174].
In C-NHEJ, DSB ends are repaired without the need for homology, and although the resulting junctions are mostly accurate or have small deletions, free DNA may be inserted or translocations can be formed [175]. In contrast, MMEJ—the best-known a-EJ pathway—joins DSB ends based on microhomology (<25 nt), mediated by the error-prone DNA polymerase θ, which is frequently upregulated in cancer. Its involvement in DSB repair is associated with chromosome rearrangements and small deletions between the microhomologous sequences, giving rise to a typical genomic pattern at the predicted breakpoints [17,18,172]. Rearrangements of larger sections, long-range template switching, and MMBIR are thought to involve the action of other replicative polymerases such as DNA polymerase δ. Other polymerases, including translesion synthesis polymerases—which allow lesion bypass—have also been associated with the induction of local template switching [174].
Although most of the mechanisms underlying copy number gains involve replication-based mechanisms, segment amplifications can also arise in a non-replicative manner via breakage-fusion-bridge (BFB) cycles, in which the loss of extensively homologous (sub)telomeric regions induces unstable dicentric chromosomes. It is believed that BFB cycles play a major role in cancer, and the same process has been invoked to explain segmental duplications frequently observed in breast and ovarian cancer [172,175].
5.2. Repair Signatures in Cancer Genomes
For many years, patterns of single nucleotide variants (SNVs) have helped uncover distinct mutational signatures in cancer genomes, which has led to improved understanding of the underlying causative processes in certain cancer types. Increasingly, a similar approach is applied to multi-nucleotide structural rearrangements or copy number changes in cancer genomes, based on the identification of junctions between genomic sequences that are not naturally together in the reference genome [179]. Although the computational analyses required to identify complex rearrangement patterns are challenging, successful reconstruction of the genomic junctions in a cancer genome can reveal repair signatures composed of gains, losses, amplifications and rearrangements. As these repair signatures represent scars of impaired repair pathways, studying cancer genomes on a large scale using whole genome sequencing approaches can help to improve our understanding of the molecular history of various cancer types. As certain repair signatures are largely cancer type-specific, they have been harnessed to classify cancers and improve stratification of specific cancer subtypes, as well as to study clonal relationships among metastases and the corresponding primary tumors [172,180,181,182].
Although genome-wide profiling of structural rearrangements enables investigation of repair signatures and errors—based on identification of non-linear junctions—these approaches typically reveal past events that have occurred at some point in the history of the cell. In the next section of this review, we change gears back towards the lesions that underlie most of the rearrangements described here, and provide an overview of the various methods that are available for the profiling of genome-wide DSB landscapes.
Part II—Studying Genome-Wide Fragility Landscapes
6. Methods for Genome-Wide DSB Profiling
With DSBs structurally underlying most types of structural genomic alterations, insight into DNA fragility landscapes and their associated processes—such as transcription and replication, but also recruitment of repair proteins—is essential to broaden our understanding of the genome, particularly in the context of cancer. Over the past few years, various methods for genome-wide DSB detection and identification have been developed, with the aim of obtaining insight into genome fragility and its molecular basis. These methods can be broadly classified into indirect and direct DSB detection methods (Table 1), depending on whether they probe directly for DSBs or for DSB proxies, such as signaling or repair proteins that accumulate at genomic regions hit by DSBs, or products of DSB repair.

Table 1.
Methods for genome-wide nucleotide-resolution DSB identification.
6.1. Indirect Identification Based on Association of Recruited or Responsible Proteins
The first genome-wide DSB landscape was mapped in yeast, using chromatin immunoprecipitation (ChIP) on a tiled microarray (ChIP-chip) based on antibodies against the phosphorylated form of histone variant H2AX, γH2AX [183]—a ubiquitous component of DSB signaling in eukaryotes [184,185,186] (Figure 2; top). This pioneering work revealed γH2AX enrichment at loci prone to replication fork stalling and breakage, of which half mapped to repressed protein-coding genes [183]. Around the same time, ChIP-chip and ChIP-qPCR [187], as well as ChIP followed by next-generation sequencing (NGS) (ChIP-seq) [188] for γH2AX were applied to human AsiSI-ER cells engineered to conditionally express the AsiSI endonuclease for genome-wide induction of sequence-specific DSBs [187]. These studies revealed that DSB formation triggers large γH2AX domains spreading up to two megabases around the induced DSBs, with active genes protected from γH2AX spreading [187,188]. More recently, ChIP-seq for TOP2B binding was used in addition to γH2AX to indirectly detect DSBs in NSPCs upon exogenous activation. This revealed that TOP2B-dependent DSBs accumulate in the promoter region of early-response genes and are required for their transcription [127]. In recent experiments based on ChIP-exo—in which an exonuclease trims ChIP-ed DNA up to the site of the actual protein-DNA crosslink [189]—TOP2 was found to be positioned at accessible regulatory regions and CTCF/cohesin-bound sites [73].
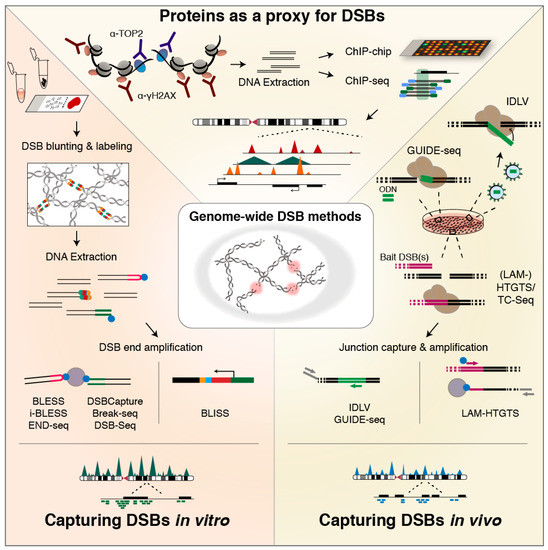
Figure 2.
Methods for genome-wide DSB profiling. (TOP) Proteins recruited to DSB sites—or associated with DSB formation—serve as a proxy for DSB formation. Chromatin containing the protein of choice is pulled down, and the extracted DNA—representing the underlying genomic regions—can then be analyzed by microarray (ChIP-chip) or high-throughput sequencing (ChIP-seq) [183,187]. The resolution of the generated binding profiles typically depends on the chosen protein. (RIGHT) Methods for in vivo capturing of DSBs utilize the non-homologous end-joining (NHEJ) repair machinery of the cell to either incorporate short dsDNA oligos (ODN) (in genome-wide unbiased identification of DSBs enabled by sequencing, GUIDE-seq [194]), or integration-deficient lentiviral vectors (in IDLV capture [195,196]) at the genomic sites of DSBs, or to generate translocation junctions between emerging DSB ends and a bait DSB, exogenously introduced and then induced in the cell (translocation-capture sequencing (TC-Seq) and high-throughput genome-wide translocation mapping (HTGTS) or linear amplification-mediated (LAM)-HTGTS, and derived methods [197,198,199,200]). Afterwards, cells are lysed and DNA is isolated, followed by method-specific approaches for specific amplification or capture of integration or translocation junctions. Subsequently, sequencing libraries are prepared, and sequence reads are aligned to the genome, typically revealing breakpoint clusters genome-wide. (LEFT) In vitro methods for genome-wide DSB identification directly label DSB ends with a dedicated adapter—with or without prior DSB end processing—in fixed cells immobilized on a surface (Breaks Labeling In Situ and Sequencing, BLISS [201]) or fixed cell suspensions (Breaks Labeling, Enrichment on Streptavidin, and Sequencing, BLESS [122]), in unfixed cells embedded in agarose plugs or beads (END-seq [202] and i-BLESS [203], respectively), in isolated DNA (DSB-Seq [126]), or isolated DNA in agarose plugs (Break-seq [204]). After labeling, DSB ends are selectively linearly amplified by in vitro transcription enabled by the BLISS adapter in BLISS. In the other methods, DSB ends are captured onto streptavidin beads that selectively capture the biotin-labeled DSB ends, and then amplified. Finally, sequencing libraries are prepared and the resulting mapped sequence reads reveal single DSB ends distributed genome-wide.
Although highly insightful, exploiting proteins as a proxy for DSBs has the disadvantage of being indirect, and assay outcome may be affected by unspecific binding [190] and spreading of the chosen protein around DSB sites. For example, γH2A.X not only accumulates around DSB sites, but is also recruited to regions with SSBs and to sites undergoing nucleotide excision repair in G1 [186,191,192,193]. To overcome these limitations and enable precise DSB mapping in a genome-wide manner, various approaches to more directly capture and identify DSB ends have been developed, and to comprehensively review these approaches, we distinguish between methods that capture DSBs based on in vivo integration or translocation events mediated by the cell’s DSB repair efforts (Figure 2; right), and methods that directly tag unrepaired DSB ends by in vitro ligation of dedicated adapter sequences (Figure 2; left).
6.2. Methods for In Vivo DSB Capture
Among the earliest in vivo capture approaches for genome-wide identification of DSBs is integration-defective lentiviral vector (IDLV) capture. IDLV employs in vivo NHEJ-assisted incorporation of lentiviral vectors into DSB sites, which are then amplified by linear amplification-mediated PCR (LAM-PCR) and sequenced [195,196]. IDLV has been applied to detect DSBs introduced by zinc finger nucleases [195], transcription activator-like effector nucleases (TALENs) [196], and CRISPR/Cas9 nucleases [196,205]. Limitations of IDLV capture include integration of the IDLV at varying distances from the actual break site, low detection frequency, sequence bias, low numbers of informative reads, and high costs [194,199].
