Abstract
Background: Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by limited interests, difficulties in social interactions, repetitive behaviors, and impairments in social communication. ASD tends to run in families, and twin studies suggest a strong genetic basis for the disorder. However, the definition of a genetic profile that indicates a risk for ASD remains unclear. Methods: This analysis includes an investigation (Autism Dataset 4 from the NIMH repository, n = 2890) and a replication (Autism Dataset 3 from the NIMH repository, n = 1233) of trio samples with GWAS data. In Phase 1, a molecular pathway analysis is conducted on the investigation sample to test for the enrichment of specific Gene Ontology (GO) terms associated with autism. In Phase 2, the identified pathways are tested for enrichment in the replication sample. Permutation tests are performed to reduce the risk of false-positive findings. Quality assessment is conducted using QQ-plots and λ values, with Plink and R utilized for the Transmission Disequilibrium Test (TDT) and permutation tests. Results: The GO term GO:0007417 was found to be enriched in both the investigation and replication samples. SNPs associated with this pathway were observed at a frequency higher than expected in the replication sample. Conclusions: The GO term GO:0007417 (development of the nervous system) was associated with autism in both trio samples. Variations in the genes TMPRSS4, TRPC4, and PCDH9 were consistently linked to autism across the two independent samples, highlighting the role of calcium signaling and cell adhesion molecules in the risk of autism-related disorders. The pathways and variations associated with autism are described in detail, which can contribute to the engineering of new pharmacological treatments for ASD.
1. Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder that manifests early in life. It is characterized by limited interests, difficulties in social interactions, repetitive behaviors, and impairments in social communication. Additionally, individuals with ASD often experience motor and intellectual deficits, as well as mood and sleep disorders, along with sensory and gastrointestinal abnormalities [1]. ASD affects up to 1% of the general population [2] and has a genetic component, with an estimated concordance rate of 60–70% in identical twins and 5–30% in siblings [3,4].
In May 2014, the sixty-seventh World Health Assembly adopted a resolution entitled comprehensive and coordinated efforts for the management of autism spectrum disorders, which was supported by more than 60 countries. ASD is associated with approximately 3.6 million USD in lifetime social costs [5].
A recent review on the molecular aspects of ASD can be found in [6].
The GWAS Atlas (https://atlas.ctglab.nl/) accessed 15 July 2024 reports on two large GWAS focused on autism [7,8]. In the first study [7], including 7387 cases and 8567 controls, only one SNP reached GWAS significance, rs7026354, a G/A variation with no previous recorded clinical significance and which is not associated with a specific gene. In the second study, which included 18,381 cases and 27,969 controls, three SNPs reached GWAS significance: rs10099100, rs71190156, and rs910805 [8]. Rs10099100 G > C SNP is located in chr 8, and it is not reported to have clinical significance, rs71190156 is a MACROD2, intron variant, and rs910805 is located on chr 20. None of them have known clinical significance.
