1. Introduction
Phaseolus vulgaris appears to have split from its nearest relatives approximately 1.3 million years ago (MYA) based on ribosomal internal transcribed spacer sequence data [
1]. The available evidence strongly points towards Middle America as the ultimate origin of
P. vulgaris where all the nearest relatives of
P. vulgaris are also localized. This origin hypothesis has been complicated by new evidence that
P. vulgaris may have originated in Middle America but the ancestral form of the species underwent further speciation through allopatry in the Amotape–Huancabamba Depression of the Andes [
2]. Several complicated movements of germplasm followed this speciation event culminating in the development of two separate gene pools in the Andes and Middle America at ~165,000 BP [
3]. Humans then domesticated
P. vulgaris within each of these gene pools nearly simultaneously in a dual event at ~8000 BP. These concurrent domestications of the same species in two geographically distinct regions has resulted in a great phylogenetic divide in domesticated beans whose existence is supported by evidence from multiple sources. The consequences of this separation are numerous, including distinct adaptations and traits in each gene pool, and genetic incompatibilities between the gene pools. Seed size, heliotropism, leaf size, and phaseolin storage protein type are some of the characteristics strongly influenced by ancestry. Plant breeders have also observed that characteristics critical to breeding, such as higher yields and stress tolerances, are highly modulated by center of domestication [
4].
One consequence of this divide is the production of so called bean races distinctive to each gene pool associated with its respective center of domestication that were first described by Singh and colleagues in 1991 [
5]. This work to define finer scale gene pools within the larger Andean-Middle American framework began with observations of phenotypic characteristics but was later supported by molecular and biochemical evidence. Specifically, differences in the leaf morphology, seed size, seed shape, pod morphology, stem thickness, internode length, plant habit, allozyme type, and phaseolin type could be used to identify three Mesoamerican races (Durango, Jalisco, and Mesoamerica) and three Andean races (Peru, Chile, and Nueva Granada). The six races could not be identified with absolute reliability and exceptions to both molecular characterization and phenotypic traits did occur in the data, but at a relatively low rate. A seventh race (Guatemala) within the Middle American gene pool, was later distinguished using random amplified polymorphic DNA (RAPD) data and geographic and morphological differences [
6]. An additional four studies have shown distinctive clustering of the races using microsatellite data, although only the RAPD work of Beebe has clearly separated race Jalisco from race Durango [
7,
8,
9,
10]. All of this molecular research has shown variable overlap between the races, thus underlining the need for further research into bean races.
The single most important event after the speciation and domestication of
P. vulgaris has been the Columbian Exchange and the dispersal of domesticated beans around the globe forming secondary centers of diversity. The Columbian Exchange brought
P. vulgaris and other crops to the Iberian Peninsula, Italy, and to the far-flung colonies of Spain and Portugal, such as the Philippines, and sub-Saharan and North Africa. An early Spanish colony that received goods from the Americas was the Philippines, and an obvious port of call near the Philippines would have been the Portuguese colony of Macau, which is an autonomous state in modern day China. As for the remainder of Europe, it is evident that multiple introductions took place over time and a simple lineage through the Iberian Peninsula cannot be derived [
11]. One result of this early dispersal of
P. vulgaris worldwide, however it may have happened, is the development of potential secondary centers of diversity in China and Europe [
11,
12]
The place of snap beans in this history of domestication and dispersal is unclear as to its initial origins. Superficially, snap beans are dry beans that have fleshier pods that are low in fiber where immature pods are eaten as a vegetable. However, there is a wide range in variation in what are classified as “snap beans”. This variation is reflected in the names that are used for this class around the world. Terminology includes “French bean”, often used in Europe to denote this type, “string bean”, referring to types where the pod suture strings must be removed, and “green bean”—vegetable forms with succulent green pods as opposed to “wax bean” with yellow pods [
13]. Those showing the least amount of change from dry beans are the large flat podded types represented by Romano beans in European culture. These have thin pod walls with low fiber at maturity (
Table 1).
In addition to low pod wall fiber, many forms of string and snap beans have thick and fleshy pod walls. Contemporary cultivars have pods that are elongated and cylindrical in shape, which also affects seed shape. Another trait now nearly ubiquitous in snap beans is a lack of a “string” (fiber) in the abaxial and adaxial pod sutures. Because both stringy and stringless types occur, genetic control of strings is thought to be independent of pod wall fiber. In this paper, we use the term “snap bean” to generally designate common bean types with low fiber that are used as a vegetable. Contemporary snap beans have been selected for additional traits that adapt them to particular production systems and uses. These include determinant bush (type I) growth habit, prolific flower production combined with concentration of pod set, pods of different sizes with pod cross-sections ranging from oval to round, variation in pod color from yellow (wax beans) to very dark green and white flower and seed color. One hypothesis is that snap beans arose from dry beans through a process of stepwise mutation—the first being reduction in pod fiber followed by changes in pod shape and eventually development of stringless types.
The belief that snap beans have been selected out of dry beans after the Columbian Exchange is based in part on the fact that accessions with snap characteristics are very rare in North, Central and South American landrace collections [
4,
14]. This rarity may be due in part to the low nutritional density of snap beans in comparison to the protein and carbohydrate content of dry beans, along with the storability of dry beans that led most pre-Columbian peoples of the Americas to prefer dry beans. In addition, traits that reduce pod fiber may reduce fitness of snap relative to dry bean. Because loss of pod fiber prevents normal pod dehiscence, snap beans are more difficult to thresh and clean for mature seed. Reduction in pod fiber may also affect ability to fend off plant pathogens.
