Identification and Characterization of the Efbzip Gene Family in Erianthus fulvus and Exploration of Functional Genes Involved in Sucrose Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification of Gene Family and Physicochemical Analysis of Proteins
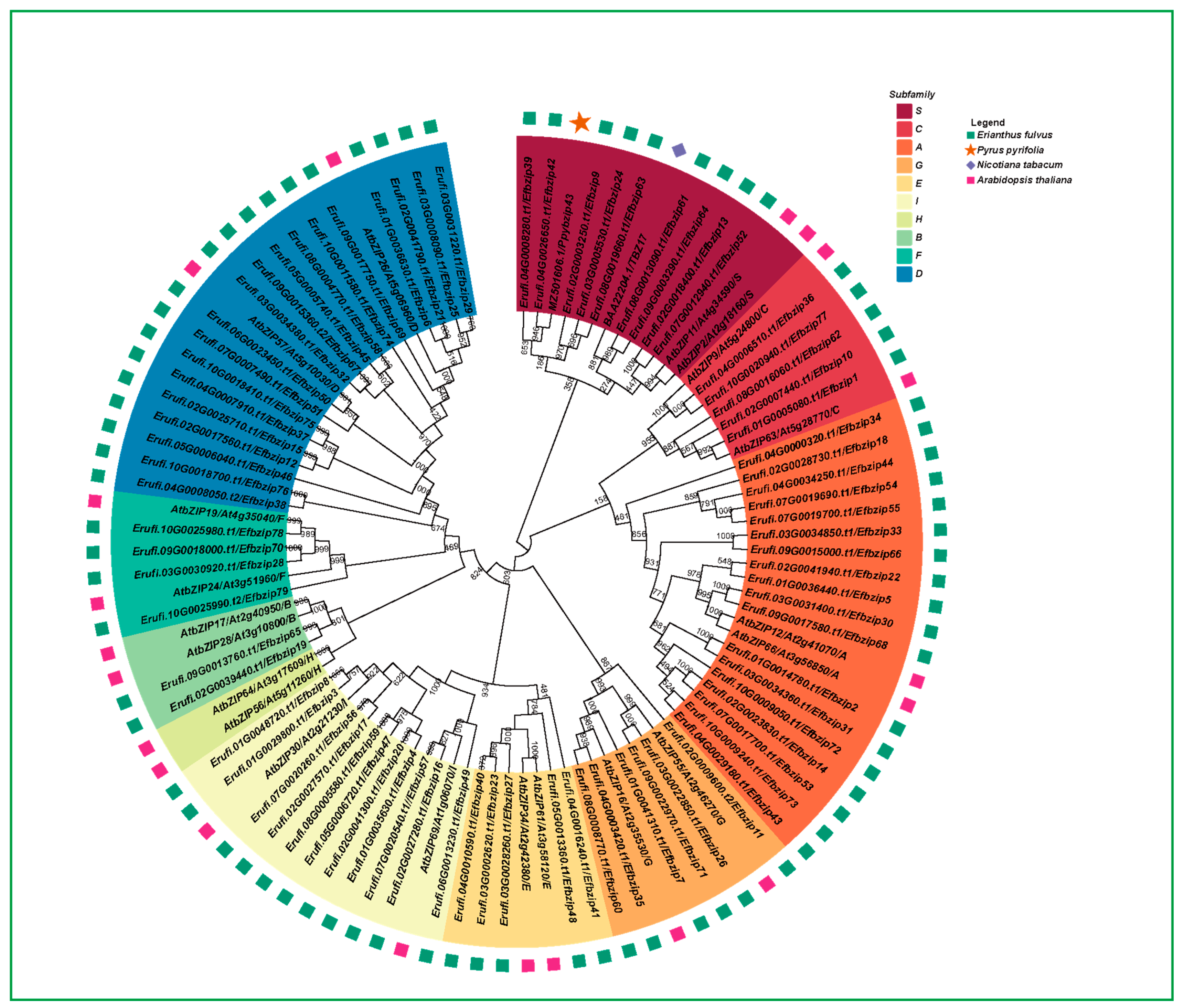
2.3. Phylogenetic Analysis of Efbzip Protein Family
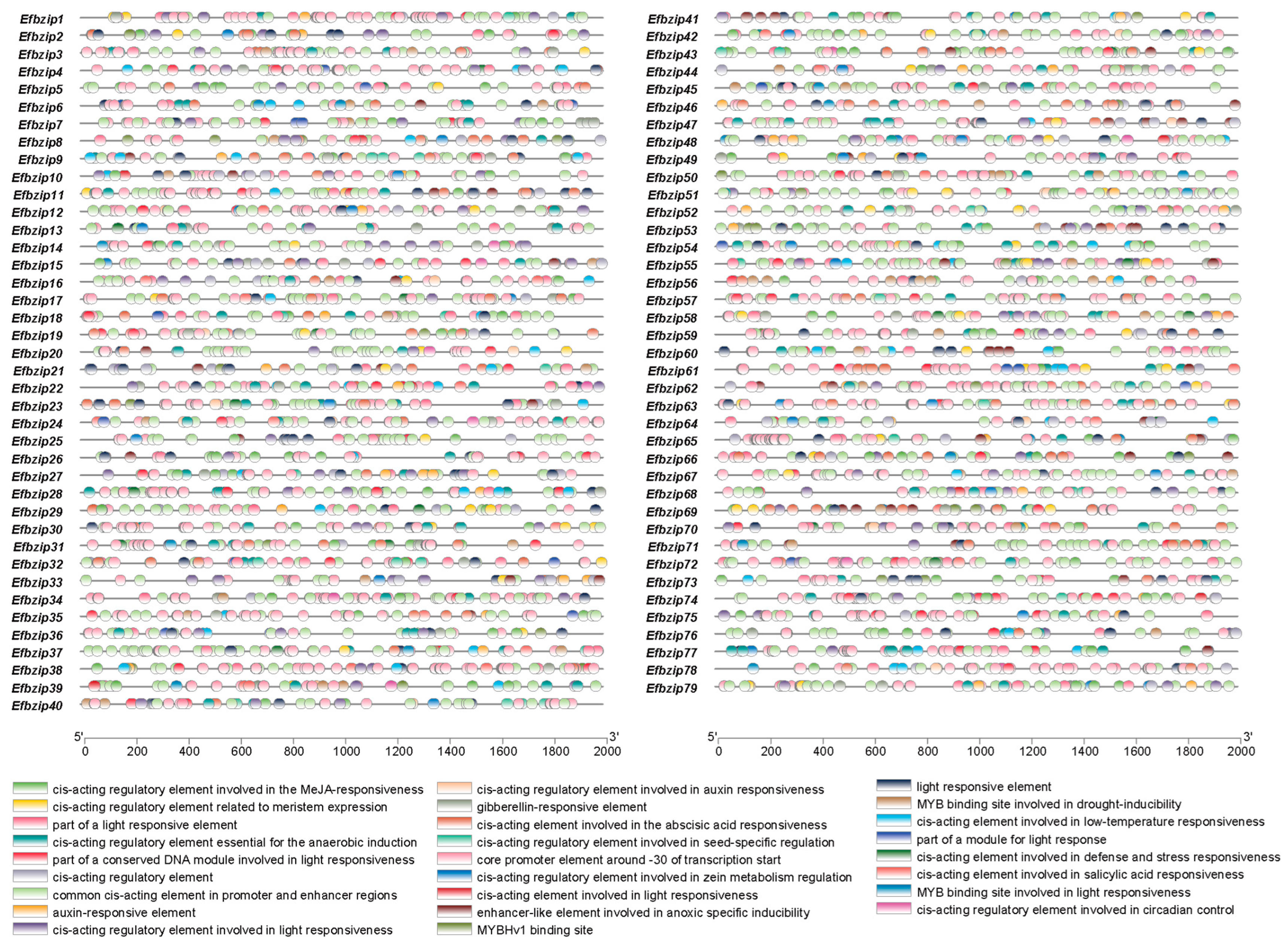
2.4. Promoter Cis-Acting Element Analysis of Efbzip Genes
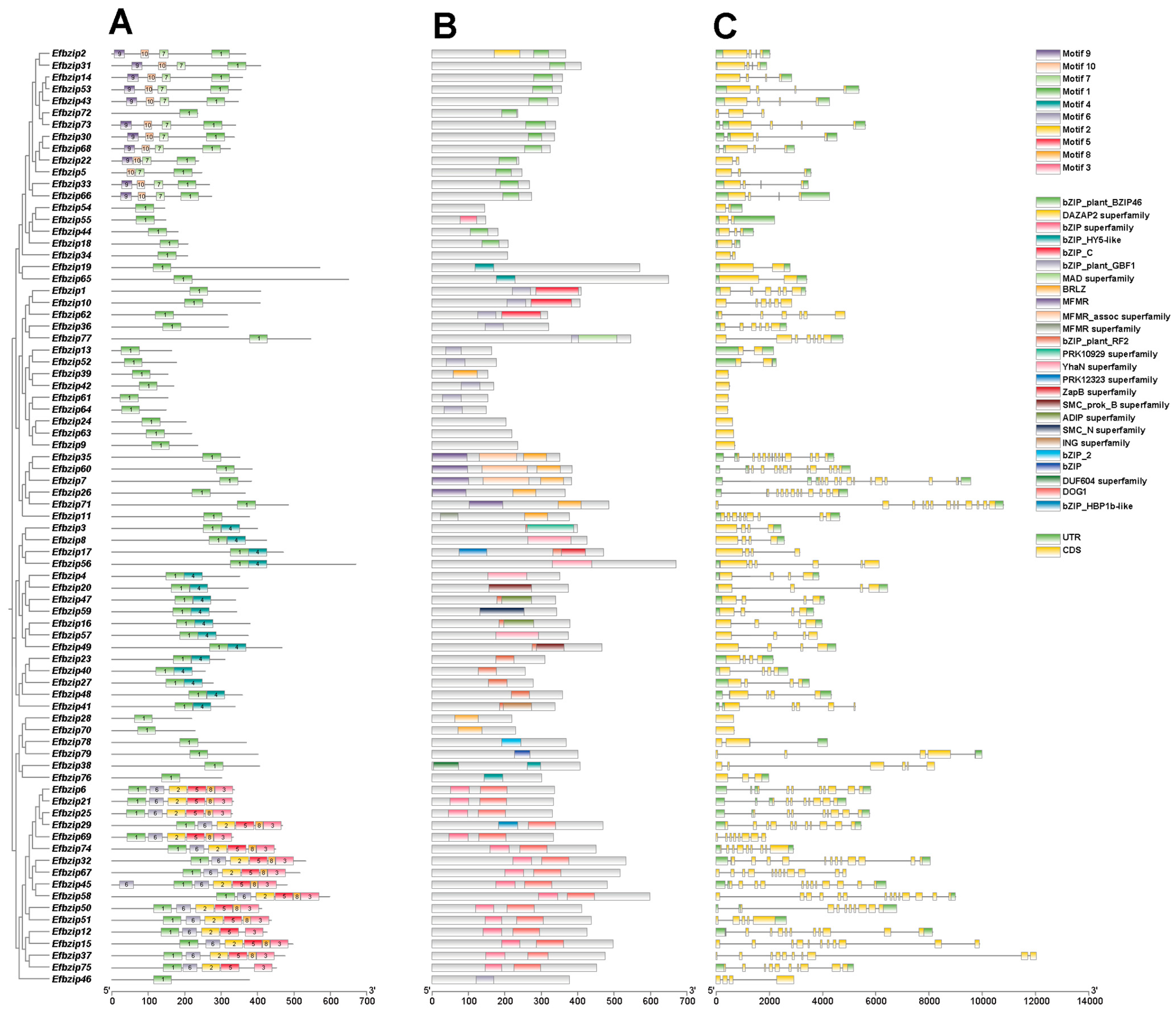
2.5. Analysis of Gene Structure, Conserved Protein Motifs, and Domains in Efbzip Family Members
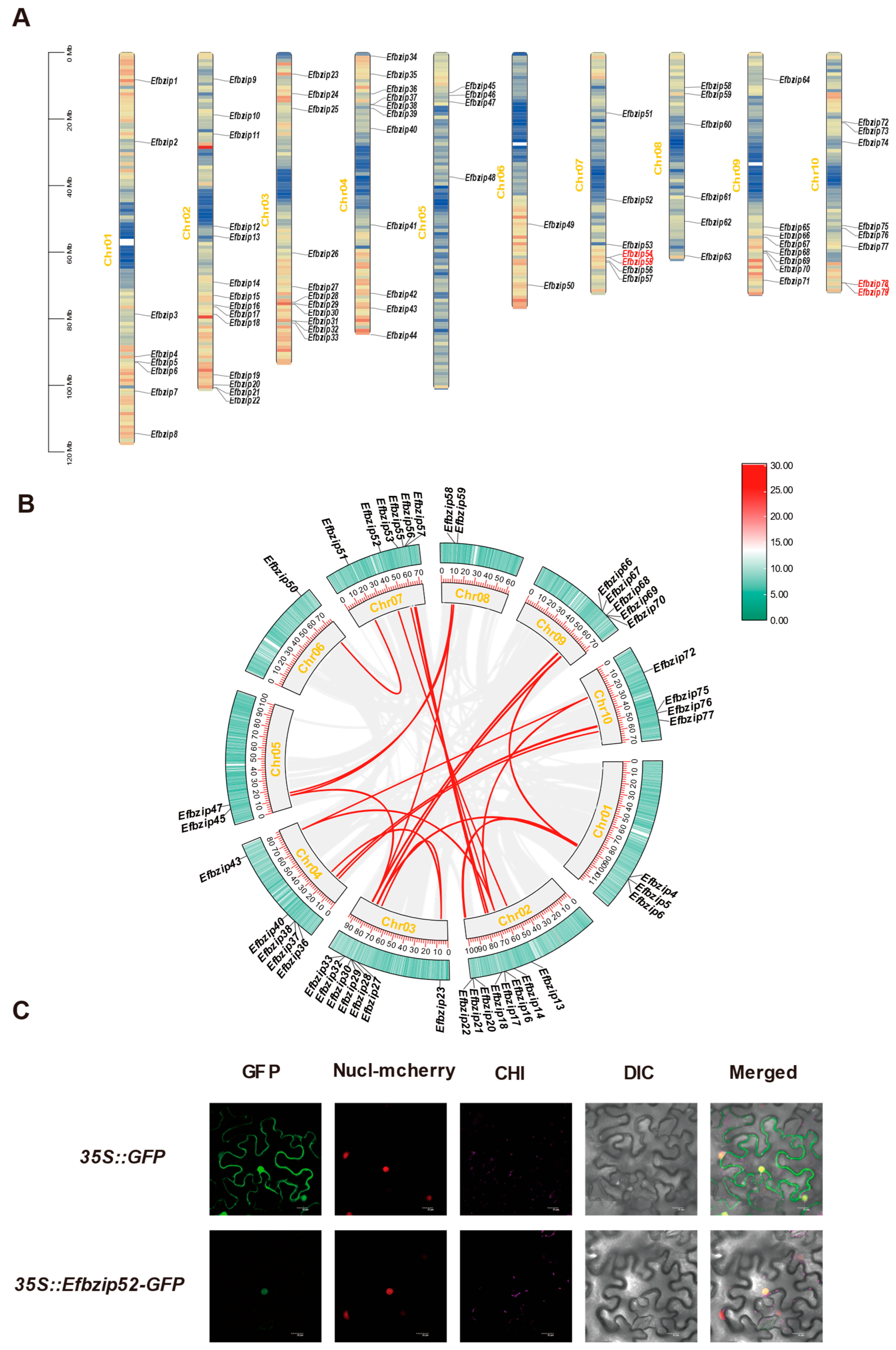
2.6. Synteny Analysis of Efbzip Genes
2.7. Expression Pattern Analysis of Efbzip Genes in Different Tissues
2.8. RNA Extraction, cDNA Synthesis, Gene Cloning, Vector Construction, and qRT-PCR Assays
2.9. Subcellular Localization Assay
2.10. Identification of Efbzip Family Members Regulating Sucrose Metabolism
2.11. Data Analysis
3. Results
3.1. Identification of Efbzip Gene Family Members and Their Physicochemical Properties
3.2. Gene Family Evolution Analysis
3.3. Analysis of Promoter Cis-Acting Elements in Efbzip Gene Family Members
3.4. Analysis of Gene Structure, Conserved Motifs, and Domains in Efbzip Family Members
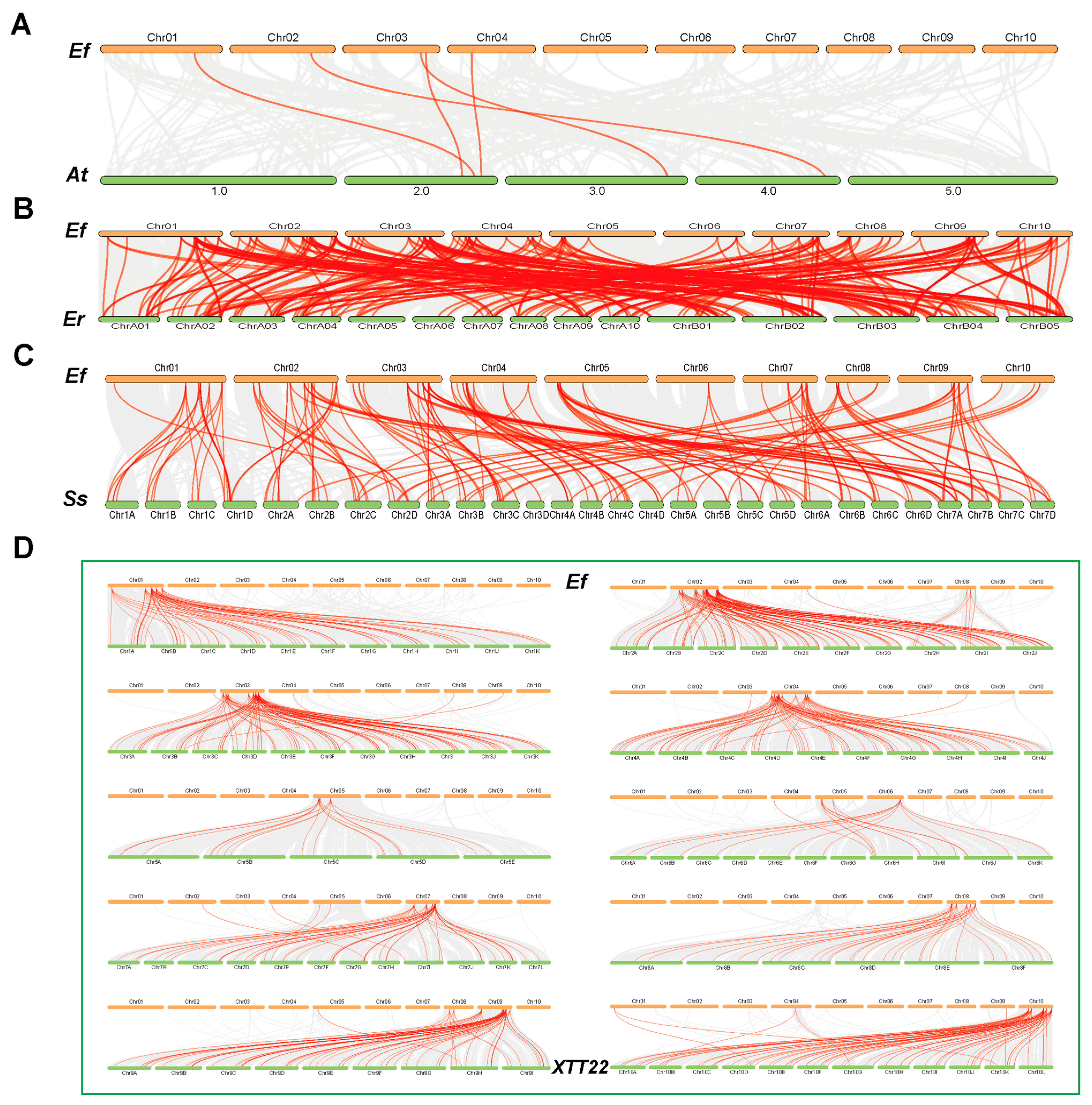
3.5. Synteny Analysis of the Efbzip Gene Family
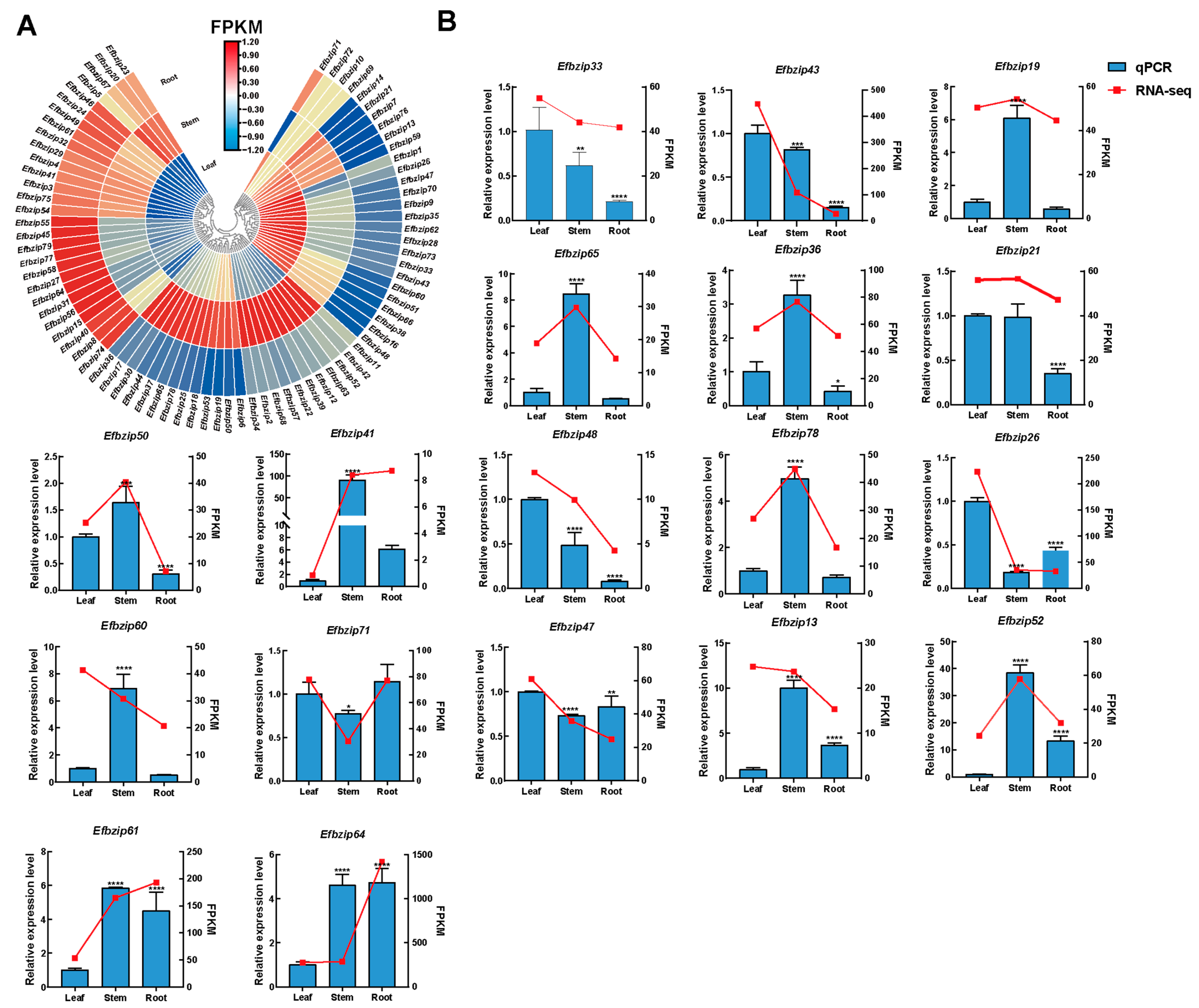
3.6. Expression Patterns of the Efbzip Gene Family in Different Tissues
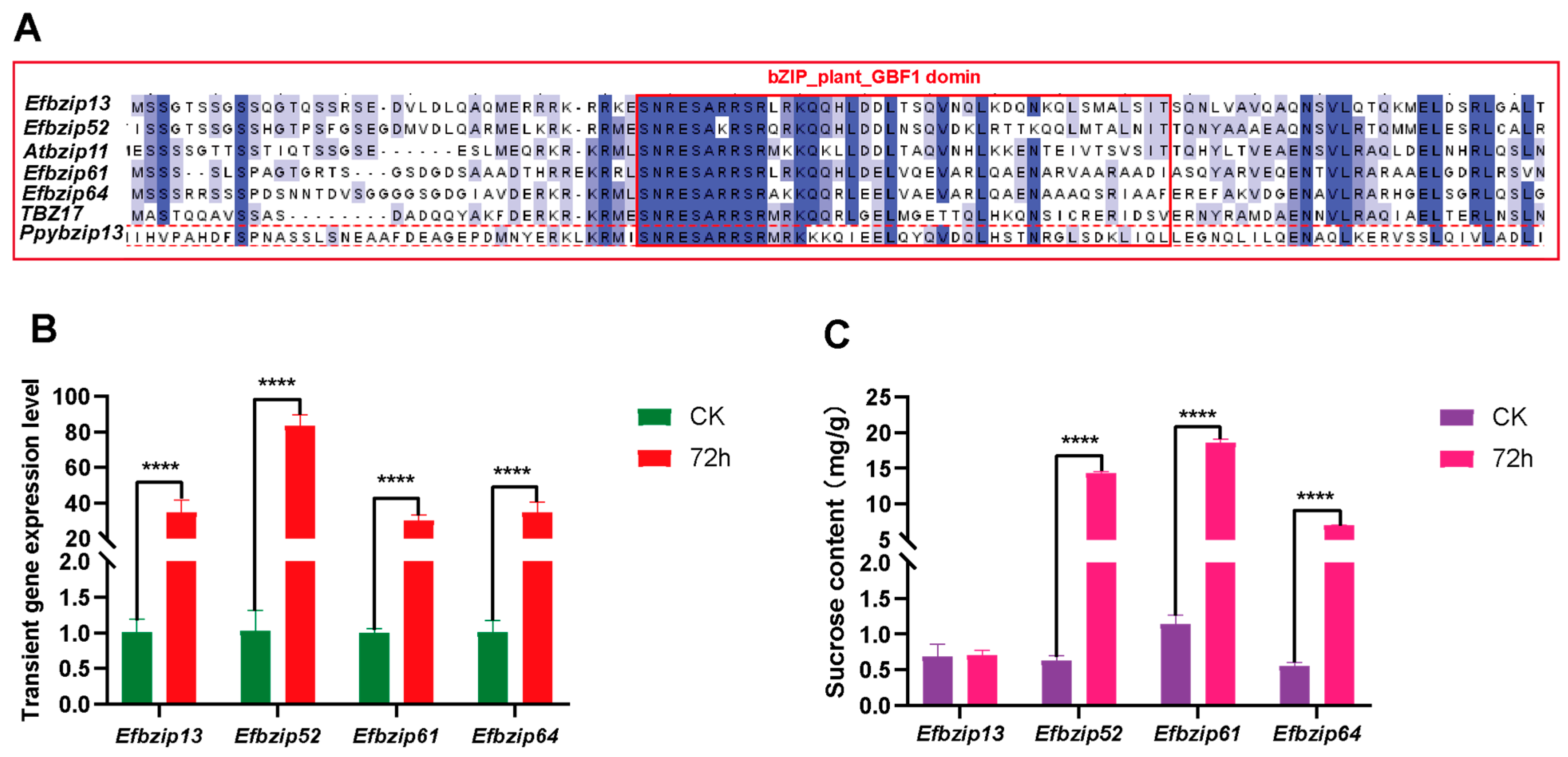
3.7. Identification of Genes Regulating Sucrose Metabolism Among Efbzip Family Members
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, K.; Liu, L.; Huang, S. Genome-Wide Identification and Functional Analysis of the bZIP Transcription Factor Family in Rice Bakanae Disease Pathogen, Fusarium fujikuroi. Int. J. Mol. Sci. 2022, 23, 6658. [Google Scholar] [CrossRef]
- Niu, S.; Gu, X.; Zhang, Q.; Tian, X.; Chen, Z.; Liu, J.; Wei, X.; Yan, C.; Liu, Z.; Wang, X.; et al. Grapevine bZIP transcription factor bZIP45 regulates VvANN1 and confers drought tolerance in Arabidopsis. Front. Plant Sci. 2023, 14, 1128002. [Google Scholar] [CrossRef]
- Choi, J.; Lim, C.W.; Lee, S.C. Role of pepper bZIP transcription factor CaADBZ1 in abscisic acid signalling and drought stress response. Physiol. Plant. 2025, 177, e70159. [Google Scholar] [CrossRef]
- Siberil, Y.; Doireau, P.; Gantet, P. Plant bZIP G-box binding factors. Modular structure and activation mechanisms. Eur. J. Biochem. 2001, 268, 5655–5666. [Google Scholar] [CrossRef]