Environmental factors that may increase the risk of ASD are estimated to account for 50% of the risk and can lead to epigenetic modifications of key genes [9]. Epigenetic modifications are not only involved in the biological processes associated with the gene–environment interactions but also play a crucial role in orchestrating the various protein and genetic events that drive neurodevelopment [10]. Consistently, ASD is thought to be a neurodevelopmental disorder. A number of genetic variations have been inconsistently associated with an increased risk of ASD [9], and GWAS investigations have not consistently identified the single critical variations associated with this disorder [11]. This contrasts with evidence indicating that the heritability of ASD due to common variants is estimated to be as high as 80%, as recently reviewed in [12]. Limited power of the analysis, small sample sizes, and a critical phenotype definition may be limitations of the studies conducted so far [11]. Moreover, de novo mutations may explain a part of the missing heritability, as it was consistently shown that they may play a role in ASD [13,14,15,16]. Nevertheless, the impact of de novo mutations is less significant than that of inherited variants. It is estimated that 49% of the genetic architecture of ASD is associated with common inherited variants, while de novo mutations account for 3%, and rare inherited variants also account for 3% [17]. De novo mutations can lead to syndromes that resemble ASD, differing primarily in certain epidemiological details, such as a higher incidence of dysmorphic features and a similar prevalence between males and females. Examples of such genetic conditions include Fragile X syndrome, Tuberous Sclerosis Complex, Dup15q syndrome, deletions in the 16p11.2 region, Rett syndrome, and neurofibromatosis. These conditions are estimated to account for up to 5% of ASD cases observed in clinical practice [18]. There is currently no conclusive evidence for a limited number of critical common variations that confer risk for most cases of ASD. However, it is estimated that an additive effect of single variations, each having a small impact on the phenotype, may explain 60% of the genetic liability in multiplex families and 40% in simplex families [19]. An updated list of the genetic variations and genes associated with ASD can be downloaded at https://gene.sfari.org/autdb/GS_Home.do (accessed 15 July 2024). The number of genes and variations associated with ASD is extensive, and the replication rate for individual genes or mutations does not support conclusive conclusions about the neuropathology of the disorder. A potential solution to this sparse genetic evidence is the “many genes, common pathways” hypothesis, which posits that the effects of different genes may converge on common pathways and that impaired function in these pathways results in the core symptoms of ASD. Analyzing molecular pathways can enhance our understanding of complex genetic diseases by identifying gene sets that exhibit an enriched profile of association with the phenotype in question. This approach aligns with the polygenic nature of complex disorders and considers the cumulative small effects of individual genetic variations on complex phenotypes, as seen in [20,21]. Previous investigations into genetic networks which disruption could lead to ASD have identified several molecular pathways that may be involved, including protein synthesis and metabolism, the modulation of transcription processes, chromatin remodeling, calcium signaling, and the oxytocin pathway [10,22]. Nevertheless, those findings are generated by systematic reviews, and to the best of our knowledge, a molecular pathway analysis on GWAS data derived from ASD trios has yet to be conducted. It must be noted that the following investigation is conducted on the information derived from the distribution of SNPs (single nucleotide polymorphisms). SNPs are probably the most prevalent form of genetic variation in the human genome (3 to 4 million). Consequently, SNPs serve as an extensive and valuable source of information [23].
Molecular pathway analysis may be a promising approach to GWAS data, but it also has limits. One of the main limits of the genome-wide molecular pathway analysis is that it relies on known molecular pathways and known genetic functions [24]. Moreover, this technique is limited by the number of available SNPs included in GWAS, which means that the risk of false-negative findings due to the inadequate coverage of specific genes cannot be ruled out. Moreover, European and North American populations are overrepresented in the international populations. There is a clear need for non-European and North American samples to be investigated. With this in mind, it is possible to combine the results of molecular pathway analysis with the current published evidence regarding specific phenotypes, thereby helping to define the genetic makeup that increases the risk for a disease or group of diseases. In the present investigation, a molecular pathway analysis is applied to two available GWAS databases on autism disorder (investigation and replication samples). The genes harboring variations significantly associated with autism in one sample are tested for their involvement in specific molecular pathways associated with the disorder in both samples. As a result of this investigation, the identification of a molecular pathway associated with increased risk for autism is expected.
2. Methods
2.1. Dataset
Genetic data were available from NIMH genetics (https://www.nimhgenetics.org/) accessed 15 July 2024. The Autism Dataset 4 (Study 65/TASC GWAS Data) sample was chosen for the investigation sample. The Autism Dataset 3 (GWAS Data on 1264 Non-AGRE Samples) served as a replication sample. GWAS were conducted with Affymetrix 5.0 in both samples.