In order to overcome some of these limitations, genome-wide unbiased identification of DSBs enabled by sequencing (GUIDE-seq) was developed [194]. Similar to IDLV capture, GUIDE-seq labels DSBs in vivo via NHEJ-mediated integration of short blunt dsDNA oligodeoxynucleotides (ODNs) at the site of a DSB, after which ODN-labeled genomic regions are amplified and sequenced. GUIDE-seq was established to assess Cas9 and Cpf1 specificity, and is considered to be a highly sensitive and precise method that enables the identification of DSBs that form during a period of several days [194,206]. In GUIDE-seq, identification of a DSB end critically depends on the cell’s NHEJ repair machinery to ligate the blunt ODN to a given DSB end, as well as on the transfection efficiency.
Rather than identifying DSBs genome-wide via local insertion of an ectopic sequence, several groups have developed techniques that exploit the inherent threat of DSBs being converted into genomic translocations. Translocation-capture sequencing (TC-Seq) [200,207] and high-throughput genome-wide translocation mapping (HTGTS) [197] were developed around the same time, and both methods indirectly identify DSB ends through translocation junctions formed in vivo with a bait DSB, introduced at an ectopically integrated I-SceI endonuclease recognition site. After amplification by conventional PCR or LAM-PCR [198,199], the bait–prey junctions are sequenced. HTGTS and TC-Seq were both applied to identify translocation junctions in B lymphocytes induced for IgH class-switching [197,200], while LAM-HTGTS was also used to assess Cas9 specificity [198]. Moreover, LAM-HTGTS has been harnessed to uncover transcription-associated DSBs in neuronal cells upon mild replicative stress [124,129], and the method has been tailored to uncover antibody repertoires generated during V(D)J recombination in B cells [218]. While the sensitivity of HTGTS methods is biased by the distance of a DSB to the bait—with nearby DSBs translocating with higher efficiencies—LAM-HTGTS was recently further improved by introducing DSB baits on twenty different mouse chromosomes via CRISPR/Cas9. This enabled identification of a more complete set of DSBs as well as appreciation of translocation preferences [208].
Although more direct and with higher resolution compared to ChIP-seq, the applicability of IDLV, GUIDE-seq, and HTGTS-based methods is limited by the need for transfection or transduction to enable integration or translocation. This can become especially disadvantageous when aiming to map DSB landscapes in primary cells or tissues, with poor or unknown transfection efficiency and possibly toxic effects of transfection or transduction. Furthermore, as these methods critically depend on active DSB repair, their applicability may be limited when working with cells with impaired DSB repair pathways, such as cancer cells. In IDLV, GUIDE-seq, and HTGTS-based methods, all identified DSB sites have undergone active repair, either locally, with ODN or IDLV sequences being integrated into the broken site, or over larger distances, with translocations onto the DSB bait in HTGTS. This suggests that the identified DSB sites only represent a subgroup of all the DSBs present at a given moment—those that are actively but not faithfully repaired. DSBs that are mostly perfectly and rapidly repaired, for example those induced by TOP2 at promoters, may be largely missed. Furthermore, translocation-based DSB identification methods such as HTGTS can only uncover DSBs in genomic regions that are prone to translocate, and can thus underestimate the actual DSB frequency and the propensity of certain DSBs to give rise to genomic alterations different from translocations. In line with this, the dependency of DSB detection methods on NHEJ repair implies that these methods can miss DSBs repaired through a different pathway [199].
6.3. Methods for In Vitro Tagging of DSBs
The first method to directly capture and identify DSBs in fixed cells in situ was Breaks Labeling, Enrichment on Streptavidin, and Sequencing (BLESS) [122]. BLESS captures DSBs by ligating short biotinylated hairpin-like adapters to blunted DSB ends, followed by capture on streptavidin beads, second adapter ligation, PCR and sequencing. BLESS was applied to identify endogenous and replication stress-induced DSBs [122], and to determine the specificity of CRISPR endonucleases [210,212]. Although BLESS does not depend on transfection or NHEJ repair, its labor-intensive protocol requires large amounts of input material and cell fixation, which has been related to the observed high background levels of DSBs [194,199,201]. Recently, two improvements to the original BLESS protocol were described: i-BLESS for highly sensitive unbiased DSB labeling in yeast immobilized in agarose beads [203], and qDSB-seq, enabling normalization of DSB frequency in a sample by spiking in cells in which DSBs were introduced at I-SceI sites [219].
Two years prior to BLESS, a method for direct labeling of DNA ends in yeast was published. In contrast to BLESS, damaged DNA immunoprecipitation (dDIP) [214]—among the first methods to generate a nucleotide-resolution map of DNA strand breaks in yeast—does not label DSBs in situ, but in extracted genomic DNA (gDNA). dDIP encompasses end-labeling with biotinylated nucleotides, followed by DNA fragmentation, IP with anti-biotin antibodies, and read-out by qPCR although NGS was also proposed. DNA break immunocapture (DBrIC) applies a similar strategy for immunocapture of DSB ends from gDNA and followed by NGS [215]. Initially, DBrIC was used to demonstrate locus-specific and genome-wide DSBs in human cancer cells. More recently, the approach was harnessed to profile DSB hotspots during the transient genome fragmentation that accompanies post-mitotic chromatin remodeling in mouse spermatids [215,216].
Biotin labeling of the 3’ ends of DSBs in isolated high-molecular-weight (HMW) gDNA is also performed in DSB-Seq [126,213], but rather than capture via biotin IP as in dDIP, DSB-Seq exploits the high biotin-streptavidin affinity to capture the labeled ends, which is followed by NGS. Unlike BLESS and most other methods, DSB-Seq can be complemented with the identification of SSBs via SSB-Seq, in which SSBs are tagged by nick translation, captured by immunoprecipitation, and sequenced [126,213]. DSB-Seq and SSB-Seq were applied to map DSBs induced by replication stress using the TOP2 poison etoposide. A major limitation of dDIP, DBrIC, and DSB-Seq is that tagging DSB ends in extracted gDNA may result in increased levels of artificial breaks. This is bypassed in Break-seq, in which isolated chromosomal DNA is embedded into agarose prior to DSB end-repair and biotin labeling [204], after which labeled extracted DNA is captured with streptavidin and sequenced. Break-seq was applied to identify DSB peaks overlapping with DNA replication origins, and to relate DNA fragility to replication fork progression and transcription during and after replication inhibition in yeast [204].
Aiming to overcome some of the limitations of BLESS, DSB-seq, and Break-seq, DSBCapture [125] and END-seq [202] were developed. Both methods feature an A-tailing step after blunting of DSB ends, and incorporate the Illumina RA5 adapter sequence directly into the ligated adapter. DSBCapture was applied to identify AsiSI-digested sites as well as endogenous DSBs at regulatory sites and G-rich regions, and revealed that persistent DSBs can be identified under physiological conditions, despite the presence of an intact DDR [125]. In END-seq, besides the adapter improvements mentioned above, DSB labeling is carried out in cells embedded and lysed inside agarose plugs, avoiding cell fixation, which can allegedly cause artificial DSBs [202,220], although a recent study reported very low noise levels in gently fixed samples [203]. END-seq was applied to map AsiSI-induced DSBs and to determine the specificity of DSBs introduced by zinc-finger nucleases, RAG endonucleases involved in V(D)J recombination, and CRISPR-Cas enzymes. Compared to BLESS, END-seq is reported to be more specific and sensitive, and to better preserve the structure of DSB ends [202].
To improve the sensitivity and specificity of BLESS, Breaks Labeling In Situ and Sequencing (BLISS) was recently developed [201]. In BLISS, the DSB-labeling adapter contains a T7 promoter sequence, allowing linear amplification of labeled DSB ends by in vitro transcription, and a sample barcode and unique molecular identifier (UMI) to enable high-throughput multiplexing and quantification of DSB ends. T7-mediated amplification helps to overcome the need for streptavidin-mediated pull-down of biotinylated DSB ends, which reduces the required amount of input material. As BLISS can label DSBs in cells or tissue sections immobilized and fixed onto a solid surface, the input requirements are further decreased, making BLISS particularly suitable for studies of genome fragility in clinical specimens [201,221]. BLISS was applied to identify endogenous and exogenous DSBs in cultured cells and tissue sections, as well as to chart the specificity of the RNA-guided endonucleases Cas9 and Cpf1 [201], and Cpf1 variants engineered to recognize alternative protospacer adjacent motifs [222]. Furthermore, BLISS has been used to assess AsiSI-induced DSBs and their repressive effect on gene expression in the DSB inducible via AsiSI (DIvA) human cell line [211].
Since the structure of DSB ends is believed to affect DSB repair pathway choice [223], DSB identification approaches that apply DSB end blunting prior to adapter ligation, and especially those that apply formaldehyde fixation [202], are at risk of altering the original and potentially informative structure of DSB ends. To specifically study DSB end structures and to unravel the activities of DSB repair pathways in protecting DSB ends, hairpin capture of DNA end structures (HCoDES) was developed [217], in which DSB ends are treated with ssDNA ligase to form hairpins that allow PCR-based amplification and subsequent sequencing to analyze the precise 5′ end and 3′ end position of both strands.
6.4. On Assay Choice
From the previous two sections it becomes clear that a variety of methods is now available for generating genome-wide DSB maps, with DSB ends identified at (near-)nucleotide resolution (Table 1). However, navigating the different methods and their differences and advantages can be complicated and overwhelming. Ultimately, assay choice should be steered by the specific research question and the characteristics of the sample to be profiled. To map endogenous DSBs, the differences between mapping DSBs over a period of several hours and based on repair, versus generating a snapshot of all DSBs including very transient ones, as well as intermediates of DSB repair or replication fork remodeling, should be considered. As discussed above, IDLV and especially GUIDE-seq or HTGTS can be great choices for mapping of induced DSB sites, recurrent DSBs and DSB hotspots, but their applicability for the profiling of endogenous genome-wide fragility or fragility related to transcription stress may be somewhat limited by the need for transfection and the time-range required for introduction of the IDLV or ODN sequence or translocation. In relation to this, when repair speed and accuracy at endogenous DSB sites are expected to be high, these methods may specifically miss a considerable amount of endogenous DSB events. Yet, even though genome-wide methods for in vitro tagging of DSB ends are less biased by differences in the type and outcome of the pathways that engage in repair, they can be affected by genome-wide differences in accessibility for DSB blunting and end labeling or adapter ligation, for example.