- Soucek, L.; Helmer-Citterich, M.; Sacco, A.; Jucker, R.; Cesareni, G.; Nasi, S. Design and properties of a Myc derivative that efficiently homodimerizes. Oncogene 1998, 17, 2463–2472. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Qiao, Y.; Pan, X.; Chen, X.; Su, W.; Li, A.; Li, X.; Liao, W. Genome-Wide identification and expression analysis of CsABF/AREB gene family in cucumber (Cucumis sativus L.) and in response to phytohormonal and abiotic stresses. Sci. Rep. 2025, 15, 15757. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Mao, B.; Ou, S.; Wang, W.; Liu, L.; Wu, Y.; Chu, C.; Wang, X. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 2014, 84, 19–36. [Google Scholar] [CrossRef]
- Zhao, P.; Ye, M.; Wang, R.; Wang, D.; Chen, Q. Systematic identification and functional analysis of potato (Solanum tuberosum L.) bZIP transcription factors and overexpression of potato bZIP transcription factor StbZIP-65 enhances salt tolerance. Int. J. Biol. Macromol. 2020, 161, 155–167. [Google Scholar] [CrossRef]
- Jin, Z.; Xu, W.; Liu, A. Genomic surveys and expression analysis of bZIP gene family in castor bean (Ricinus communis L.). Planta 2014, 239, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, Y.; Wang, Q.; Tao, X.; Fang, J.; Zheng, W.; Zhu, L.; Jia, B.; Heng, W.; Li, S. Identification of bZIP transcription factors and their responses to brown spot in pear. Genet. Mol. Biol. 2022, 45, e20210175. [Google Scholar] [CrossRef]
- Wang, X.; Lu, X.; Malik, W.A.; Chen, X.; Wang, J.; Wang, D.; Wang, S.; Chen, C.; Guo, L.; Ye, W. Differentially expressed bZIP transcription factors confer multi-tolerances in Gossypium hirsutum L. Int. J. Biol. Macromol. 2020, 146, 569–578. [Google Scholar] [CrossRef]
- Guan, R.; Xu, S.; Lu, Z.; Su, L.; Zhang, L.; Sun, W.; Zhang, Y.; Jiang, C.; Liu, Z.; Duan, L.; et al. Genomic characterization of bZIP transcription factors related to andrographolide biosynthesis in Andrographis paniculata. Int. J. Biol. Macromol. 2022, 223, 1619–1631. [Google Scholar] [CrossRef]
- Jia, X.; Gao, H.; Zhang, L.; Tang, W.; Wei, G.; Sun, J.; Xiong, W. Expression of Foxtail Millet bZIP Transcription Factor SibZIP67 Enhances Drought Tolerance in Arabidopsis. Biomolecules 2024, 14, 958. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Z.; Ji, A.; Luo, H.; Song, J. Genomic survey of bZIP transcription factor genes related to tanshinone biosynthesis in Salvia miltiorrhiza. Acta Pharm. Sin. B 2018, 8, 295–305. [Google Scholar] [CrossRef]