2.2. Toolset
Plink [25] served for the TDI GWAS analysis and genetic annotations. Plink was chosen because it is the open-source gold standard for GWAS analysis. R [26] and dedicated packages served for the permutations analysis and QQ-plot creation. R was chosen along with dedicated packages because it is an open-source widely used platform for statistical analysis. Haploview [27] served for the identification of the known available SNPs for each gene. The Gene Ontology Consortium (http://geneontology.org/) accessed 15 July 2024 was interrogated to detect enriched GO annotations. Given a list of genes, the Gene Ontology Consortium will provide a list of GO terms that identify the molecular pathways in which the genes are embedded. This process is made possible by the implementation of the R packages that were used for the analysis. Annotations of single variations were manually curated, and the source of information was an international database on genetic variations (www.gwascentral.org, accessed on 15 July 2024). This information is also accessed through the R dedicated packages. The enrichment analysis was conducted using the R software (R-4.3) suite through Bioconductor [28] and the package ReactomePA [29]. Reactome [30] is a manually curated database the includes chemical reactions, biological processes, and molecular pathways. Further investigations on elements belonging to the enriched pathways were performed through Cytoscape software 3.10 [31] and the plugins String 1.5.1, GeneMania 1.0, ClueGO 0.3-66, and CluePedia 0.3-66. Here, the physical and genetic interactions between the elements were investigated to produce an interaction network between the six enriched pathways. Further, the same approach was used to extend the network to include drugs interactions (using the Cytargetlinker 4.1.0 plugin and Drugbank Database (ver. 5.6.1)).
Chart 1 shows the protocol of the analysis.
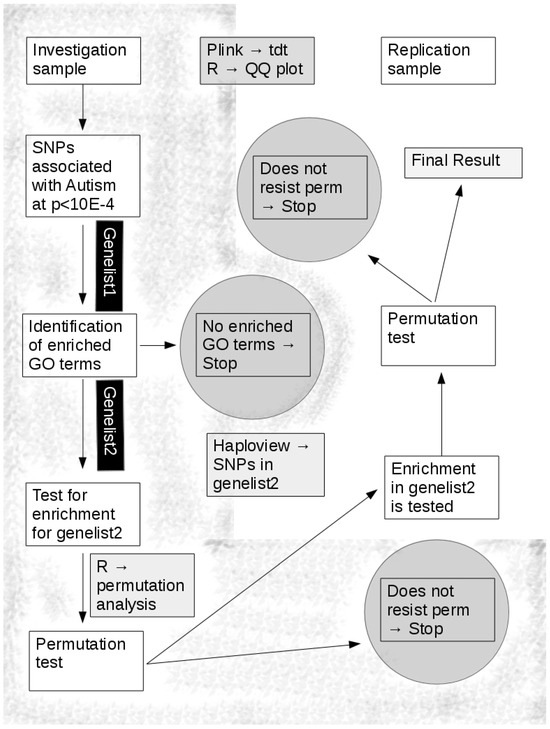
Chart 1.
The analysis flow is shown. The shadowed areas refer to the analysis of the investigation sample. Circles indicate the points in which the analysis is meant to stop, and a negative result is given. Arrows show the direction of the analysis. Enrichment in genelist2 in the replication sample is conducted while accounting for p thresholds < 0.01 and a consistent direction of association between the investigation and the replication sample (OR> or <1).
2.3. Analysis Flow
2.3.1. Investigation Sample
TDI (trait-dependent interaction—this analysis investigates how the interaction between genetics variants can influence the phenotype under analysis) associations were run with Plink, and the results were annotated. The standard quality thresholds were applied to the original sample before pruning and imputing. The minor allele frequency was set at 0.05, the genotype rate was set at 0.95, and the Hardy–Weinberg equilibrium was set at 0.00001. Pruning was set at the standard—indep 50 5 2, where 50 is the number of SNPs considered at every step, 5 is the number of SNPs to be shifted at every step, and 2 is the VIF threshold (1/(1 − R2), where R2 is the multiple correlation coefficient). A R^2 equal to 10 implies that two SNPs carry the same signal. A R2 equal to 1 implies that two SNPs are completely independent. The quality of the results was checked (QQ-plot and λ values; please refer to Figure 1 and Figure 2). QQ-plots and lambda values are essential tools for excluding inflation, which refers to the presence of structural false-positive findings in a dataset. QQ-plots illustrate the deviation from expected to observed significant associations, while a lambda value close to one suggests the absence of inflation in the dataset. Variations associated with autism with a significance threshold inferior to 10 × 10−4 were selected and the corresponding genes—genes that harbor the selected variations—identified (genelist1). SNP characteristics and corresponding data are shown in Table 1. Enriched GO (Gene Ontology) terms were searched in genelist1: the GO Consortium served for the analysis (http://geneontology.org/, accessed on 15 July 2024). Such an analysis investigates whether specific terms from the GO database are statistically overrepresented in a given set of genes. Genes within the enriched GO terms were selected (thus, creating genelist2) and their SNPs identified (www.gwascentral.com, accessed on 15 July 2024) through interrogation of the international online databases that pair, when possible, single genetic variations to genes and searched for in the available NIMH GWAS dataset. SNPs belonging to genetlis2 available for the analysis were annotated.