In pre-Columbian times, there was very likely consumption of immature dry bean pods [
5] when pod fiber is low as is found in subsistence cultures around the world today. This consumption typically happens during the “hunger season” when the crop has been planted and is growing, but stored food reserves are depleted. Because the primary use remained the mature and dry seed, little selection for vegetable characteristics has been practiced. Linguistic evidence suggests the vegetable use of common bean, such as the word, exotl, from Nahuatl language of the Aztecs referring to green pods and the word, chaucha, from the Quechua language also referring to green pods [
15,
16]. This linguistic evidence, may reflect consumption of immature dry bean pods, or, as other authors have observed, it “(…) may show a long-term knowledge of snap beans that was not introduced or re-introduced as modified dry bean germplasm from outside the region (…)” [
16]. In addition, one potential Native North American snap bean, ‘Cherokee Trail of Tears’, has been identified [
17]. Based on the low-level presence of snap traits in American landrace material and the linguistic knowledge of snap beans by some pre-Columbian people of the Americas, it is possible that snap traits existed in a few of the materials coming from the Americas, but these traits were highly amplified by selection in Europe where snap beans were developed. Snap beans were returned to the Americas with migration of immigrants to the New World. European farmer selected snap materials form the basis of commercial snap beans today, although snap beans may have also been developed separately in China.
Another aspect of the origin of snap beans that remains unclear is the relationship of snap beans to the Middle American and Andean gene pools and races. The original work on bean races categorized all snap beans (gene pools 11 and 12) as races Nueva Granada and Chile of Andean origin [
4,
5]. Gepts and Bliss [
18] found that the majority of beans with low fiber pods from Europe had T phaseolin (associated with the Andean gene pool) but some landraces had S phaseolin (associated with the Middle American gene pool). In the Americas, all snap beans had either T or C phaseolin [
19]. Later work using microsatellite data and RAPDs showed apparent hybridization between Middle American and Andean gene pools in snap beans [
15,
20,
21]. More recently, a phylogenetic analysis of both dry and snap beans using a single nucleotide polymorphism (SNP) array placed several snap bean cultivars in an intermediate region, although the snap beans were more heavily skewed towards the Andean gene pool [
22]. These data suggest that snap beans may have originated in more than one gene pool, and have undergone extensive intermating.
Historically, an important region for snap bean production in the 20th and 21st centuries has been the Willamette Valley of Oregon, where a distinctive lineage of snap bean is grown that is generally referred to as “Blue Lake”. This snap bean lineage was brought to Oregon from the Blue Lakes region of California early in the 20th century, but its ultimate origin is unclear. It is known for excellent canning and processing characteristics and quality traits that are desirable to consumers as well as the ability to achieve high yields in the field. The origin of this distinctive lineage has been purported to be ‘Scotia’, ‘Genuine Cornfield’, or ‘White Creaseback’ [