- Schlogl, P.S.; Nogueira, F.T.; Drummond, R.; Felix, J.M.; De Rosa, V.J.; Vicentini, R.; Leite, A.; Ulian, E.C.; Menossi, M. Identification of new ABA- and MEJA-activated sugarcane bZIP genes by data mining in the SUCEST database. Plant Cell Rep. 2008, 27, 335–345. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Droge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Casaretto, J.; Ho, T.D. The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. Plant Cell 2003, 15, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Wiese, A.; Elzinga, N.; Wobbes, B.; Smeekens, S. Sucrose-induced translational repression of plant bZIP-type transcription factors. Biochem. Soc. Trans. 2005, 33, 272–275. [Google Scholar] [CrossRef]
- Liu, H.; Tang, X.; Zhang, N.; Li, S.; Si, H. Role of bZIP Transcription Factors in Plant Salt Stress. Int. J. Mol. Sci. 2023, 24, 7893. [Google Scholar] [CrossRef]
- Banerjee, A.; Roychoudhury, A. Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 2017, 254, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Liu, Y.; Chen, S.; Shityakov, S. Meta-Analysis of the Effects of Overexpressed bZIP Transcription Factors in Plants under Drought Stress. Plants 2024, 13, 337. [Google Scholar] [CrossRef]
- Hwang, I.; Jung, H.; Park, J.; Yang, T.; Nou, I. Transcriptome analysis of newly classified bZIP transcription factors of Brassica rapa in cold stress response. Genomics 2014, 104, 194–202. [Google Scholar] [CrossRef]
- Alves, M.S.; Dadalto, S.P.; Goncalves, A.B.; De Souza, G.B.; Barros, V.A.; Fietto, L.G. Plant bZIP transcription factors responsive to pathogens: A review. Int. J. Mol. Sci. 2013, 14, 7815–7828. [Google Scholar] [CrossRef]
- Sagor, G.H.M.; Berberich, T.; Tanaka, S.; Nishiyama, M.; Kanayama, Y.; Kojima, S.; Muramoto, K.; Kusano, T. A novel strategy to produce sweeter tomato fruits with high sugar contents by fruit-specific expression of a single bZIP transcription factor gene. Plant Biotechnol. J. 2016, 14, 1116–1126. [Google Scholar] [CrossRef]
- Meng, D.; Cao, H.; Yang, Q.; Zhang, M.; Borejsza-Wysocka, E.; Wang, H.; Dandekar, A.M.; Fei, Z.; Cheng, L. SnRK1 kinase-mediated phosphorylation of transcription factor bZIP39 regulates sorbitol metabolism in apple. Plant Physiol. 2023, 192, 2123–2142. [Google Scholar] [CrossRef]
- D’Haeseleer, K.; De Keyser, A.; Goormachtig, S.; Holsters, M. Transcription factor MtATB2: About nodulation, sucrose and senescence. Plant Cell Physiol. 2010, 51, 1416–1424. [Google Scholar] [CrossRef]