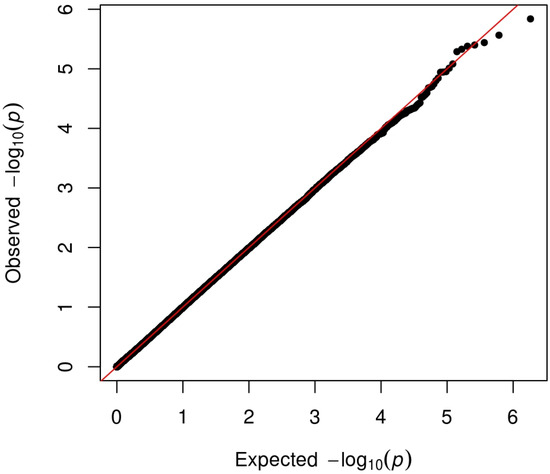
Figure 1.
QQ-plot of the TDI association test in the primary sample. The picture shows the QQ-plot analysis. The presence of inflation is excluded from the database under analysis. x-axis: Represents the expected p-values under the null hypothesis (no significant difference between cases and controls). These values are what you would expect if there were no true associations. y-axis: Represents the observed p-values from the GWAS analysis, which indicate the significance of the associations found in the data. If the observed p-values closely align with the expected p-values (i.e., points fall along the diagonal line), it suggests that there is no significant genetic stratification in the sample, supporting the null hypothesis. A deviation from this line, particularly where observed p-values are significantly lower (indicating higher significance), suggests the presence of true genetic associations. This could indicate genetic stratification, where the cases and controls differ in ways that affect the results. A large number of observed p-values that are more significant than expected could indicate that the sample is genetically stratified. This stratification can lead to inflated type I error rates, meaning that the study may falsely identify associations that do not exist. The presence of a small fraction of observed p-values that are significantly lower than expected may indicate true genetic associations, which are of interest for further investigation.
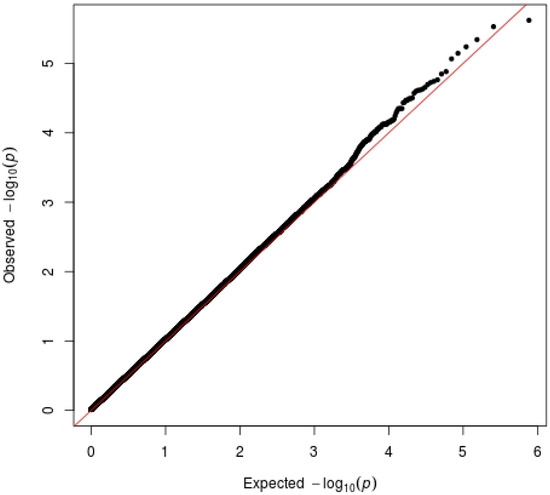
Figure 2.
QQ-plot in the secondary sample. The picture shows the QQ-plot analysis. The presence of inflation is excluded from the database under analysis. x-axis: Represents the expected p-values under the null hypothesis (no significant difference between cases and controls). These values are what you would expect if there were no true associations. y-axis: Represents the observed p-values from the GWAS analysis, which indicate the significance of the associations found in the data. If the observed p-values closely align with the expected p-values (i.e., points fall along the diagonal line), it suggests that there is no significant genetic stratification in the sample, supporting the null hypothesis. A deviation from this line, particularly where observed p-values are significantly lower (indicating higher significance), suggests the presence of true genetic associations. This could indicate genetic stratification, where the cases and controls differ in ways that affect the results. A large number of observed p-values that are more significant than expected could indicate that the sample is genetically stratified. This stratification can lead to inflated type I error rates, meaning that the study may falsely identify associations that do not exist. The presence of a small fraction of observed p-values that are significantly lower than expected may indicate true genetic associations, which are of interest for further investigation.