When resources permit and when not restricted by cell number or the nature of a clinical specimen, the combination of a genome-wide assay with nucleotide resolution, such as BLISS, with a repair-based assay such as HTGTS, could help to place the results in a broader context. Together, these assays may provide a complementary picture in which both the fragility at a certain moment and then those DSBs able to engage in translocations or insertions over a period of time can be assessed. However, when working with clinical specimens where cell numbers are generally limited and transfectability may be suboptimal and variable between successive samples, it may be favorable to work with cell fixation and in vitro tagging of DSBs.
Another important aspect worth considering is how the generated data will be analyzed to grasp the factors that shape the genome-wide landscape of DSB frequency and distribution in a particular cell type. Fragility scores can for example be correlated to maps of replication timing, transcriptional activity, or chromatin accessibility, preferentially generated in parallel on the same cell type. Furthermore, the frequency of DSB events can be analyzed in light of genome-wide maps of R-loops generated with DNA-RNA immunoprecipitation combined with sequencing (DRIP-seq) [224], binding of transcription or architectural factors created with (exo-)ChIP-seq, and 3D genome folding or nuclear organization, for instance assessed with (capture) Hi-C [225,226] or DamID technology [227].
7. Concluding Remarks and Outlook
7.1. Conclusions and Additional Remarks
In this review, we have highlighted several endogenous processes that can lead to the formation of DSBs—either planned or as a result of local disturbances. Furthermore, although some endogenous DSBs play vital physiological roles—for example, in the context of planned genomic recombination, transcription, or replication fork rescue—any type of DSB represents a threat to the stability of the genome when faithful repair fails. The variety of methods to profile DSBs discussed in Part II, as well as their diverse applications, illustrates the complexity and diversity of the field of DSB identification, especially when one considers that genome-wide nucleotide-resolution methods only represent one of several angles to approach DSB biology.
7.1.1. Integrative Approaches and Confounding Factors
In the years to come, to deepen our understanding of variation in DSB susceptibility and cellular implications, integrative approaches will be needed to help decipher how genome-wide landscapes of endogenous DSBs, mapped through one of the methods outlined above (Table 1), are shaped by and/or shape the underlying transcriptome, epigenome, 3D genome, and, possibly, the compartmentalized context of the nucleus. While the genome-wide DSB frequency, distribution [122], and repair rate [228] differ between underlying causative processes, these will in turn also be affected by the spatiotemporally varying architecture of the genome and cell cycle dynamics. Importantly, many factors associated with genome-wide fragility, including DNA replication and transcription, are inherently interconnected, and correlate to underlying genomic and epigenomic features across multiple genomic scales, such as gene density, GC levels, and DNase I hypersensitivity [229]. Hence, understanding the relative impact of a given feature on DSB susceptibility is complicated by potential confounding factors [230], and fragility should be approached as a probabilistic outcome. In the next paragraphs, we will briefly touch upon two features that relate to the observed fragility landscapes and to the possible long-term consequences of repair outcome: clustering of DSBs and repair pathway choice.
7.1.2. Compartmentalized DSB repair
In addition to certain genomic regions being more prone than others to form DSBs under specific conditions, the possibility of compartmentalized DSB repair and the sequestering of some DSBs to dedicated nuclear regions in mammalian cells is now emerging [231,232,233,234]. For example, using capture Hi-C [225,235] to assess the genomic surroundings and clustering of DSBs induced by the AsiSI endonuclease, clustering was observed in the case of DSBs induced in transcriptionally active genes during G1, and cluster formation was associated with delayed DSB repair [233,236]. Large 53BP1 and γH2AX clusters have also been observed upon mild replication stress in human fibroblasts [237,238], and re-localization of DSBs in heterochromatin to the nuclear periphery—to enable safe HR repair—was recently further elucidated in Drosophila [239,240].
DSB repair compartmentalization might allow for spatiotemporal uncoupling of different DSB repair pathways in a cell cycle phase-dependent manner [231,241], potentially acting to suppress genomic rearrangements between juxtaposed repetitive chromosomal domains. Related to this, the choice of DSB repair pathway is controlled by multiple factors, including cell cycle phase and proliferative state, as well as local chromatin composition [209], and by the severity and nature of the DSBs [242]. Interestingly, a recent attempt to quantify the kinetics and fidelity of repair of DSBs induced by the Cas9 nuclease revealed that—in this context—DSB repair is variably slow and surprisingly error-prone [228]. Although these findings may not be directly representative of endogenous DSBs, the presented approach can serve as an inspiration for future studies of repair pathway choice, rate, and fidelity.
7.1.3. Studying DSB Biology at Ectopically Induced Genome-Wide DSB Sites
Although endogenous DSBs can be induced by cell stimulation with hormones or transcription factors, or by applying mild replication stress, the effects on fragility are generally not homogenous throughout the cell population, which makes it challenging to draw general conclusions about their effects on transcription or repair. While not valid as endogenous DSBs, the frequently used cell system DIvA [187] and its auxin-inducible degron variant (AID-AsiSI-ER, to rapidly degrade AsiSI and allow DSB repair) [243], have elegantly enabled assessment of the chromatin surrounding the over one thousand AsiSI cut sites, and have helped to pave the way towards better understanding of DSB biology. For instance, a recent study uncovered preferences of different DSB repair pathways as well as chromatin alterations around the most frequently digested AsiSI sites using ChIP-seq [209,243]. While this system presents a powerful tool to study genome-wide effects of DSB formation on transcription of nearby genes, repair, 3D genome organization and many more nuclear processes, a downside is that many of the AsiSI sites in the genome cannot be efficiently cut, and even for the most frequently successfully digested sites, digestion frequency is probabilistic and not uniform across the cell population.
To our knowledge there is, at present, no method that allows for 100% efficiency of DSB induction at defined sites in the human genome. Although RNA-guided endonucleases are an arguable exception, it has been suggested—as mentioned above—that the cellular response to CRISPR/Cas9-induced DSBs may not be indicative of regular repair of endogenous DSBs [228]. In line with this, also the events observed at the AsiSI sites introduced in DIvA cells may not fully recapitulate the same processes that occur upon endogenous DSB formation. Yet, both of these approaches are highly valuable to broaden our knowledge of DSB biology in the context of the nucleus.
Furthermore, we envision methods that combine identification of endogenous DSBs with, for example, concurrent assessment of local features such as protein identity or 3D genomic neighborhoods. Another venue of future technology development will be the implementation of combinatorial assays to simultaneously map DSBs and key epigenomic features in the same sample and even single cells, over time. In line with this, single-cell approaches to DSB identification will not only be key to deciphering cell-to-cell variation in fragility landscapes, repair outcomes, and genome folding, but also to helping advance our knowledge of the heterogeneous nature of DSB landscapes in cancer cells and tumor populations underlying tumor evolution.
7.2. Open Questions in the Field and Outlook
Despite the variety of assays available to map genome-wide DSB landscapes, several issues have not been solved so far. As pointed out previously [121], due to the genome-wide nature of these assays, the focus is mostly on sites that form recurrent DSBs. Even when single-cell DSB profiling could be performed robustly, single DSBs would not be easily identified above the background. Furthermore, whether DSB profiling is applied to endogenous or exogenous DSBs, the effect of noise—as well as the absence of a proper notion of what may be considered as background signal—is hard to work around, and low-abundance yet recurrent or pathological DSBs may be masked. Although different assays have their specific caveats, the community would benefit from a reference standard to which all mapped landscapes of DSBs can be compared, and on which novel and existing assays can be tested in addition to regular validation assays [121].
In line with the emerging relevance of repair signatures in cancer genome studies, exploring the levels of ongoing genome fragility by mapping DSBs directly in clinical specimens might provide new insights into the mechanisms that shape cancer genomes and their evolution. Ongoing fragility underlies most of the future CNAs and SVs, and especially in the context of a cancer cell, DSB landscapes may hold clues on highly fragile regions that underlie rearrangements frequently associated with a tendency to metastasize. In line with that, comparing DSB profiles with the landscape of rearrangements at a later timepoint during tumor evolution, as recently attempted [244], will be instrumental for a better understanding of how DSBs are converted into rearrangements that fuel tumor progression.