- Thalor, S.K.; Berberich, T.; Lee, S.S.; Yang, S.H.; Zhu, X.; Imai, R.; Takahashi, Y.; Kusano, T. Deregulation of sucrose-controlled translation of a bZIP-type transcription factor results in sucrose accumulation in leaves. PLoS ONE 2012, 7, e33111. [Google Scholar] [CrossRef]
- Zhang, H.; Tao, X.; Fan, X.; Zhang, S.; Qin, G. PpybZIP43 contributes to sucrose synthesis in pear fruits by activating PpySPS3 expression and interacts with PpySTOP1. Physiol. Plant. 2022, 174, e13732. [Google Scholar] [CrossRef]
- Araujo, M.A.; Melo, A.; Silva, V.M.; Reis, A. Selenium enhances ROS scavenging systems and sugar metabolism increasing growth of sugarcane plants. Plant Physiol. Biochem. 2023, 201, 107798. [Google Scholar] [CrossRef]
- Akbar, S.; Yao, W.; Qin, L.; Yuan, Y.; Powell, C.A.; Chen, B.; Zhang, M. Comparative Analysis of Sugar Metabolites and Their Transporters in Sugarcane Following Sugarcane mosaic virus (SCMV) Infection. Int. J. Mol. Sci. 2021, 22, 13574. [Google Scholar] [CrossRef]
- Sun, N.; Xu, X.; Zhu, Z.; Zhou, X.; Liu, Y.; Li, D.; Cao, F.; Wang, L.; Zhang, H. Tonoplast sugar transporter ZmTST1 positively regulates plant growth, salt and drought tolerance. Plant Physiol. Biochem. 2025, 229, 110380. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Li, Y.P.; Gai, P.Z.; Gao, J.; Xu, L. Exogenously applied ABA alleviates dysplasia of maize (Zea mays L.) ear under drought stress by altering photosynthesis and sucrose transport. Plant Signal. Behav. 2025, 20, 2462497. [Google Scholar] [CrossRef]
- Zhao, J.; Li, S.; Xu, Y.; Ahmad, N.; Kuang, B.; Feng, M.; Wei, N.; Yang, X. The subgenome Saccharum spontaneum contributes to sugar accumulation in sugarcane as revealed by full-length transcriptomic analysis. J. Adv. Res. 2023, 54, 1–13. [Google Scholar] [CrossRef]
- Wang, M.; Li, A.; Liao, F.; Chen, Z.; Qin, C.; Zhang, B.; Li, X.; Su, Z.; Pan, Y.; Huang, D. Sugarcane microRNA shy-miR164 regulates sugar metabolism through direct cleavage of the transcription factor ScNAC mRNA. Plant Physiol. 2025, 198, kiaf354. [Google Scholar] [CrossRef]
- Chen, M.; Liu, P.; An, R.; He, X.; Zhao, P.; Huang, D.; Yang, X. Sugarcane Pan-Transcriptome Identifying a Master Gene ScHCT Regulating Lignin and Sugar Traits. J. Agric. Food. Chem. 2025, 73, 1739–1755. [Google Scholar] [CrossRef]
- Ling, K.; Yi-Ning, D.; Majeed, A.; Zi-Jiang, Y.; Jun-Wen, C.; Li-Lian, H.; Xian-Hong, W.; Lu-Feng, L.; Zhen-Feng, Q.; Dan, Z.; et al. Evaluation of genome size and phylogenetic relationships of the Saccharum complex species. 3 Biotech 2022, 12, 327. [Google Scholar] [CrossRef] [PubMed]