Table 1.
SNPs associated with autism with p < 10 × 10−4.
2.3.2. Replication Sample
SNPs were derived from genetlist2, and the incidence of observed vs. expected significant associations was checked in the replication sample.
2.4. Selection of Significant Thresholds
Genelist1 was created after selecting SNPs which strength of association with the investigated phenotype was stronger than 10 × 10−4. This threshold was chosen in order to prioritize the best available SNPs after the genetic association analysis while retaining a sufficient number of SNPs to identify a list of candidate genes also available in the replication sample. In the following steps of the analysis, the p thresholds were set to a less punitive level in order to identify the possible positive associations while abating the risk for false-positive associations with permutations (see Section 2.6). We were interested in analyzing the hypothesis that the prevalence of SNPs from genelist2 associated with the phenotype under analysis was higher—and not lesser—than the prevalence of the same SNPs in genelist2. Since we were not interested in one side of the distribution of probabilities (we were only interested in “more represented” and not “less represented”), a more punitive 0.025 (instead for 0.05) and 0.005 (instead for 0.01) threshold was chosen.
2.5. Index Pathway Definition
The index pathway is defined as the molecular pathway that is significantly overrepresented in genelist1. Genes belonging to the overrepresented molecular pathways are stored in genelist2.
2.6. Simulation
A subset of SNPs was selected in the replication sample while respecting the following characteristics:
- should be associated with autism at a p threshold < 0.001 in the investigation sample;
- should be available in the replication sample;
- should have the same direction of association (OR> or <1 as in the investigation sample).
Permutations were run until a monotone number of different outputs resulted from the process. The permutated molecular pathways had the same length (number of SNPs) of the pathway in list1 in order to avoid an overrepresentation of SNPs associated with the phenotype under analysis due to the difference in lengths of the permutated molecular pathways (more genes equals to higher probability to analyze more SNPs). The probability of having a random pathway presenting with a number of significant (p < 0.05) associations superior to or equal to the index pathway (extracted from the investigation sample and tested in the replication sample) was then calculated.
Therefore, 10 × 106 permutations were conducted in the investigation sample, creating a number of molecular pathways similar to the index pathway in terms of the number of missense splicing variations and variations without annotation. The probability of having a random pathway presenting with a number of significant (p < 0.05 and p < 0.01) associations superior to or equal to the index pathway was then calculated. Permutations were instrumental in accepting or rejecting the hypothesis that a statistically significant enrichment was observed for the same pathway in both the investigation and replication sample.
2.6.1. Acceptance of H0 (Null Hypothesis)
Under the null hypothesis, enrichment of the index pathway is to be considered a false-positive finding.
Acceptance of the null hypothesis was to take place in case of more than 2.5% and 0.05% of the simulated pathways showing the same or an increased number of SNPs significantly (p < 0.05 and p < 0.01) associated with the phenotype under analysis.
2.6.2. Acceptance of H1 (Enrichment Hypothesis)
Under H1, enrichment of the index pathway is to be considered a true-positive finding.
Acceptance of H1 was to take place in case of less than 2.5% and 0.5% of the simulate pathways showing the same or an increased number of SNPs significantly (p < 0.05 and p < 0.01) associated with the phenotype under analysis. In case of the acceptance of H1, SNPs in the replication sample that are harbored by genes in genelist2 were identified and their association (p-values) and direction of association (OR> or <1) with autism recorded. SNPs and OR were recorded in list1.
3. Results
3.1. Investigation Sample
The database under analysis was the Autism Dataset 4 (Study 65/TASC GWAS Data) sample. The phenotype under analysis was the “nimh_autism_dataset4_altpheno_plink.txt”, as downloaded from the NIMH genetics dataset. Therefore, 2890 individuals (1824 males, 1069 females) with non-missing phenotypes were found: 936 cases, 1954 controls, and 3 missing. Additionally,1930 founders and 963 non-founders were found, and the total genotyping rate was 0.93. Also, 965 nuclear families were detected, two founder singletons were found, and 963 non-founders with two parents in 963 nuclear families were included in the analysis. Finally, 934 affected offspring trios were identified, and 1,160,305 markers were available for the analysis.