Funding
This work was supported by a Rubicon Postdoctoral Fellowship from the Netherlands Organisation for Scientific Research (NWO, the Netherlands) to B.B. and by funding from the Swedish Research Council (521-2014-2866), the Swedish Cancer Research Foundation (CAN 2015/585), the Ragnar Söderberg Foundation (Fellows in Medicine 2016), and the Strategic Research Programme in Cancer (StratCan) at Karolinska Institutet to N.C.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Garinis, G.A.; van der Horst, G.T.J.; Vijg, J.; Hoeijmakers, J.H.J. DNA damage and ageing: new-age ideas for an age-old problem. Nat. Cell Biol. 2008, 10, 1241–1247. [Google Scholar] [CrossRef] [PubMed]
- White, R.R.; Vijg, J. Do DNA Double-Strand Breaks Drive Aging? Mol. Cell 2016, 63, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lindahl, T. Maintenance of Genome Stability. Genomics Proteomics Bioinform. 2016, 14, 119–121. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef] [PubMed]
- Vilenchik, M.M.; Knudson, A.G. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. USA 2003, 100, 12871–12876. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA damage response: making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Cannan, W.J.; Pederson, D.S. Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. J. Cell. Physiol. 2016, 231, 3–14. [Google Scholar] [CrossRef] [PubMed]
- van Gent, D.C.; Hoeijmakers, J.H.; Kanaar, R. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2001, 2, 196–206. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, P.J.; Caldecott, K.W. DNA strand break repair and human genetic disease. Annu. Rev. Genomics Hum. Genet. 2007, 8, 37–55. [Google Scholar] [CrossRef] [PubMed]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, G.; d’Adda di Fagagna, F. Transcription and DNA Damage: Holding Hands or Crossing Swords? J. Mol. Biol. 2017, 429, 3215–3229. [Google Scholar] [CrossRef] [PubMed]
- Borde, V.; de Massy, B. Programmed induction of DNA double strand breaks during meiosis: Setting up communication between DNA and the chromosome structure. Curr. Opin. Genet. Dev. 2013, 23, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Her, J.; Bunting, S.F. How cells ensure correct repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10502–10511. [Google Scholar] [CrossRef] [PubMed]
- Alt, F.W.; Schwer, B. DNA double-strand breaks as drivers of neural genomic change, function, and disease. DNA Repair 2018. [Google Scholar] [CrossRef] [PubMed]
- Burma, S.; Chen, B.P.C.; Chen, D.J. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair 2006, 5, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- McVey, M.; Lee, S.E. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. TIG 2008, 24, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Frit, P.; Barboule, N.; Yuan, Y.; Gomez, D.; Calsou, P. Alternative end-joining pathway(s): Bricolage at DNA breaks. DNA Repair 2014, 17, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Sallmyr, A.; Tomkinson, A.E. Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J. Biol. Chem. 2018, 293, 10536–10546. [Google Scholar] [CrossRef] [PubMed]
- Little, R.D.; Platt, T.H.; Schildkraut, C.L. Initiation and termination of DNA replication in human rRNA genes. Mol. Cell. Biol. 1993, 13, 6600–6613. [Google Scholar] [CrossRef] [PubMed]
- Petryk, N.; Kahli, M.; d’Aubenton-Carafa, Y.; Jaszczyszyn, Y.; Shen, Y.; Silvain, M.; Thermes, C.; Chen, C.-L.; Hyrien, O. Replication landscape of the human genome. Nat. Commun. 2016, 7, 10208. [Google Scholar] [CrossRef] [PubMed]
- Fachinetti, D.; Bermejo, R.; Cocito, A.; Minardi, S.; Katou, Y.; Kanoh, Y.; Shirahige, K.; Azvolinsky, A.; Zakian, V.A.; Foiani, M. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol. Cell 2010, 39, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Dewar, J.M.; Walter, J.C. Mechanisms of DNA replication termination. Nat. Rev. Mol. Cell Biol. 2017, 18, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Gelot, C.; Magdalou, I.; Lopez, B.S. Replication stress in Mammalian cells and its consequences for mitosis. Genes 2015, 6, 267–298. [Google Scholar] [CrossRef] [PubMed]
- Petrakis, T.G.; Komseli, E.-S.; Papaioannou, M.; Vougas, K.; Polyzos, A.; Myrianthopoulos, V.; Mikros, E.; Trougakos, I.P.; Thanos, D.; Branzei, D.; et al. Exploring and exploiting the systemic effects of deregulated replication licensing. Semin. Cancer Biol. 2016, 37–38, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Shima, N.; Pederson, K.D. Dormant origins as a built-in safeguard in eukaryotic DNA replication against genome instability and disease development. DNA Repair 2017, 56, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Hills, S.A.; Diffley, J.F.X. DNA replication and oncogene-induced replicative stress. Curr. Biol. CB 2014, 24, R435–444. [Google Scholar] [CrossRef] [PubMed]
- Dellino, G.I.; Pelicci, P.G. Next-generation sequencing and DNA replication in human cells: The future has arrived. Future Oncol. Lond. Engl. 2014, 10, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Macheret, M.; Halazonetis, T.D. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 2015, 10, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.L.; Orr-Weaver, T.L. Replication fork instability and the consequences of fork collisions from rereplication. Genes Dev. 2016, 30, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Halazonetis, T.D.; Gorgoulis, V.G.; Bartek, J. An oncogene-induced DNA damage model for cancer development. Science 2008, 319, 1352–1355. [Google Scholar] [CrossRef] [PubMed]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic instability--an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Di Micco, R.; Fumagalli, M.; Cicalese, A.; Piccinin, S.; Gasparini, P.; Luise, C.; Schurra, C.; Garre, M.; Nuciforo, P.G.; Bensimon, A.; et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006, 444, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Lin, A.W.; McCurrach, M.E.; Beach, D.; Lowe, S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997, 88, 593–602. [Google Scholar] [CrossRef]
- Zeman, M.K.; Cimprich, K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D.; Foiani, M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 2010, 11, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Cortez, D. Preventing replication fork collapse to maintain genome integrity. DNA Repair 2015, 32, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Mirkin, E.V.; Mirkin, S.M. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. MMBR 2007, 71, 13–35. [Google Scholar] [CrossRef] [PubMed]
- Maizels, N.; Gray, L.T. The G4 genome. PLoS Genet. 2013, 9, e1003468. [Google Scholar] [CrossRef] [PubMed]
- Valton, A.-L.; Prioleau, M.-N. G-Quadruplexes in DNA Replication: A Problem or a Necessity? Trends Genet. TIG 2016, 32, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Westhorpe, F.G.; Straight, A.F. Functions of the centromere and kinetochore in chromosome segregation. Curr. Opin. Cell Biol. 2013, 25, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Langston, L.D.; O’Donnell, M. DNA replication: Keep moving and don’t mind the gap. Mol. Cell 2006, 23, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.R.G.; Yeeles, J.T.P. The Initial Response of a Eukaryotic Replisome to DNA Damage. Mol. Cell 2018, 70, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, A.M.; Newlon, C.S. DNA replication fork pause sites dependent on transcription. Science 1996, 272, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Azvolinsky, A.; Giresi, P.G.; Lieb, J.D.; Zakian, V.A. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell 2009, 34, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; García-Muse, T. Causes of genome instability. Annu. Rev. Genet. 2013, 47, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; Gómez-González, B. Genome instability: A mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008, 9, 204–217. [Google Scholar] [CrossRef] [PubMed]
- Prado, F.; Aguilera, A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 2005, 24, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Samarabandu, J.; Devdhar, R.S.; Siegel, A.J.; Acharya, R.; Berezney, R. Segregation of transcription and replication sites into higher order domains. Science 1998, 281, 1502–1506. [Google Scholar] [CrossRef] [PubMed]
- Meryet-Figuiere, M.; Alaei-Mahabadi, B.; Ali, M.M.; Mitra, S.; Subhash, S.; Pandey, G.K.; Larsson, E.; Kanduri, C. Temporal separation of replication and transcription during S-phase progression. Cell Cycle Georget. Tex 2014, 13, 3241–3248. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.M.; Ryan, M.; Kim, R.; Zakas, A.L.; Fu, H.; Lin, C.M.; Reinhold, W.C.; Davis, S.R.; Bilke, S.; Liu, H.; et al. Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 2011, 21, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- Helmrich, A.; Ballarino, M.; Nudler, E.; Tora, L. Transcription-replication encounters, consequences and genomic instability. Nat. Struct. Mol. Biol. 2013, 20, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Huvet, M.; Nicolay, S.; Touchon, M.; Audit, B.; d’Aubenton-Carafa, Y.; Arneodo, A.; Thermes, C. Human gene organization driven by the coordination of replication and transcription. Genome Res. 2007, 17, 1278–1285. [Google Scholar] [CrossRef] [PubMed]