- Kui, L.; Majeed, A.; Wang, X.; Yang, Z.; Chen, J.; He, L.; Di, Y.; Li, X.; Qian, Z.; Jiao, Y.; et al. A chromosome-level genome assembly for Erianthus fulvus provides insights into its biofuel potential and facilitates breeding for improvement of sugarcane. Plant Commun. 2023, 4, 100562. [Google Scholar] [CrossRef]
- Qian, Z.; Zhao, C.; Wan, H.; He, L.; Wang, X.; Li, F. Research Progress and Utilization Potential of Saccharum hortensis, a Wild Relative of Sugarcane. J. Plant Genet. Resour. 2025, 26, 1485–1498. [Google Scholar] [CrossRef]
- Qian, Z.; Zhao, C.; Wan, H.; Rao, X.; Luo, Z.; He, L.; Li, F. Genome-wide identification of DREB1 transcription factors in Erianthus fulvus and the functional role of EfDREB1C in regulating cold tolerance of transgenic Arabidopsis and sugarcane. Int. J. Biol. Macromol. 2025, 317, 144859. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lin, W.; He, S. Reflections on Developing and Utilizing Wild Sugarcane Germplasm Resources. Resour. Dev. Mark. 2004, 20, 266–270. [Google Scholar] [CrossRef]
- Zhang, J.; Qi, Y.; Hua, X.; Wang, Y.; Wang, B.; Qi, Y.; Huang, Y.; Yu, Z.; Gao, R.; Zhang, Y.; et al. The highly allo-autopolyploid modern sugarcane genome and very recent allopolyploidization in Saccharum. Nat. Genet. 2025, 57, 242–253. [Google Scholar] [CrossRef]
- Wang, T.; Wang, B.; Hua, X.; Tang, H.; Zhang, Z.; Gao, R.; Qi, Y.; Zhang, Q.; Wang, G.; Yu, Z.; et al. A complete gap-free diploid genome in Saccharum complex and the genomic footprints of evolution in the highly polyploid Saccharum genus. Nat. Plants 2023, 9, 554–571. [Google Scholar] [CrossRef]
- Blum, M.; Andreeva, A.; Florentino, L.C.; Chuguransky, S.R.; Grego, T.; Hobbs, E.; Pinto, B.L.; Orr, A.; Paysan-Lafosse, T.; Ponamareva, I.; et al. InterPro: The protein sequence classification resource in 2025. Nucleic Acids Res. 2025, 53, D444–D456. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.T.; Jakovlic, I.; Wang, W. In silico characterisation, homology modelling and structure-based functional annotation of blunt snout bream (Megalobrama amblycephala) Hsp70 and Hsc70 proteins. J. Anim. Sci. Technol. 2015, 57, 44. [Google Scholar] [CrossRef] [PubMed]
- Long, W.; Zhao, L.; Yang, H.; Yang, X.; Bai, Y.; Xue, X.; Wang, D.; Han, S. Genome-Wide Characterization of Wholly Disordered Proteins in Arabidopsis. Int. J. Mol. Sci. 2025, 26, 1117. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Li, X.; Li, F.; Li, D.; Dong, Y.; Fan, Y. Bioinformatics Analysis of WRKY Family Genes in Erianthus fulvus Ness. Genes 2022, 13, 2102. [Google Scholar] [CrossRef]