Genomic inflation was excluded (λ = 1.002) in Figure 1.
The analysis of the molecular pathways associated with genetlis1 resulted in the identification of a single molecular pathway showing a significant overrepresentation. The molecular pathway extracted from genelist1 based on its overrepresentation was the central nervous system development pathway (GO:0007417, index pathway). The results are reported in Table 2. Table 3 shows the genes that belong to GO:0007417 and were found to harbor SNPs associated with autism. Additionally, 1124 SNPs belonging to the index pathway were identified and were available for the analysis in the investigation sample.

Table 2.
Analysis of the overrepresentation of specific molecular pathways.

Table 3.
GO terms enrichment analysis. Genes belonging to the enriched GO pathway are detailed.
The prevalence of significant associations within the pathway was 6.4% at a p level of 0.05 (28% more than expected) and 1.3% at a p level of 0.01 (30% more than expected).
The number of simulated pathways that had the same or an increased number of SNPs significantly associated with the phenotype under investigation in the investigated sample was 2.2% at a p level of 0.05. The null hypothesis was then rejected, and the index pathway was deemed significantly enriched after the TDT in the sample of autistic patients.
3.2. Replication Sample
The Autism Dataset 3 (GWAS Data on 1264 Non-AGRE Samples) was the replication sample, and 1233 individuals were available from the dataset: 789 males, 444 females, and 588 cases. Additionally, 333 nuclear families and three founder singletons were detected. Also, 579 non-founders with two parents in 321 nuclear families, 14 non-founders without two parents in 9 nuclear families, and 579 affected offspring trios were detected. Finally, 393,763 markers were included in the analysis.
Genomic inflation was excluded (λ = 1.03) in Figure 2.
The SNPs belonging to the index pathway in the investigation sample (n = 1124) were identified in the replication sample, and 519 SNPs were detected that belonged to the index pathway from the investigation sample and represented in the replication sample and had the same direction of association with autism as in the investigation sample. The prevalence of SNPs associated with autism in the replication sample was 5.4% at a p level of 0.05 and 1.9% at a p level of 0.01 (Figure 3). After the permutation analysis, the prevalence of SNPs associated with autism was confirmed at a p level of <0.01.
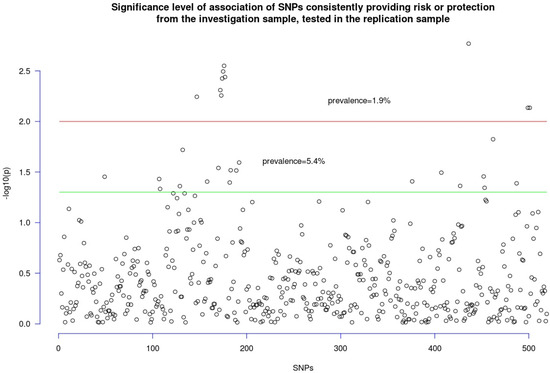
Figure 3.
Replication findings. SNPs associated with autism in the investigation sample, available in the replication sample, and having the same direction of association in both samples are shown. The prevalence of SNPs belonging to the index pathway is slightly more superior than expected at a p threshold < 0.05 and double the expected at a p threshold < 0.01. The latter enriched resisted the permutation test.
As a final result, the hypothesis that the enriched pathway in the investigation sample could be replicated in the replication sample was accepted after the risk for false positives was abated by 10 × 105 permutations. The main results are reported in Table 4 and Table 5.

Table 4.
SNPs which associations with autism were replicated at a p level < 0.01.

Table 5.
Genes which associations with autism were replicated in the present investigation (at least one SNP associated at a p level < 0.01).