23].
The objective of the present study of the genetic history and diversity of snap beans was to elucidate the origins, intermixing, gene pools, and, where possible, the bean races associated with snap beans.
4. Discussion
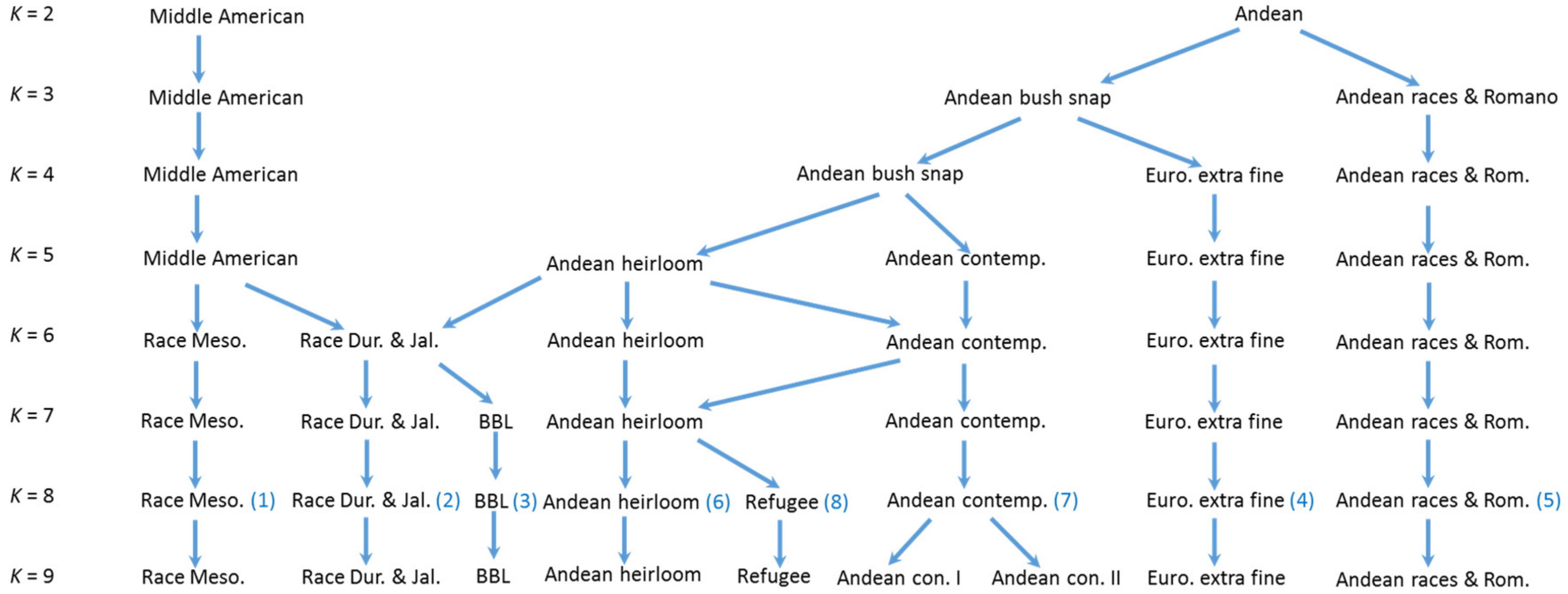
4.1. K = 2 Structure Analysis
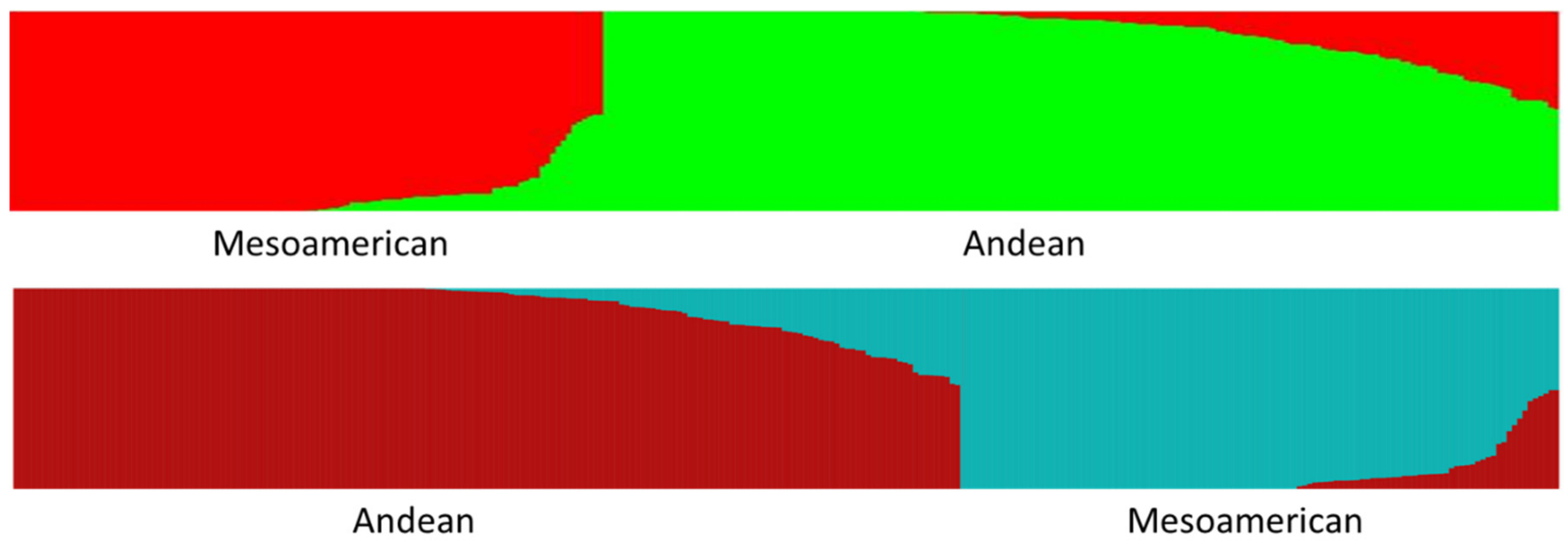
The
K = 2 Structure analysis provides a perspective that is strongly focused on the two overarching Andean and Middle American gene pools present in common bean. This perspective can shed light upon the differences and similarities between the geographic arenas of China, The Iberian Peninsula, Europe, and North America. The
K = 2 Structure analysis shows that the commercial beans in the BeanCAP diversity panel are both skewed towards the Andean center and heavily intermixed with nearly half containing an admixture to their predominate type of greater than 10% and about a third containing an admixture greater than 20% (
Table 2). This likely reflects the work of plant breeders to intercross lines for selection of new trait combinations, but a small portion of this intermixing could also reflect accidental outcrosses in an on farm context, although outcrossing rates in common bean are known to be extremely low. In comparison to the BeanCAP, the rates of intermixing were significantly lower for heirloom pole beans of Europe and North America and for Spanish bean lines, and were lower still for Chinese bean lines and dry beans. This indicates that these bean lines have undergone much less intercrossing by breeders. The Chinese and American heirlooms beans also contained much higher percentages of predominately Middle American germplasm, whereas commercial materials from Europe and North America as well as the Spanish collection are predominately Andean. The high degree of Middle American genetic background in American snap heirlooms may reflect the proximity of Central America to places such as the Southeastern United States.
4.2. Expected Heterozygosity
Expected heterozygosity was also analyzed to gain a sense of the genetic diversity present in these germplasm collections. This analysis showed that rare alleles were present at higher rates in the dry beans than in any other collection. This result is consistent with the notion that snap beans were derived from dry beans and that snap beans may have undergone a genetic bottleneck. But it also showed that the cumulative diversity of all the snap beans, including Chinese, Spanish, American/European pole beans, and the BeanCAP were nearly as diverse as our selected dry bean subset containing both centers of domestication and all bean races. This shows that a broader collection of snap beans or a carefully chosen core collection at a gene bank could capture a significant amount of genetic diversity in snap beans. There is a difference in the size of n between the dry beans and all snap beans combined, and this difference in n would tend to reduce the HET value for dry beans, but this is counterbalanced by the fact that the dry beans were handpicked to be as diverse as possible with all bean races represented. An interesting side-light to this analysis is the surprising diversity of European and American heirloom pole beans (all other pole snap) which was higher than any other subset of snap beans.