- Qian, Z.; Rao, X.; Zhang, R.; Gu, S.; Shen, Q.; Wu, H.; Lv, S.; Xie, L.; Li, X.; Wang, X.; et al. Genome-Wide Identification, Evolution, and Expression Analyses of AP2/ERF Family Transcription Factors in Erianthus fulvus. Int. J. Mol. Sci. 2023, 24, 7102. [Google Scholar] [CrossRef]
- E, Z.G.; Zhang, Y.P.; Zhou, J.H.; Wang, L. Mini review roles of the bZIP gene family in rice. Genet. Mol. Res. 2014, 13, 3025–3036. [Google Scholar] [CrossRef]
- Zhao, K.; Chen, S.; Yao, W.; Cheng, Z.; Zhou, B.; Jiang, T. Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 2021, 21, 122. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, W.; Li, H.; Wang, Y.; Li, D.; Xue, C.; Liu, Z.; Liu, M.; Zhao, J. Genome-wide analysis of the bZIP gene family in Chinese jujube (Ziziphus jujuba Mill.). BMC Genom. 2020, 21, 483. [Google Scholar] [CrossRef]
- Gao, H.; Cao, X.; Ma, Y.; Qin, X.; Bai, X.; Zhang, X.; Xiong, A.; Yin, Y.; Zheng, R. Genome-Wide Identification of bZIP Gene Family in Lycium barbarum and Expression During Fruit Development. Int. J. Mol. Sci. 2025, 26, 4665. [Google Scholar] [CrossRef]
- Li, H.; Li, L.; Shangguan, G.; Jia, C.; Deng, S.; Noman, M.; Liu, Y.; Guo, Y.; Han, L.; Zhang, X.; et al. Genome-wide identification and expression analysis of bZIP gene family in Carthamus tinctorius L. Sci. Rep. 2020, 10, 15521. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Hou, Z.; He, Q.; Zhang, X.; Yan, K.; Han, R.; Liang, Z. Genome-Wide Characterization and Expression Analysis of bZIP Gene Family Under Abiotic Stress in Glycyrrhiza uralensis. Front. Genet. 2021, 12, 754237. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Gao, T.; Bian, K.; Meng, C.; Tang, X.; Mao, Y. Genome-wide analysis and expression profile of the bZIP gene family in Neopyropia yezoensis. Front. Plant Sci. 2024, 15, 1461922. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Wang, J.; Gao, T.; Qu, C.; Mo, X.; Zhang, X. Systematic analysis of bZIP gene family in Suaeda australis reveal their roles under salt stress. BMC Plant Biol. 2024, 24, 816. [Google Scholar] [CrossRef]
- Ye, W.; Wang, Y.; Dong, S.; Tyler, B.M.; Wang, Y. Phylogenetic and transcriptional analysis of an expanded bZIP transcription factor family in Phytophthora sojae. BMC Genom. 2013, 14, 839. [Google Scholar] [CrossRef]