4. Discussion
ASD is a frequent condition in the general population [32] that is characterized by impaired social abilities, restricted interests, and repetitive behavior. The disorder has consistently been shown to have a genetic basis, with the number of involved genes potentially reaching into the hundreds (source: SFARI Gene). This suggests a polygenic nature, indicating that different genetic backgrounds may converge on the same phenotype. To investigate the convergence of specific molecular pathways related to autism spectrum disorder (ASD), a metabolic pathway analysis was conducted using both an investigation sample and a replication sample of autistic trios. The inclusion of a replication sample and the use of permutations were implemented to reduce the risk of false-positive findings. The analysis of complex disorders through a molecular pathway analysis is a well-known and implemented type of analysis, used both in psychiatry and in non-psychiatric disorders [33,34,35]. Research into the genetics of autism has been ongoing for decades, indicating a long-standing interest in understanding its hereditary components. Autism, like many complex psychiatric disorders, is influenced by numerous genetic factors. Individual common genetic variations often have minimal effects on the overall genetic architecture of autism, making it challenging to pinpoint specific risk factors. Genome-wide epigenetic investigations have not yielded conclusive results, suggesting that the interplay between genetics and environmental factors in autism is complex and not fully understood [36]. Increasing sample sizes in genetic studies is a common strategy to enhance the power of detecting associations. Larger samples can help identify variants with small effects that might otherwise go unnoticed. While this approach has led to some interesting findings [37], interpreting these results can be difficult due to the multifactorial nature of autism. A promising strategy to address the limitations of focusing solely on single common variations is the molecular pathway approach. This method combines the effects of multiple genetic variations that contribute to specific biological pathways, potentially providing a more comprehensive understanding of the genetic risk for autism. This approach has shown success in other psychiatric disorders [38,39,40,41], suggesting it could be beneficial for autism research as well. However, its application in autism studies has not been widespread, indicating a gap in the current research methodologies. This finding can be instrumental in developing genetic screening tools for early diagnosis, bridging the gap between psychiatry and other specialties [42,43,44,45]. An interesting approach may be Mendelian randomization, which uses genetic variants that are associated with a modifiable exposure as proxies to infer the causal effect of that exposure on the risk of being diagnosed with autism [46,47].
As a result of this hypothesis-free approach, the GO:0007417 pathway (development of the nervous system) was found to be enriched in the investigation sample. Several variations within this pathway were associated with autism in both samples, as detailed in Table 3 and Table 4. The risk of false positives in the replication analysis was mitigated through permutations, and the findings remained significant at a nominal p-value of 0.01, as illustrated in Figure 3. This supports the theory that autism is a neurodevelopmental disorder and is further confirmed by previous studies [48,49].
The present investigation offers a list of SNPs and identifies a GO molecular pathway associated with an increased risk of autism. This identification enhances our understanding of the molecular mechanisms involved in autism, potentially paving the way for new treatments. In this context, we recommend further analysis of the following genes, as reported in Table 5: PCDH9 (Protocadherin 9), also previously found to be associated with autism [50]; TRPC4 (Transient Receptor Potential Cation Channel Subfamily C Member 4), also previously associated with autism [51]; and TMPRSS4 (Transmembrane Serine Protease 4), also previously found to be potentially associated with autism [52].
This aspect is of prime relevance, in that most patients with autism are pharmacologically treated in order to target non-core symptoms of their disease, including self-injury, aggression, irritability, and hyperactivity [53]. The molecular pathways that are targets for such treatments are represented in Figure 4. Some drugs have been used in order to treat the target core of the disorder, including growth factors [54], glutamatergic drugs [55,56,57], GABAergic drugs [58,59], and oxytocin [60]. The results are encouraging, but further research is required [61].
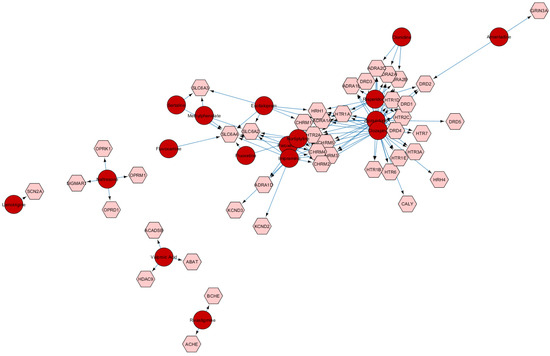
Figure 4.