- Helmrich, A.; Ballarino, M.; Tora, L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell 2011, 44, 966–977. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Jinks-Robertson, S. Transcription as a source of genome instability. Nat. Rev. Genet. 2012, 13, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Berretta, J.; Morillon, A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 2009, 10, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; van Oudenaarden, A. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell 2008, 135, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Nudler, E. RNA polymerase backtracking in gene regulation and genome instability. Cell 2012, 149, 1438–1445. [Google Scholar] [CrossRef] [PubMed]
- Core, L.J.; Waterfall, J.J.; Lis, J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 2008, 322, 1845–1848. [Google Scholar] [CrossRef] [PubMed]
- Westover, K.D.; Bushnell, D.A.; Kornberg, R.D. Structural basis of transcription: Separation of RNA from DNA by RNA polymerase II. Science 2004, 303, 1014–1016. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; García-Muse, T. R loops: From transcription byproducts to threats to genome stability. Mol. Cell 2012, 46, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Sanz, L.A.; Hartono, S.R.; Lim, Y.W.; Steyaert, S.; Rajpurkar, A.; Ginno, P.A.; Xu, X.; Chédin, F. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol. Cell 2016, 63, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Ginno, P.A.; Lott, P.L.; Christensen, H.C.; Korf, I.; Chédin, F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 2012, 45, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Hamperl, S.; Bocek, M.J.; Saldivar, J.C.; Swigut, T.; Cimprich, K.A. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 2017, 170, 774–786.e19. [Google Scholar] [CrossRef] [PubMed]
- Ginno, P.A.; Lim, Y.W.; Lott, P.L.; Korf, I.; Chédin, F. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013, 23, 1590–1600. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Chedin, F.; Hsieh, C.-L.; Wilson, T.E.; Lieber, M.R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003, 4, 442–451. [Google Scholar] [CrossRef] [PubMed]
- García-Muse, T.; Aguilera, A. Transcription-replication conflicts: How they occur and how they are resolved. Nat. Rev. Mol. Cell Biol. 2016, 17, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Skourti-Stathaki, K.; Proudfoot, N.J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014, 28, 1384–1396. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Guan, Z.; Liu, J.; Gui, T.; Shen, K.; Manley, J.L.; Li, X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011, 25, 2041–2056. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S.K. A critical role for topoisomerase IIb and DNA double strand breaks in transcription. Transcription 2016, 7, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Sun, Y.; Huang, S.-Y.N.; Nitiss, J.L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016, 17, 703–721. [Google Scholar] [CrossRef] [PubMed]
- Uusküla-Reimand, L.; Hou, H.; Samavarchi-Tehrani, P.; Rudan, M.V.; Liang, M.; Medina-Rivera, A.; Mohammed, H.; Schmidt, D.; Schwalie, P.; Young, E.J.; et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016, 17, 182. [Google Scholar] [PubMed]
- Canela, A.; Maman, Y.; Jung, S.; Wong, N.; Callen, E.; Day, A.; Kieffer-Kwon, K.-R.; Pekowska, A.; Zhang, H.; Rao, S.S.P.; et al. Genome Organization Drives Chromosome Fragility. Cell 2017, 170, 507–521.e18. [Google Scholar] [CrossRef] [PubMed]
- Saunders, A.; Core, L.J.; Lis, J.T. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 2006, 7, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.X.; Smith, E.R.; Shilatifard, A. Born to run: Control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2018, 19, 464–478. [Google Scholar] [CrossRef] [PubMed]
- Petermann, E.; Helleday, T. Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 2010, 11, 683–687. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.; Ashley, A.K.; Hromas, R.; Nickoloff, J.A. More forks on the road to replication stress recovery. J. Mol. Cell Biol. 2011, 3, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, N.; Maehara, K.; Yoshida, K.; Ohkawa, Y.; Fujita, M. Genome-wide analysis of the spatiotemporal regulation of firing and dormant replication origins in human cells. Nucleic Acids Res. 2018, 46, 6683–6696. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D.; Foiani, M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008, 9, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D.; Foiani, M. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 2005, 17, 568–575. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.; Carr, A.M. Impediments to replication fork movement: Stabilisation, reactivation and genome instability. Chromosoma 2013, 122, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, M.; Rossiello, F.; Clerici, M.; Barozzi, S.; Cittaro, D.; Kaplunov, J.M.; Bucci, G.; Dobreva, M.; Matti, V.; Beausejour, C.M.; et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat. Cell Biol. 2012, 14, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Lundin, C.; Erixon, K.; Arnaudeau, C.; Schultz, N.; Jenssen, D.; Meuth, M.; Helleday, T. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 2002, 22, 5869–5878. [Google Scholar] [CrossRef] [PubMed]
- Truong, L.N.; Li, Y.; Shi, L.Z.; Hwang, P.Y.-H.; He, J.; Wang, H.; Razavian, N.; Berns, M.W.; Wu, X. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. USA 2013, 110, 7720–7725. [Google Scholar] [CrossRef] [PubMed]
- Sollier, J.; Stork, C.T.; García-Rubio, M.L.; Paulsen, R.D.; Aguilera, A.; Cimprich, K.A. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 2014, 56, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Marnef, A.; Cohen, S.; Legube, G. Transcription-Coupled DNA Double-Strand Break Repair: Active Genes Need Special Care. J. Mol. Biol. 2017, 429, 1277–1288. [Google Scholar] [CrossRef] [PubMed]
- Geijer, M.E.; Marteijn, J.A. What happens at the lesion does not stay at the lesion: Transcription-coupled nucleotide excision repair and the effects of DNA damage on transcription in cis and trans. DNA Repair 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yuan, F.; Wu, X.; Rechkoblit, O.; Taylor, J.S.; Geacintov, N.E.; Wang, Z. Error-prone lesion bypass by human DNA polymerase eta. Nucleic Acids Res. 2000, 28, 4717–4724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yuan, F.; Wu, X.; Wang, M.; Rechkoblit, O.; Taylor, J.S.; Geacintov, N.E.; Wang, Z. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000, 28, 4138–4146. [Google Scholar] [CrossRef] [PubMed]
- Neelsen, K.J.; Lopes, M. Replication fork reversal in eukaryotes: From dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015, 16, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Ji, F.; Helleday, T.; Ying, S. Mechanisms for stalled replication fork stabilization: New targets for synthetic lethality strategies in cancer treatments. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Guirouilh-Barbat, J.; Lambert, S.; Bertrand, P.; Lopez, B.S. Is homologous recombination really an error-free process? Front. Genet. 2014, 5, 175. [Google Scholar] [CrossRef] [PubMed]
- Quinet, A.; Lemaçon, D.; Vindigni, A. Replication Fork Reversal: Players and Guardians. Mol. Cell 2017, 68, 830–833. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D.; Foiani, M. The checkpoint response to replication stress. DNA Repair 2009, 8, 1038–1046. [Google Scholar] [CrossRef] [PubMed]
- Ait Saada, A.; Lambert, S.A.E.; Carr, A.M. Preserving replication fork integrity and competence via the homologous recombination pathway. DNA Repair 2018. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Ledesma, F.; Aguilera, A. Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep. 2006, 7, 919–926. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.; Foiani, M.; Sogo, J.M. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 2006, 21, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Balestrini, A.; Ristic, D.; Dionne, I.; Liu, X.Z.; Wyman, C.; Wellinger, R.J.; Petrini, J.H.J. The Ku heterodimer and the metabolism of single-ended DNA double-strand breaks. Cell Rep. 2013, 3, 2033–2045. [Google Scholar] [CrossRef] [PubMed]
- Glover, T.W.; Berger, C.; Coyle, J.; Echo, B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 1984, 67, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Durkin, S.G.; Glover, T.W. Chromosome fragile sites. Annu. Rev. Genet. 2007, 41, 169–192. [Google Scholar] [CrossRef] [PubMed]
- Glover, T.W.; Wilson, T.E.; Arlt, M.F. Fragile sites in cancer: More than meets the eye. Nat. Rev. Cancer 2017, 17, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Georgakilas, A.G.; Tsantoulis, P.; Kotsinas, A.; Michalopoulos, I.; Townsend, P.; Gorgoulis, V.G. Are common fragile sites merely structural domains or highly organized “functional” units susceptible to oncogenic stress? Cell. Mol. Life Sci. CMLS 2014, 71, 4519–4544. [Google Scholar] [CrossRef] [PubMed]
- Franchitto, A. Genome instability at common fragile sites: Searching for the cause of their instability. BioMed Res. Int. 2013, 2013, 730714. [Google Scholar] [CrossRef] [PubMed]
- Fungtammasan, A.; Walsh, E.; Chiaromonte, F.; Eckert, K.A.; Makova, K.D. A genome-wide analysis of common fragile sites: What features determine chromosomal instability in the human genome? Genome Res. 2012, 22, 993–1005. [Google Scholar] [CrossRef] [PubMed]
- Barlow, J.H.; Faryabi, R.B.; Callén, E.; Wong, N.; Malhowski, A.; Chen, H.T.; Gutierrez-Cruz, G.; Sun, H.-W.; McKinnon, P.; Wright, G.; et al. Identification of early replicating fragile sites that contribute to genome instability. Cell 2013, 152, 620–632. [Google Scholar] [CrossRef] [PubMed]