- Yang, X.; Gao, C.; Hu, Y.; Ma, Q.; Li, Z.; Wang, J.; Li, Z.; Zhang, L.; Li, D. Identification and expression analysis of bZIP transcription factors in Setaria italica in response to dehydration stress. Front. Genet. 2024, 15, 1466486. [Google Scholar] [CrossRef]
- Liu, M.; Wen, Y.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification, phylogeny, evolutionary expansion and expression analyses of bZIP transcription factor family in tartaty buckwheat. BMC Genom. 2019, 20, 483. [Google Scholar] [CrossRef]
- Liu, X.; Chu, Z. Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genom. 2015, 16, 227. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, C.; Li, Z.; Sun, J.; Wang, D.; Xu, L.; Li, X.; Guo, Y. Identification and Analysis of bZIP Family Genes in Potato and Their Potential Roles in Stress Responses. Front. Plant Sci. 2021, 12, 637343. [Google Scholar] [CrossRef]
- Pophaly, S.D.; Tellier, A. Population Level Purifying Selection and Gene Expression Shape Subgenome Evolution in Maize. Mol. Biol. Evol. 2015, 32, 3226–3235. [Google Scholar] [CrossRef]
- Balakrishnan, S.; Bhasker, R.; Ramasamy, Y.; Dev, S.A. Genome-wide analysis of cellulose synthase gene superfamily in Tectona grandis L.f. 3 Biotech 2024, 14, 86. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chen, T.; Wu, Y.; Tang, H.; Yu, J.; Dai, X.; Zheng, Y.; Wan, X.; Yang, Y.; Tan, X. Genome-wide analysis of the peanut CaM/CML gene family reveals that the AhCML69 gene is associated with resistance to Ralstonia solanacearum. BMC Genom. 2024, 25, 200. [Google Scholar] [CrossRef]
- Gao, Z.; Wu, Y.; Li, M.; Ding, L.; Li, J.; Liu, Y.; Cao, Y.; Hua, Y.; Jia, Q.; Wang, D. The auxin response factor (ARF) gene family in Cyclocarya paliurus: Genome-wide identification and their expression profiling under heat and drought stresses. Physiol. Mol. Biol. Plants 2024, 30, 921–944. [Google Scholar] [CrossRef]
- Feng, Y.; Bakari, A.; Guan, H.; Wang, J.; Zhang, L.; Xu, M.; Nyoni, M.; Cao, S.; Zhang, Z. An Investigation into the Evolutionary Characteristics and Expression Patterns of the Basic Leucine Zipper Gene Family in the Endangered Species Phoebe bournei Under Abiotic Stress Through Bioinformatics. Plants 2025, 14, 2292. [Google Scholar] [CrossRef]
- Ma, F.; Zhou, H.; Xu, Y.; Huang, D.; Wu, B.; Xing, W.; Chen, D.; Xu, B.; Song, S. Comprehensive analysis of bZIP transcription factors in passion fruit. iScience 2023, 26, 106556. [Google Scholar] [CrossRef]