4.3. Phylogenetic Analysis
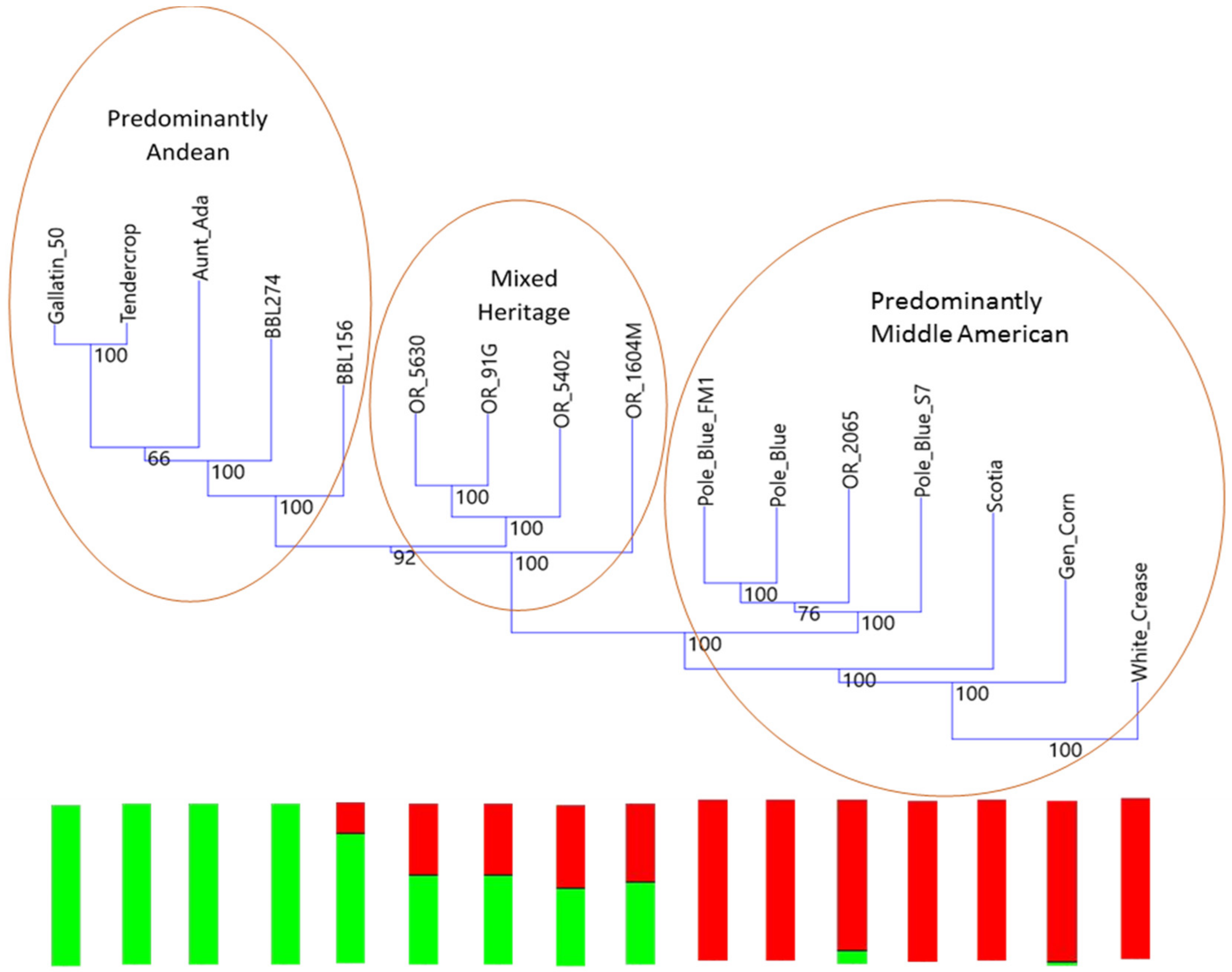
The Blue Lake phylogenetic tree shows the degree to which the original Blue Lake genetic background has been altered by breeding efforts over the years. This represents two opportunities for future breeding in the Blue Lake background. First, it suggests a relatively straightforward method for monitoring changes to the genome by gauging the level of admixture. Second, it suggests one path forward for improving Blue Lake, namely returning to the original pole habit materials for the original quality traits that first made Blue Lake beans popular.
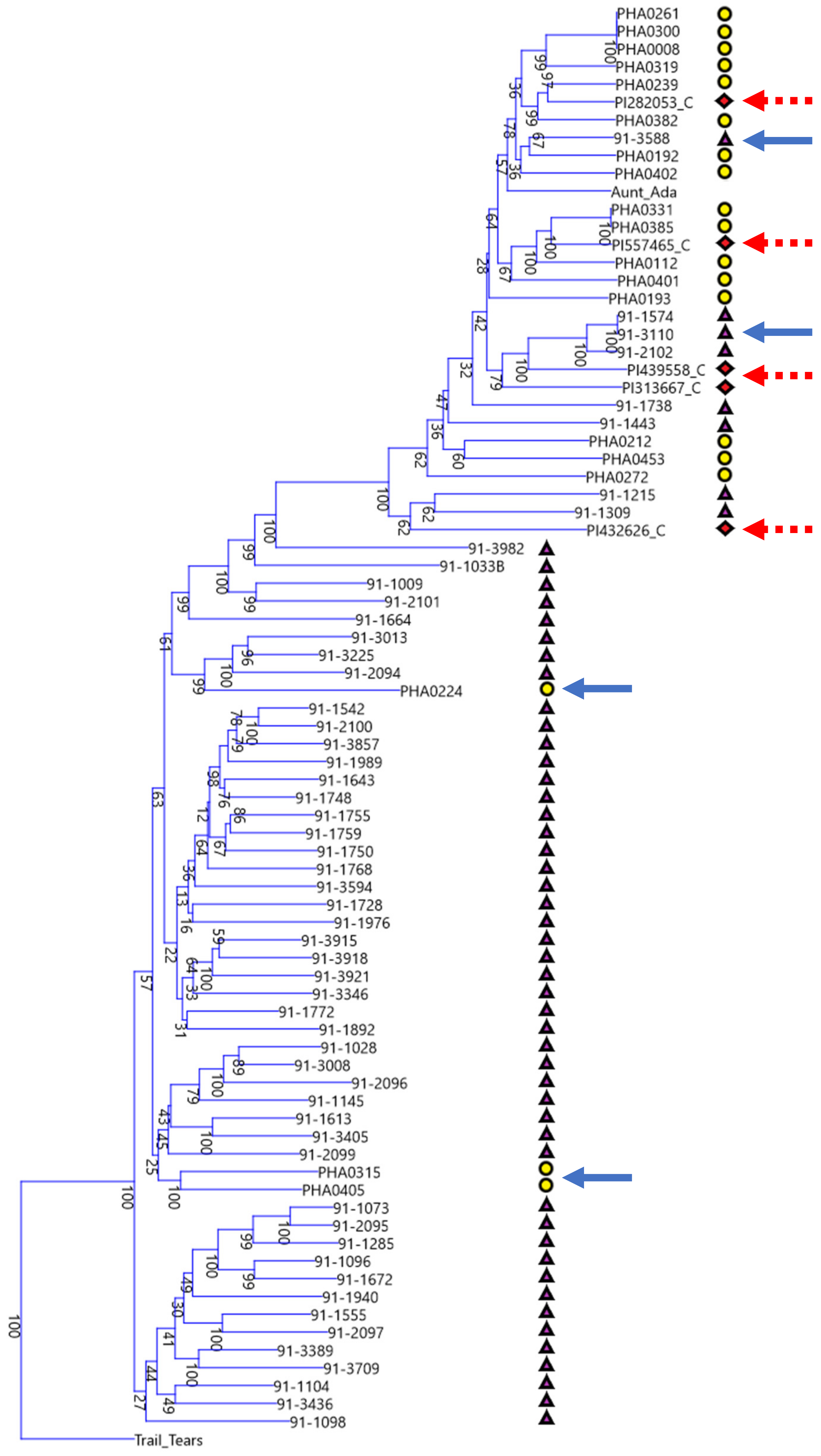
The phylogenetic tree of Chinese, Spanish, and race Chile bean lines shows that these disparate geographic centers of secondary diversity (i.e., the Iberian Peninsula and China) are actually connected genetically as can be seen in the multiple shared clades in both the Middle American and Andean gene pools. Despite the geographic distance, this should not be entirely surprising. There are plausible trade routes out of the Americas during the early years of the Columbian