- De, S.; Michor, F. DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat. Biotechnol. 2011, 29, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Janoueix-Lerosey, I.; Hupé, P.; Maciorowski, Z.; La Rosa, P.; Schleiermacher, G.; Pierron, G.; Liva, S.; Barillot, E.; Delattre, O. Preferential occurrence of chromosome breakpoints within early replicating regions in neuroblastoma. Cell Cycle Georget. Tex 2005, 4, 1842–1846. [Google Scholar] [CrossRef] [PubMed]
- Stamatoyannopoulos, J.A.; Adzhubei, I.; Thurman, R.E.; Kryukov, G.V.; Mirkin, S.M.; Sunyaev, S.R. Human mutation rate associated with DNA replication timing. Nat. Genet. 2009, 41, 393–395. [Google Scholar] [CrossRef] [PubMed]
- Sima, J.; Gilbert, D.M. Complex correlations: Replication timing and mutational landscapes during cancer and genome evolution. Curr. Opin. Genet. Dev. 2014, 25, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Pope, B.D.; Ryba, T.; Dileep, V.; Yue, F.; Wu, W.; Denas, O.; Vera, D.L.; Wang, Y.; Hansen, R.S.; Canfield, T.K.; et al. Topologically associating domains are stable units of replication-timing regulation. Nature 2014, 515, 402–405. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Mulia, J.C.; Gilbert, D.M. Replication timing and transcriptional control: Beyond cause and effect-part III. Curr. Opin. Cell Biol. 2016, 40, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Mesner, L.D.; Valsakumar, V.; Cieslik, M.; Pickin, R.; Hamlin, J.L.; Bekiranov, S. Bubble-seq analysis of the human genome reveals distinct chromatin-mediated mechanisms for regulating early- and late-firing origins. Genome Res. 2013, 23, 1774–1788. [Google Scholar] [CrossRef] [PubMed]
- Guilbaud, G.; Rappailles, A.; Baker, A.; Chen, C.-L.; Arneodo, A.; Goldar, A.; d’Aubenton-Carafa, Y.; Thermes, C.; Audit, B.; Hyrien, O. Evidence for sequential and increasing activation of replication origins along replication timing gradients in the human genome. PLoS Comput. Biol. 2011, 7, e1002322. [Google Scholar] [CrossRef] [PubMed]
- Finn, K.J.; Li, J.J. Single-stranded annealing induced by re-initiation of replication origins provides a novel and efficient mechanism for generating copy number expansion via non-allelic homologous recombination. PLoS Genet. 2013, 9, e1003192. [Google Scholar] [CrossRef] [PubMed]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Roukos, V.; Misteli, T. The biogenesis of chromosome translocations. Nat. Cell Biol. 2014, 16, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Wijchers, P.J.; de Laat, W. Genome organization influences partner selection for chromosomal rearrangements. Trends Genet. TIG 2011, 27, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Norton, H.K.; Phillips-Cremins, J.E. Crossed wires: 3D genome misfolding in human disease. J. Cell Biol. 2017, 216, 3441–3452. [Google Scholar] [CrossRef] [PubMed]
- Haffner, M.C.; De Marzo, A.M.; Meeker, A.K.; Nelson, W.G.; Yegnasubramanian, S. Transcription-induced DNA double strand breaks: Both oncogenic force and potential therapeutic target? Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 3858–3864. [Google Scholar] [CrossRef] [PubMed]
- Vitelli, V.; Galbiati, A.; Iannelli, F.; Pessina, F.; Sharma, S.; d’Adda di Fagagna, F. Recent Advancements in DNA Damage-Transcription Crosstalk and High-Resolution Mapping of DNA Breaks. Annu. Rev. Genomics Hum. Genet. 2017, 18, 87–113. [Google Scholar] [PubMed]
- Crosetto, N.; Mitra, A.; Silva, M.J.; Bienko, M.; Dojer, N.; Wang, Q.; Karaca, E.; Chiarle, R.; Skrzypczak, M.; Ginalski, K.; et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods 2013, 10, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Mourad, R.; Ginalski, K.; Legube, G.; Cuvier, O. Predicting double-strand DNA breaks using epigenome marks or DNA at kilobase resolution. Genome Biol. 2018, 19, 34. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.-C.; Chang, A.N.; Kao, J.; Du, Z.; Meyers, R.M.; Alt, F.W.; Schwer, B. Long Neural Genes Harbor Recurrent DNA Break Clusters in Neural Stem/Progenitor Cells. Cell 2016, 164, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Lensing, S.V.; Marsico, G.; Hänsel-Hertsch, R.; Lam, E.Y.; Tannahill, D.; Balasubramanian, S. DSBCapture: In situ capture and sequencing of DNA breaks. Nat. Methods 2016, 13, 855–857. [Google Scholar] [CrossRef] [PubMed]
- Baranello, L.; Kouzine, F.; Wojtowicz, D.; Cui, K.; Przytycka, T.M.; Zhao, K.; Levens, D. DNA break mapping reveals topoisomerase II activity genome-wide. Int. J. Mol. Sci. 2014, 15, 13111–13122. [Google Scholar] [CrossRef] [PubMed]
- Madabhushi, R.; Gao, F.; Pfenning, A.R.; Pan, L.; Yamakawa, S.; Seo, J.; Rueda, R.; Phan, T.X.; Yamakawa, H.; Pao, P.-C.; et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell 2015, 161, 1592–1605. [Google Scholar] [CrossRef] [PubMed]
- Suberbielle, E.; Sanchez, P.E.; Kravitz, A.V.; Wang, X.; Ho, K.; Eilertson, K.; Devidze, N.; Kreitzer, A.C.; Mucke, L. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat. Neurosci. 2013, 16, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Schwer, B.; Wei, P.-C.; Chang, A.N.; Kao, J.; Du, Z.; Meyers, R.M.; Alt, F.W. Transcription-associated processes cause DNA double-strand breaks and translocations in neural stem/progenitor cells. Proc. Natl. Acad. Sci. USA 2016, 113, 2258–2263. [Google Scholar] [CrossRef] [PubMed]
- Pankotai, T.; Bonhomme, C.; Chen, D.; Soutoglou, E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat. Struct. Mol. Biol. 2012, 19, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Shanbhag, N.M.; Rafalska-Metcalf, I.U.; Balane-Bolivar, C.; Janicki, S.M.; Greenberg, R.A. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 2010, 141, 970–981. [Google Scholar] [CrossRef] [PubMed]
- Capozzo, I.; Iannelli, F.; Francia, S.; d’Adda di Fagagna, F. Express or repress? The transcriptional dilemma of damaged chromatin. FEBS J. 2017, 284, 2133–2147. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.A.; Ali, S.O. Physiological Roles of DNA Double-Strand Breaks. J. Nucleic Acids 2017, 2017, 6439169. [Google Scholar] [CrossRef] [PubMed]
- Puc, J.; Kozbial, P.; Li, W.; Tan, Y.; Liu, Z.; Suter, T.; Ohgi, K.A.; Zhang, J.; Aggarwal, A.K.; Rosenfeld, M.G. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell 2015, 160, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Hartung, M.L.; Gruber, D.C.; Koch, K.N.; Grüter, L.; Rehrauer, H.; Tegtmeyer, N.; Backert, S.; Müller, A. H. pylori-Induced DNA Strand Breaks Are Introduced by Nucleotide Excision Repair Endonucleases and Promote NF-κB Target Gene Expression. Cell Rep. 2015, 13, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Toller, I.M.; Neelsen, K.J.; Steger, M.; Hartung, M.L.; Hottiger, M.O.; Stucki, M.; Kalali, B.; Gerhard, M.; Sartori, A.A.; Lopes, M.; et al. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc. Natl. Acad. Sci. USA 2011, 108, 14944–14949. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.-G.; Lunyak, V.V.; Perissi, V.; Garcia-Bassets, I.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 2006, 312, 1798–1802. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Panova, T.; Miller, G.; Volkov, A.; Porter, A.C.G.; Russell, J.; Panov, K.I.; Zomerdijk, J.C.B.M. Topoisomerase IIα promotes activation of RNA polymerase I transcription by facilitating pre-initiation complex formation. Nat. Commun. 2013, 4, 1598. [Google Scholar] [CrossRef] [PubMed]
- Madabhushi, R. The Roles of DNA Topoisomerase IIβ in Transcription. Int. J. Mol. Sci. 2018, 19, 1917. [Google Scholar] [CrossRef] [PubMed]
- Schaefer-Klein, J.L.; Murphy, S.J.; Johnson, S.H.; Vasmatzis, G.; Kovtun, I.V. Topoisomerase 2 Alpha Cooperates with Androgen Receptor to Contribute to Prostate Cancer Progression. PLoS ONE 2015, 10, e0142327. [Google Scholar] [CrossRef] [PubMed]
- Bar-Ziv, R.; Voichek, Y.; Barkai, N. Chromatin dynamics during DNA replication. Genome Res. 2016, 26, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Kouzine, F.; Gupta, A.; Baranello, L.; Wojtowicz, D.; Ben-Aissa, K.; Liu, J.; Przytycka, T.M.; Levens, D. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 2013, 20, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Naughton, C.; Avlonitis, N.; Corless, S.; Prendergast, J.G.; Mati, I.K.; Eijk, P.P.; Cockroft, S.L.; Bradley, M.; Ylstra, B.; Gilbert, N. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 2013, 20, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Corless, S.; Gilbert, N. Effects of DNA supercoiling on chromatin architecture. Biophys. Rev. 2016, 8, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Ashour, M.E.; Atteya, R.; El-Khamisy, S.F. Topoisomerase-mediated chromosomal break repair: An emerging player in many games. Nat. Rev. Cancer 2015, 15, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Herreros, F.; Zagnoli-Vieira, G.; Ntai, I.; Martínez-Macías, M.I.; Anderson, R.M.; Herrero-Ruíz, A.; Caldecott, K.W. TDP2 suppresses chromosomal translocations induced by DNA topoisomerase II during gene transcription. Nat. Commun. 2017, 8, 233. [Google Scholar] [CrossRef] [PubMed]
- Sasanuma, H.; Tsuda, M.; Morimoto, S.; Saha, L.K.; Rahman, M.M.; Kiyooka, Y.; Fujiike, H.; Cherniack, A.D.; Itou, J.; Callen Moreu, E.; et al. BRCA1 ensures genome integrity by eliminating estrogen-induced pathological topoisomerase II-DNA complexes. Proc. Natl. Acad. Sci. USA 2018. [Google Scholar] [CrossRef] [PubMed]
- Nitiss, J.L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 2009, 9, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Allan, J.M.; Travis, L.B. Mechanisms of therapy-related carcinogenesis. Nat. Rev. Cancer 2005, 5, 943–955. [Google Scholar] [CrossRef] [PubMed]
- Wright, R.L.; Vaughan, A.T.M. A systematic description of MLL fusion gene formation. Crit. Rev. Oncol. Hematol. 2014, 91, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rowley, J.D. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair 2006, 5, 1282–1297. [Google Scholar] [CrossRef] [PubMed]