- Sprenger-Haussels, M.; Weisshaar, B. Transactivation properties of parsley proline-rich bZIP transcription factors. Plant J. 2000, 22, 1–8. [Google Scholar] [CrossRef]
- Rook, F.; Gerrits, N.; Kortstee, A.; van Kampen, M.; Borrias, M.; Weisbeek, P.; Smeekens, S. Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J. 1998, 15, 253–263. [Google Scholar] [CrossRef]
- Fox, R.M.; Andrew, D.J. Transcriptional regulation of secretory capacity by bZip transcription factors. Front. Biol. 2015, 10, 28–51. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.W.; Hsueh, A.J. Genomic analyses of the evolution of LGR genes. Change Gung Med. J. 2006, 29, 2–8. [Google Scholar]
- Bizotto, F.M.; Ceratti, R.S.; Braz, A.; Masuda, H.P. Evolutionary history of Mo25 gene in plants, a component of RAM/MOR signaling network. Mech. Dev. 2018, 153, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Nong, Q.; Xie, J.; Wang, Z.; Liang, Q.; Solanki, M.K.; Malviya, M.K.; Liu, X.; Li, Y.; Htun, R.; et al. Molecular Characterization and Co-expression Analysis of the SnRK2 Gene Family in Sugarcane (Saccharum officinarum L.). Sci. Rep. 2017, 7, 17659. [Google Scholar] [CrossRef] [PubMed]
- Babula-Skowronska, D. Functional divergence of Brassica napus BnaABI1 paralogs in the structurally conserved PP2CA gene subfamily of Brassicaceae. Genomics 2021, 113, 3185–3197. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, C.; Nong, W.; Qian, Z.; Ding, Q.; Wang, Y.; He, L.; Li, F. Identification and Characterization of the Efbzip Gene Family in Erianthus fulvus and Exploration of Functional Genes Involved in Sucrose Metabolism. Genes 2025, 16, 1434. https://doi.org/10.3390/genes16121434
Zhao C, Nong W, Qian Z, Ding Q, Wang Y, He L, Li F. Identification and Characterization of the Efbzip Gene Family in Erianthus fulvus and Exploration of Functional Genes Involved in Sucrose Metabolism. Genes. 2025; 16(12):1434. https://doi.org/10.3390/genes16121434
Chicago/Turabian StyleZhao, Changzu, Weiyou Nong, Zhenfeng Qian, Qian Ding, Yujie Wang, Lilian He, and Fusheng Li. 2025. "Identification and Characterization of the Efbzip Gene Family in Erianthus fulvus and Exploration of Functional Genes Involved in Sucrose Metabolism" Genes 16, no. 12: 1434. https://doi.org/10.3390/genes16121434
APA StyleZhao, C., Nong, W., Qian, Z., Ding, Q., Wang, Y., He, L., & Li, F. (2025). Identification and Characterization of the Efbzip Gene Family in Erianthus fulvus and Exploration of Functional Genes Involved in Sucrose Metabolism. Genes, 16(12), 1434. https://doi.org/10.3390/genes16121434