Exchange, such as trade through the Spanish colony of the Philippines and then through the Portuguese colony of Macau in modern day China, and day length adaptations that overlap in parts of China and the Iberian Peninsula. According to Singh and colleagues [
5], bean race Chile is centered on modern day Chile at a latitude of approximately 30° to 40° in its central part, which overlaps favorably with the roughly 36° to 43° latitude of the Iberian Peninsula and with the Northern parts of China. Yet in spite of these overlaps and cladistic connections, there are also stark differences. When viewed through the lens of either/or groupings, the Chinese materials are overwhelmingly derived from the Middle American gene pool but the Spanish materials are overwhelmingly derived from the Andean gene pool. This can be seen in the disproportionate split between Chinese and Spanish accessions shown in the phylogram centered at the branch point just below ‘91-3982’. Just as shared trade routes and shared day length adaptations at similar latitudes may explain the similarities, differing trade routes and the differing day length adaptations of Southern China in comparison to Chile and the Iberian Peninsula may explain the strong influence of the Mesoamerican gene pool on the Chinese lines.
4.4. Principal Coordinates Analysis
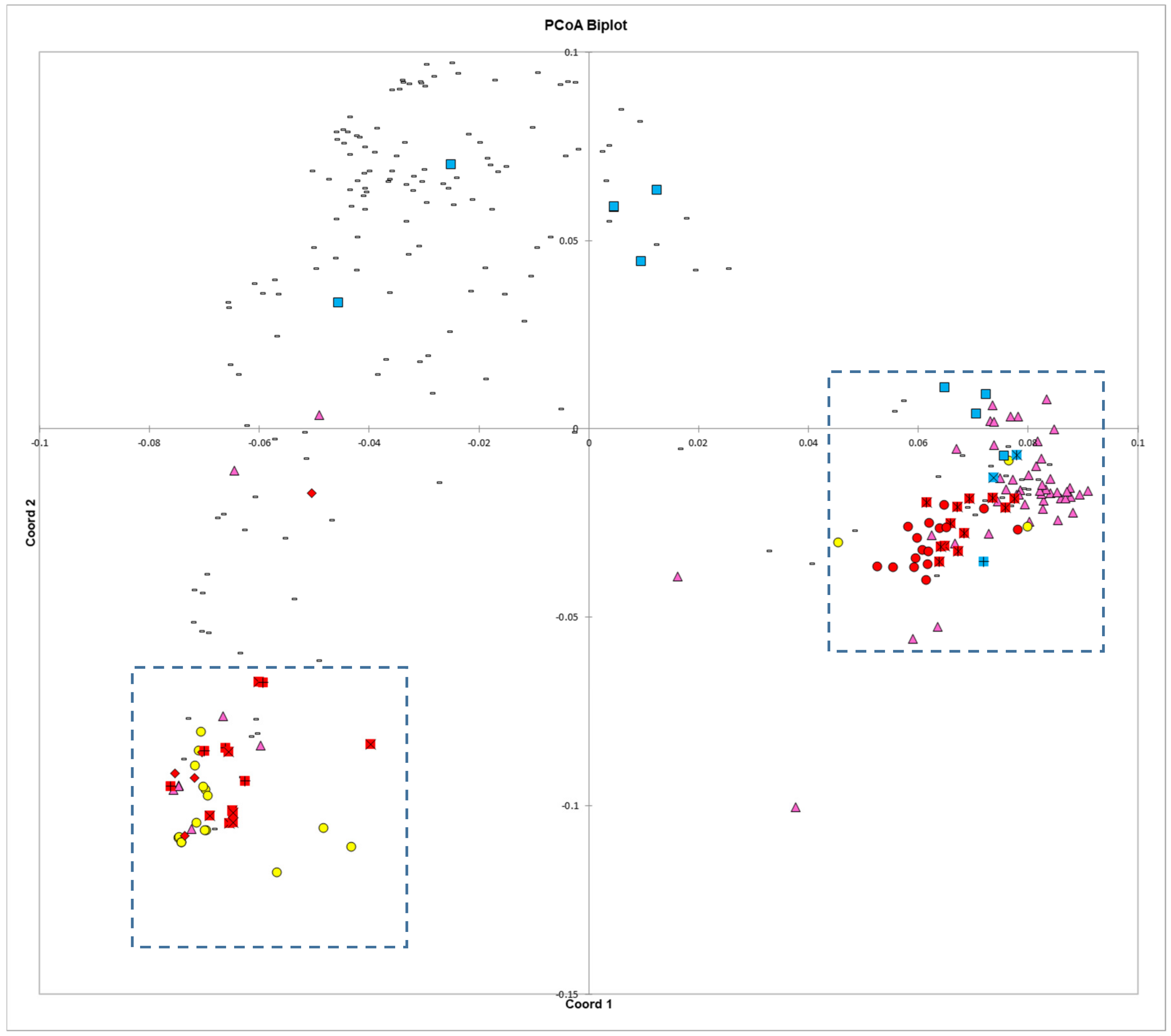
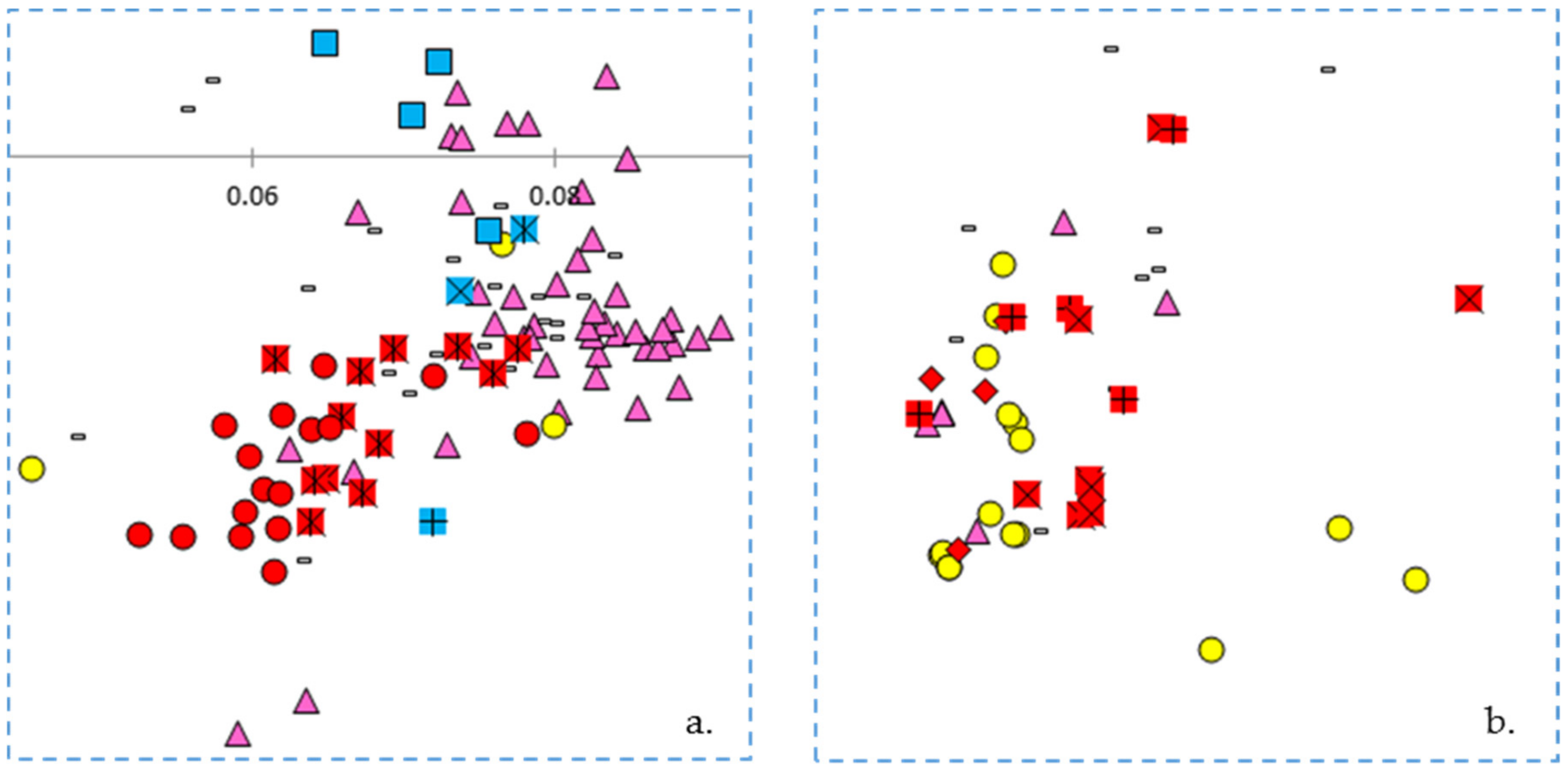
The principal coordinates analysis supports many of the findings of the phylogenetic trees. The Blue Lake lines are spread out across the PCoA biplot in a manner that mirrors the phylogenetic tree. The Blue Lake varieties that contain the most Andean admixture are furthest to the left on the phylogram where the Andean genotypes reside and also are the furthest to the left on the PCoA biplot where the Andean gene pool genotypes cluster (
Figure 4). Similarly, the mixed genotypes are found in an intermediate position and the purest genotypes from the earliest days of Blue Lake breeding are closely clustered with the genotypes of the Middle American gene pool. The two enlarged clusters from the biplot also verifies and validates that close connections between Chinese and Spanish lines apparent in the phylogram and the connections to race Chile.