- Simard, J.L.; Kircher, S.M.; Didwania, A.; Goel, M.S. Screening for Recurrence and Secondary Cancers. Med. Clin. North Am. 2017, 101, 1167–1180. [Google Scholar] [CrossRef] [PubMed]
- Habash, M.; Bohorquez, L.C.; Kyriakou, E.; Kron, T.; Martin, O.A.; Blyth, B.J. Clinical and Functional Assays of Radiosensitivity and Radiation-Induced Second Cancer. Cancers 2017, 9, 147. [Google Scholar] [CrossRef] [PubMed]
- Cowell, I.G.; Austin, C.A. Mechanism of generation of therapy related leukemia in response to anti-topoisomerase II agents. Int. J. Environ. Res. Public. Health 2012, 9, 2075–2091. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Yaffe, E.; Tanay, A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 2011, 43, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Splinter, E.; Heath, H.; Kooren, J.; Palstra, R.-J.; Klous, P.; Grosveld, F.; Galjart, N.; de Laat, W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006, 20, 2349–2354. [Google Scholar] [CrossRef] [PubMed]
- Nuebler, J.; Fudenberg, G.; Imakaev, M.; Abdennur, N.; Mirny, L.A. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc. Natl. Acad. Sci. USA 2018, 115, E6697–E6706. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.S.; Cattoglio, C.; Darzacq, X.; Tjian, R. Recent evidence that TADs and chromatin loops are dynamic structures. Nucl. Austin Tex 2018, 9, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, V.B.; Semple, C.A. Chromatin loop anchors are associated with genome instability in cancer and recombination hotspots in the germline. Genome Biol. 2018, 19, 101. [Google Scholar] [CrossRef] [PubMed]
- Krefting, J.; Andrade-Navarro, M.A.; Ibn-Salem, J. Evolutionary stability of topologically associating domains is associated with conserved gene regulation. BMC Biol. 2018, 16, 87. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Kemp, C.J.; Henikoff, S. Anthracyclines induce double-strand DNA breaks at active gene promoters. Mutat. Res. 2015, 773, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Rada-Iglesias, A.; Grosveld, F.G.; Papantonis, A. Forces driving the three-dimensional folding of eukaryotic genomes. Mol. Syst. Biol. 2018, 14, e8214. [Google Scholar] [CrossRef] [PubMed]
- Gothe, H.J.; Bouwman, B.A.M.; Gusmao, E.G.; Piccinno, R.; Sayols, S.; Drechsel, O.; Petrosino, G.; Minneker, V.; Josipovic, N.; Mizi, A.; et al. Spatial chromosome folding and active transcription drive DNA fragility and formation of oncogenic MLL translocations. bioRxiv 2018, 485763. [Google Scholar]
- Roca, J. Transcriptional inhibition by DNA torsional stress. Transcription 2011, 2, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Brackley, C.A.; Johnson, J.; Bentivoglio, A.; Corless, S.; Gilbert, N.; Gonnella, G.; Marenduzzo, D. Stochastic Model of Supercoiling-Dependent Transcription. Phys. Rev. Lett. 2016, 117, 018101. [Google Scholar] [CrossRef] [PubMed]
- Tock, A.J.; Henderson, I.R. Hotspots for Initiation of Meiotic Recombination. Front. Genet. 2018, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Alt, F.W.; Zhang, Y.; Meng, F.-L.; Guo, C.; Schwer, B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 2013, 152, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Paigen, K.; Petkov, P. Mammalian recombination hot spots: Properties, control and evolution. Nat. Rev. Genet. 2010, 11, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gostissa, M.; Hildebrand, D.G.; Becker, M.S.; Boboila, C.; Chiarle, R.; Lewis, S.; Alt, F.W. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv. Immunol. 2010, 106, 93–133. [Google Scholar] [PubMed]
- Ghosh, R.; Das, D.; Franco, S. The Role for the DSB Response Pathway in Regulating Chromosome Translocations. Adv. Exp. Med. Biol. 2018, 1044, 65–87. [Google Scholar] [PubMed]
- Willis, N.A.; Rass, E.; Scully, R. Deciphering the Code of the Cancer Genome: Mechanisms of Chromosome Rearrangement. Trends Cancer 2015, 1, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.M.B.; Lupski, J.R. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016, 17, 224–238. [Google Scholar] [CrossRef] [PubMed]
- Hastings, P.J.; Lupski, J.R.; Rosenberg, S.M.; Ira, G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009, 10, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Helleday, T.; Eshtad, S.; Nik-Zainal, S. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 2014, 15, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Stankiewicz, P.; Lupski, J.R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010, 61, 437–455. [Google Scholar] [CrossRef] [PubMed]
- Maciejowski, J.; Imielinski, M. Modeling cancer rearrangement landscapes. Curr. Opin. Syst. Biol. 2017, 1, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.K.; Bashashati, A.; Anglesio, M.S.; Cochrane, D.R.; Grewal, D.S.; Ha, G.; McPherson, A.; Horlings, H.M.; Senz, J.; Prentice, L.M.; et al. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat. Genet. 2017, 49, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.J.; Yachida, S.; Mudie, L.J.; Stephens, P.J.; Pleasance, E.D.; Stebbings, L.A.; Morsberger, L.A.; Latimer, C.; McLaren, S.; Lin, M.-L.; et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 2010, 467, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- McPherson, A.W.; Chan, F.C.; Shah, S.P. Observing Clonal Dynamics across Spatiotemporal Axes: A Prelude to Quantitative Fitness Models for Cancer. Cold Spring Harb. Perspect. Med. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Szilard, R.K.; Jacques, P.-E.; Laramée, L.; Cheng, B.; Galicia, S.; Bataille, A.R.; Yeung, M.; Mendez, M.; Bergeron, M.; Robert, F.; et al. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nat. Struct. Mol. Biol. 2010, 17, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Capetillo, O.; Allis, C.D.; Nussenzweig, A. Phosphorylation of histone H2B at DNA double-strand breaks. J. Exp. Med. 2004, 199, 1671–1677. [Google Scholar] [CrossRef] [PubMed]
- Ward, I.M.; Chen, J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 2001, 276, 47759–47762. [Google Scholar] [CrossRef] [PubMed]
- Iacovoni, J.S.; Caron, P.; Lassadi, I.; Nicolas, E.; Massip, L.; Trouche, D.; Legube, G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010, 29, 1446–1457. [Google Scholar] [CrossRef] [PubMed]
- Massip, L.; Caron, P.; Iacovoni, J.S.; Trouche, D.; Legube, G. Deciphering the chromatin landscape induced around DNA double strand breaks. Cell Cycle Georget. Tex 2010, 9, 2963–2972. [Google Scholar] [CrossRef] [PubMed]
- Rhee, H.S.; Pugh, B.F. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 2011, 147, 1408–1419. [Google Scholar] [CrossRef] [PubMed]
- Vrouwe, M.G.; Pines, A.; Overmeer, R.M.; Hanada, K.; Mullenders, L.H.F. UV-induced photolesions elicit ATR-kinase-dependent signaling in non-cycling cells through nucleotide excision repair-dependent and -independent pathways. J. Cell Sci. 2011, 124, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.-Z.; Li, B.; Huang, B.; Wang, Y.; Liu, X.-D.; Guan, H.; Zhang, S.-M.; Tang, Y.; Rang, W.-Q.; Zhou, P.-K. γH2AX foci formation in the absence of DNA damage: Mitotic H2AX phosphorylation is mediated by the DNA-PKcs/CHK2 pathway. FEBS Lett. 2013, 587, 3437–3443. [Google Scholar] [CrossRef] [PubMed]
- Marti, T.M.; Hefner, E.; Feeney, L.; Natale, V.; Cleaver, J.E. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 2006, 103, 9891–9896. [Google Scholar] [CrossRef] [PubMed]
- Turinetto, V.; Giachino, C. Multiple facets of histone variant H2AX: A DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015, 43, 2489–2498. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Q.; Zheng, Z.; Nguyen, N.T.; Liebers, M.; Topkar, V.V.; Thapar, V.; Wyvekens, N.; Khayter, C.; Iafrate, A.J.; Le, L.P.; et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015, 33, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, R.; Lombardo, A.; Arens, A.; Miller, J.C.; Genovese, P.; Kaeppel, C.; Nowrouzi, A.; Bartholomae, C.C.; Wang, J.; Friedman, G.; et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011, 29, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Wu, X.; Wang, J.; Wang, Y.; Qiu, Z.; Chang, T.; Huang, H.; Lin, R.-J.; Yee, J.-K. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat. Biotechnol. 2015, 33, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Chiarle, R.; Zhang, Y.; Frock, R.L.; Lewis, S.M.; Molinie, B.; Ho, Y.-J.; Myers, D.R.; Choi, V.W.; Compagno, M.; Malkin, D.J.; et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 2011, 147, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Frock, R.L.; Hu, J.; Meyers, R.M.; Ho, Y.-J.; Kii, E.; Alt, F.W. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015, 33, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Meyers, R.M.; Dong, J.; Panchakshari, R.A.; Alt, F.W.; Frock, R.L. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat. Protoc. 2016, 11, 853–871. [Google Scholar] [CrossRef] [PubMed]
- Klein, I.A.; Resch, W.; Jankovic, M.; Oliveira, T.; Yamane, A.; Nakahashi, H.; Di Virgilio, M.; Bothmer, A.; Nussenzweig, A.; Robbiani, D.F.; et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 2011, 147, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.X.; Mirzazadeh, R.; Garnerone, S.; Scott, D.; Schneider, M.W.; Kallas, T.; Custodio, J.; Wernersson, E.; Li, Y.; Gao, L.; et al. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun. 2017, 8, 15058. [Google Scholar] [CrossRef] [PubMed]
- Canela, A.; Sridharan, S.; Sciascia, N.; Tubbs, A.; Meltzer, P.; Sleckman, B.P.; Nussenzweig, A. DNA Breaks and End Resection Measured Genome-wide by End Sequencing. Mol. Cell 2016, 63, 898–911. [Google Scholar] [CrossRef] [PubMed]
- Biernacka, A.; Zhu, Y.; Skrzypczak, M.; Forey, R.; Pardo, B.; Grzelak, M.; Nde, J.; Mitra, A.; Kudlicki, A.; Crosetto, N.; et al. i-BLESS is an ultra-sensitive method for detection of DNA double-strand breaks. Commun. Biol. 2018, 1, 181. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, E.A.; McCulley, A.; Haarer, B.; Arnak, R.; Feng, W. Break-seq reveals hydroxyurea-induced chromosome fragility as a result of unscheduled conflict between DNA replication and transcription. Genome Res. 2015, 25, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Osborn, M.J.; Webber, B.R.; Knipping, F.; Lonetree, C.; Tennis, N.; DeFeo, A.P.; McElroy, A.N.; Starker, C.G.; Lee, C.; Merkel, S.; et al. Evaluation of TCR Gene Editing Achieved by TALENs, CRISPR/Cas9, and megaTAL Nucleases. Mol. Ther. J. Am. Soc. Gene Ther. 2016, 24, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Sánchez-Hernández, S.; Gutiérrez-Guerrero, A.; Pinedo-Gomez, J.; Benabdellah, K. Biased and Unbiased Methods for the Detection of Off-Target Cleavage by CRISPR/Cas9: An Overview. Int. J. Mol. Sci. 2016, 17, 1507. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, T.Y.; Resch, W.; Jankovic, M.; Casellas, R.; Nussenzweig, M.C.; Klein, I.A. Translocation capture sequencing: A method for high throughput mapping of chromosomal rearrangements. J. Immunol. Methods 2012, 375, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.-C.; Lee, C.-S.; Du, Z.; Schwer, B.; Zhang, Y.; Kao, J.; Zurita, J.; Alt, F.W. Three classes of recurrent DNA break clusters in brain progenitors identified by 3D proximity-based break joining assay. Proc. Natl. Acad. Sci. USA 2018, 115, 1919–1924. [Google Scholar] [CrossRef] [PubMed]
- Clouaire, T.; Rocher, V.; Lashgari, A.; Arnould, C.; Aguirrebengoa, M.; Biernacka, A.; Skrzypczak, M.; Aymard, F.; Fongang, B.; Dojer, N.; et al. Comprehensive Mapping of Histone Modifications at DNA Double-Strand Breaks Deciphers Repair Pathway Chromatin Signatures. Mol. Cell 2018, 72, 250–262.e6. [Google Scholar] [CrossRef] [PubMed]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Iannelli, F.; Galbiati, A.; Capozzo, I.; Nguyen, Q.; Magnuson, B.; Michelini, F.; D’Alessandro, G.; Cabrini, M.; Roncador, M.; Francia, S.; et al. A damaged genome’s transcriptional landscape through multilayered expression profiling around in situ-mapped DNA double-strand breaks. Nat. Commun. 2017, 8, 15656. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Baranello, L.; Kouzine, F.; Wojtowicz, D.; Cui, K.; Zhao, K.; Przytycka, T.M.; Capranico, G.; Levens, D. Mapping DNA Breaks by Next-Generation Sequencing. Methods Mol. Biol. Clifton NJ 2018, 1672, 155–166. [Google Scholar]
- Leduc, F.; Faucher, D.; Bikond Nkoma, G.; Grégoire, M.-C.; Arguin, M.; Wellinger, R.J.; Boissonneault, G. Genome-wide mapping of DNA strand breaks. PLoS ONE 2011, 6, e17353. [Google Scholar] [CrossRef] [PubMed]
- Grégoire, M.-C.; Massonneau, J.; Leduc, F.; Arguin, M.; Brazeau, M.-A.; Boissonneault, G. Quantification and genome-wide mapping of DNA double-strand breaks. DNA Repair 2016, 48, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Grégoire, M.-C.; Leduc, F.; Morin, M.H.; Cavé, T.; Arguin, M.; Richter, M.; Jacques, P.-É.; Boissonneault, G. The DNA double-strand “breakome” of mouse spermatids. Cell. Mol. Life Sci. CMLS 2018, 75, 2859–2872. [Google Scholar] [CrossRef] [PubMed]
- Dorsett, Y.; Zhou, Y.; Tubbs, A.T.; Chen, B.-R.; Purman, C.; Lee, B.-S.; George, R.; Bredemeyer, A.L.; Zhao, J.-Y.; Sodergen, E.; et al. HCoDES reveals chromosomal DNA end structures with single-nucleotide resolution. Mol. Cell 2014, 56, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.G.; Ba, Z.; Du, Z.; Zhang, Y.; Hu, J.; Alt, F.W. Highly sensitive and unbiased approach for elucidating antibody repertoires. Proc. Natl. Acad. Sci. USA 2016, 113, 7846–7851. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Dojer, N.; Biernacka, A.; Pardo, B.; Forey, R.; Skrzypczak, M.; Fongang, B.; Kengne, J.B.N.; Yousefi, R.; Pasero, P.; et al. Quantitative DSB sequencing (qDSB-Seq): A method for genome-wide accurate estimation of absolute DNA double-strand break frequencies per cell. bioRxiv 2017, 171405. [Google Scholar]
- Ross, W.E.; Shipley, N. Relationship between DNA damage and survival in formaldehyde-treated mouse cells. Mutat. Res. 1980, 79, 277–283. [Google Scholar] [CrossRef]
- Mirzazadeh, R.; Kallas, T.; Bienko, M.; Crosetto, N. Genome-Wide Profiling of DNA Double-Strand Breaks by the BLESS and BLISS Methods. Methods Mol. Biol. Clifton NJ 2018, 1672, 167–194. [Google Scholar]
- Gao, L.; Cox, D.B.T.; Yan, W.X.; Manteiga, J.C.; Schneider, M.W.; Yamano, T.; Nishimasu, H.; Nureki, O.; Crosetto, N.; Zhang, F. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 2017, 35, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Huertas, P. DNA resection in eukaryotes: Deciding how to fix the break. Nat. Struct. Mol. Biol. 2010, 17, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Halász, L.; Karányi, Z.; Boros-Oláh, B.; Kuik-Rózsa, T.; Sipos, É.; Nagy, É.; Mosolygó-L, Á.; Mázló, A.; Rajnavölgyi, É.; Halmos, G.; et al. RNA-DNA hybrid (R-loop) immunoprecipitation mapping: An analytical workflow to evaluate inherent biases. Genome Res. 2017, 27, 1063–1073. [Google Scholar] [CrossRef] [PubMed]
- Schoenfelder, S.; Furlan-Magaril, M.; Mifsud, B.; Tavares-Cadete, F.; Sugar, R.; Javierre, B.-M.; Nagano, T.; Katsman, Y.; Sakthidevi, M.; Wingett, S.W.; et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015, 25, 582–597. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; de Klein, A.; Wessels, L.; de Laat, W.; et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008, 453, 948–951. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, E.K.; Chen, T.; de Haas, M.; Holland, H.A.; Akhtar, W.; van Steensel, B. Kinetics and Fidelity of the Repair of Cas9-Induced Double-Strand DNA Breaks. Mol. Cell 2018, 70, 801–813.e6. [Google Scholar] [CrossRef] [PubMed]
- ENCODE Project Consortium A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011, 9, e1001046.
- de Wit, E.; de Laat, W. A decade of 3C technologies: Insights into nuclear organization. Genes Dev. 2012, 26, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Lemaître, C.; Soutoglou, E. DSB (Im)mobility and DNA repair compartmentalization in mammalian cells. J. Mol. Biol. 2015, 427, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Kalousi, A.; Soutoglou, E. Nuclear compartmentalization of DNA repair. Curr. Opin. Genet. Dev. 2016, 37, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Aymard, F.; Aguirrebengoa, M.; Guillou, E.; Javierre, B.M.; Bugler, B.; Arnould, C.; Rocher, V.; Iacovoni, J.S.; Biernacka, A.; Skrzypczak, M.; et al. Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nat. Struct. Mol. Biol. 2017, 24, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Marnef, A.; Legube, G. Organizing DNA repair in the nucleus: DSBs hit the road. Curr. Opin. Cell Biol. 2017, 46, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dryden, N.H.; Broome, L.R.; Dudbridge, F.; Johnson, N.; Orr, N.; Schoenfelder, S.; Nagano, T.; Andrews, S.; Wingett, S.; Kozarewa, I.; et al. Unbiased analysis of potential targets of breast cancer susceptibility loci by Capture Hi-C. Genome Res. 2014, 24, 1854–1868. [Google Scholar] [CrossRef] [PubMed]
- Guénolé, A.; Legube, G. A meeting at risk: Unrepaired DSBs go for broke. Nucl. Austin Tex 2017, 8, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Harrigan, J.A.; Belotserkovskaya, R.; Coates, J.; Dimitrova, D.S.; Polo, S.E.; Bradshaw, C.R.; Fraser, P.; Jackson, S.P. Replication stress induces 53BP1-containing OPT domains in G1 cells. J. Cell Biol. 2011, 193, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Lukas, C.; Savic, V.; Bekker-Jensen, S.; Doil, C.; Neumann, B.; Pedersen, R.S.; Grøfte, M.; Chan, K.L.; Hickson, I.D.; Bartek, J.; et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 2011, 13, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Caridi, C.P.; D’Agostino, C.; Ryu, T.; Zapotoczny, G.; Delabaere, L.; Li, X.; Khodaverdian, V.Y.; Amaral, N.; Lin, E.; Rau, A.R.; et al. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 2018, 559, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Caridi, C.P.; Delabaere, L.; Tjong, H.; Hopp, H.; Das, D.; Alber, F.; Chiolo, I. Quantitative Methods to Investigate the 4D Dynamics of Heterochromatic Repair Sites in Drosophila Cells. Methods Enzymol. 2018, 601, 359–389. [Google Scholar] [PubMed]
- Tsouroula, K.; Furst, A.; Rogier, M.; Heyer, V.; Maglott-Roth, A.; Ferrand, A.; Reina-San-Martin, B.; Soutoglou, E. Temporal and Spatial Uncoupling of DNA Double Strand Break Repair Pathways within Mammalian Heterochromatin. Mol. Cell 2016, 63, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Mladenov, E.; Magin, S.; Soni, A.; Iliakis, G. DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: Cell cycle and proliferation-dependent regulation. Semin. Cancer Biol. 2016, 37–38, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Aymard, F.; Bugler, B.; Schmidt, C.K.; Guillou, E.; Caron, P.; Briois, S.; Iacovoni, J.S.; Daburon, V.; Miller, K.M.; Jackson, S.P.; et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014, 21, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Ballinger, T.J.; Bouwman, B.; Mirzazadeh, R.; Garnerone, S.; Crosetto, N.; Semple, C. Modelling double strand break susceptibility to interrogate structural variation in cancer. bioRxiv 2018, 441832. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).