The principal coordinates analysis also goes beyond the phylograms and gives more information on the relationships between these different collections and races. Whereas the phylogram does not resolve the closest relative to ‘Pole Blue Lake’ among ‘Scotia’, ‘Genuine Cornfield’, and ‘White Creaseback’, the PCoA biplot seems to give a clear ranking of similarity with ‘White Creaseback’ as closest to ‘Pole Blue Lake’ and Scotia as the furthest. The PCoA biplot also shows a high degree of similarity between ‘Pole Blue Lake’ and Spanish line ‘PHA0315’. Furthermore, the PCoA biplot not only confirms the cladistic connections identified in
Figure 3, but also shows that race Nueva Granada is similar to at least one Chinese and Spanish line respectively as well as to one American heirloom pole bean. Also apparent in the biplot is the similarity of dry bean members of race Mesoamerica/Guatemala and race Jalisco/Durango to some Chinese lines.
4.5. Genetic Structure of Common Bean
Our analysis was only partially successful in differentiating races of common bean. Races from different centers of origin were reliably separated, but within centers, only Mesoamerican/Guatemalan races were distinct from Durango/Jalisco in the Middle American gene pool. This may be a function of the markers (SNPs) used in this study as other studies have relied on RAPD and SSR markers as well as phenotypic assessments.
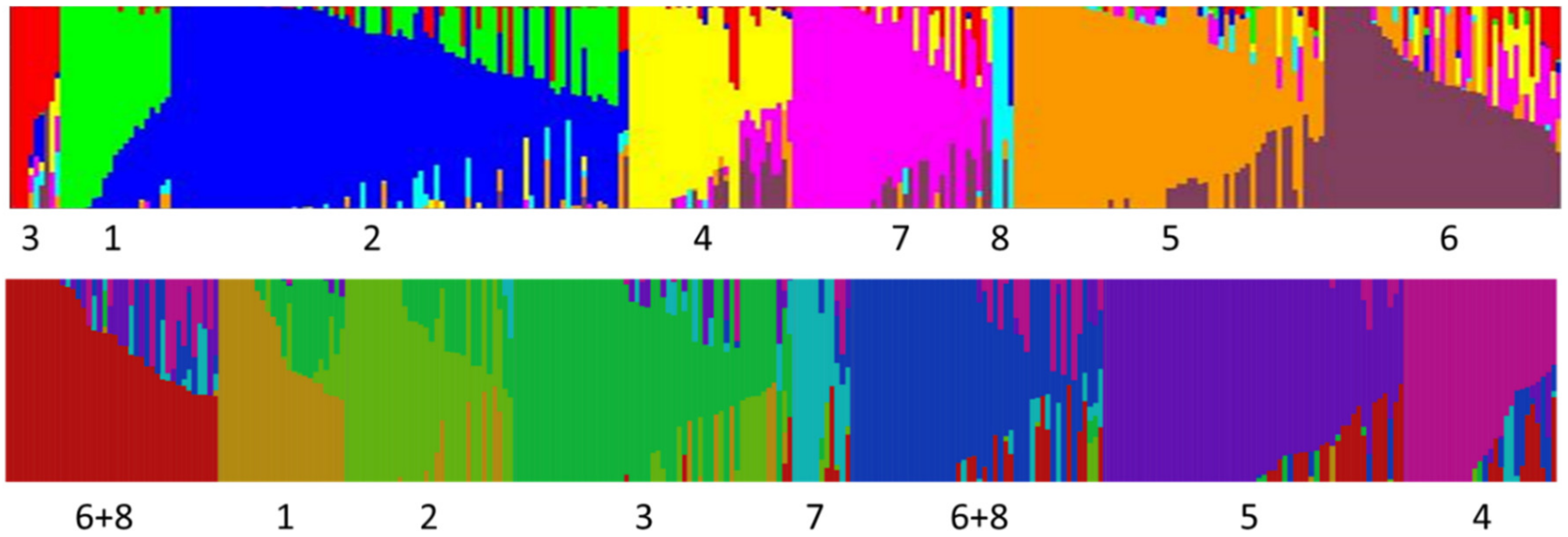
One interesting finding is that five of the snap bean groups designated by Structure appear to be unique and separate from the dry beans included in the study. In some cases, this distinctness may indicate an independent lineage and unique alleles in particular groups (such as Refugee types), but more likely, separate grouping is a function of unique combinations of alleles. We have not been able to resolve in this study whether snap beans represent a novel source of genetic variation for dry beans, but there at least appears to be unique combinations of alleles that could assist dry bean breeders in crossing center of domestication barriers.
Some dry bean races showed small, but significant admixture whereas many snap beans within the same K group showed no mixing. Based on ancestry, we would have expected the dry bean races to have distinct allelic assemblages and the snap beans belonging to these assemblages to show some admixture. The algorithm as applied in our study seems to indicate that Structure identifies and designates groups based primarily on unique combinations of alleles with less emphasis on unique alleles within populations. For example, we know the ancestry of the Bush Blue Lake types, yet at K = 8 Structure designated this group as a distinct population with some individuals having no admixture. A similar pattern can be seen with the European extra fine types. None of these have lineages that trace back to historical materials, but rather, are the result of considerable mixing between other derived snap bean groups.
One of our major findings is that among contemporary snap beans, admixtures between centers of domestication are quite widespread, especially for bush blue lake, European extra fine, and many American Andean snap bean cultivars. Older historical cultivars and landraces do not show the same degree of mixing. The impetus for crossing among different snap types appears to be varied. For example, Middle American and Andean pole types were crossed to bush cultivars to bring a particular type into a bush background more amenable to mechanical harvest. A number of crosses have been made to introgress disease resistances available in dry bean backgrounds of different centers into snap beans (documented in several Plant Variety Protection certificates). Inter genepool crosses among snap types have also been used to transfer quality traits.
4.6. Snap Bean Gene Pools
Based on the results of or genetic diversity analysis, we postulate that there have been at least two and possibly up to five independent derivations of snap beans from dry beans. First, Middle American pole beans appear to have been derived from a race Durango/Jalisco ancestor, resulting in a large number of cultivars that center around production along the eastern seaboard of the US. A second derivation occurred from Andean materials and probably race Chile, producing large flat podded types that were introduced into Europe and spread around the world early after the Colombian exchange. Third, round-podded snap beans derived from race Nueva Granada may have been developed. The Andean snap bean groups in our Structure analysis show a large degree of mixing, so it is difficult to point to particular ancestral lines. The fourth distinctive group contains the Refugee types. These are most likely of Andean origin, and in some Structure runs were grouped with other Andean accessions, but we regard them as a distinct and separate group because they are phenotypically distinct from all other snap beans. In their ancestral form, they are late maturing with light green pods on plants with a half runner (type III) growth habit (bush beans are type I and all other pole beans are type IV). In our Structure analysis, this group did not show any particular relationship to any dry bean races, so it is not possible to postulate an ancestral type. Refugee types appear to have been so named because they were brought to the North American by French Huguenots fleeing persecution in Europe [
32]. Further complicating the picture, Refugee types have been widely used in breeding in other snap bean groups. For example, ‘Corbett’s Refugee’ has been the primary source of
I gene resistance to BCMV for most contemporary snap beans. Fifth, there may have been a separate Middle American pole type derived from race Mesoamerica dry beans as typified by ‘Scotia’ and ‘Cherokee Trail of Tears’. Oral histories have revealed that the ‘Cherokee Trail of Tears’ was carried and preserved by the Cherokee during their forced removal from Georgia in the Eastern US to reservations in Oklahoma [
17].
We can think of two genetics and genomics tests that could help determine how many independent derivations of snap beans took place. Using a selected set parents that represent the different groups but show little evidence of mixing, these could be crossed in a diallel and the progeny evaluated for complementation of the various snap bean traits. Preliminary studies have indicated that complementation for pod fiber, pod cross-section and suture strings can be observed in a BBL by Andean snap bean cross [
34]. Secondly, as candidate genes for snap bean pod traits are identified, these can be sequenced in accessions from different groups to determine whether the same or different alleles are present in different groups.
4.7. Geographic Origins of Snap Beans
While there is evidence for more than one independent derivation of the snap bean type, the exact origins remain unknown. One of the main questions is whether snap beans were derived in the New World, or only after dry beans arrived in the Old World. Primary traits involved in the domestication of common bean included reduction in seed dispersal mechanisms (pod shatter), loss of seed dormancy, increase in seed size and seed color diversity, and changes in plant phenology and growth habit. The main driver for evolution in dry bean would be the primary use and consumption of dry seed, with vegetable use only a minor consideration. Only the first trait through loss of some fiber might have led to a snap bean phenotype, but even for this trait, too much fiber loss would compromise threshing ability and lead to a reduction in harvest efficiency. While most cultures around the world would regard immature bean pods as a food for times of shortage, Europe and especially Southern Europe does seem to have had a culture of using immature parts of traditional field crops as vegetables as evidenced by the pre-Columbian exchange vegetable use of peas, bottle gourd, cole crops and several leafy vegetables. Thus, Europeans may have been predisposed to adopting field crops such as dry bean and developing vegetable forms from these. Available evidence suggests that at least some snap bean derivations happened in Europe, but some derivations may have happened in the Americas and were then brought to Europe. In particular, the small subset of Middle American pole beans that appear most closely related to race Mesoamerica may have been developed in the Southeastern US by the Cherokee or related tribes. Pre-Colombian introduction routes for dry beans into North America probably varied based on bean race. Those of race Durango most likely came overland from Mexico and were carried northwards to what is now North Dakota and eastwards into the central and northeastern US. Race Mesoamerica beans originated in Central American and may have come to the US by dispersal to Cuba and hence into Florida, and up the eastern seaboard as far as present day Ontario. The Guanahatabey of Western Cuba were traditional users of small black beans, which were probably introduced directly from Middle America [
35], (J. Kelly, MSU, pers. comm.). These materials are genetically distinct, and at least in the case of ‘Cherokee Trail of Tears’, we have an oral history that indicates stewardship, if not origination of the cultivar. That said, the Colombian exchange began at least two centuries prior to the Trail of Tears events beginning in 1838, so it is possible that these types were introduced to Native Americans by early European settlers.
The large flat podded Andean types are another group that may have had its origins in South America rather than in southern Europe. Evidence for this includes the close affinity of these types to dry bean races (especially race Chile) and the fact that some Spanish and Chinese accessions show close similarity. The relatedness of accessions from these secondary centers would suggest that these were derived from the same original source.