The network below represents the target of molecular drugs used in the treatment of non-core symptoms of autism.
The network was built from the Drugbank database. The query was performed through the CyTargetLinker plugin of Cytoscape 3.10.3 software.
Dark-shadowed hexagons indicate the targeted proteins of pharmaceutical compounds. The proteins were obtained from the previous analyses on GO:0007417. Dark-shadowed circles indicate the compounds which interactions with the targeted proteins have been validated in the Drugbank Database.
The GO:0007417 pathway represents a comprehensive network that encompasses several biological subprocesses involved in the development and physiological functions of the central nervous system. A detailed view of the network and its subnetworks within GO:0007417 is illustrated in the figure below (Figure 5a,b). In particular, Figure 5b highlights various molecular interactions associated with GO:0007417, including those related to astrocyte and oligodendrocyte differentiation, the development of the limbic system, and postsynaptic transmission, among others. This representation is particularly noteworthy, as the role of glial cells in shaping neuronal networks during development may be crucial for understanding several psychiatric disorders, including autism [62]. Moreover, the involvement of the limbic system may play a major role in the pathophysiology of autism, which is characterized by a diminished ability to process affective inputs [63,64].
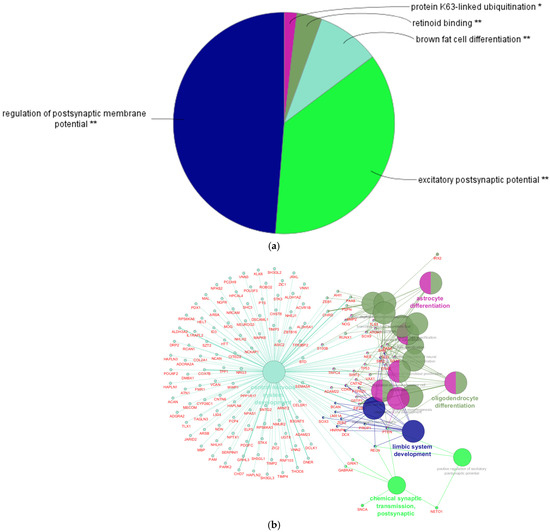
Figure 5.
(a) In the following diagram, the biologic processes related to the genes under investigation are represented. The diagram shows the subnetworks, and the genes not included in subnetworks, that belong to GO:0007417. In the network below is presented a graphic visualization of the molecular network. The asterisks indicate when the correlation between the subnetworks and the set of genes under investigation is significant (*) or highly significant (**). This was calculated by the Cluepedia plugin for Cytoscape 3.10.3 software. To calculate the degree of significance for each biological process, the software evaluates the ratio between (1) the number of genes within the network under investigation that also belong to the biological process and (2) the total number of genes composing the same biological process. (b) GO:0007417. The molecular pathway is represented, along with some of the connected molecular networks, including the astrocyte and oligodendrocyte differentiation, the development of the limbic system, and postsynaptic transmission, among others. The network was built from the GO Biological Process database (9 February 2016). The query was performed through the ClueGO plugin of Cytoscape software. Details on the query, including kappa score, the Tree interval considered, and others, are available upon request.
Radial glia cells help neurons migrate towards their final neurological destination through the developmental process, starting from the fetal period [65]. The migration from the germinal areas of the nervous system, which is dependent on the correct activity of the glia system, shapes some nervous structures later during the developing period, called cortical mini columns [66]. Those structures were found to be significantly altered in the brains of autistic patients, containing more neurons when compared to the normal controls and signs for a global GABAergic dysfunction [67]. Finally, the glia is involved in the pruning process, an event that continues in late adolescence and is central to the development of brain structures. This process is mediated in part by glia cells [68], as it was demonstrated that the glia is overrepresented in the brains of autistic patients, which shows a pattern of activation that is aberrant or consistent with a pro-inflammatory state [69].
This is consistent with the hypothesis that autism arises from disruptions in the physiological interregional connectivity between brain regions. Our findings align with previous observations in the literature and suggest an involvement of neurodevelopmental events, as well as a potential role of glial cells in shaping these processes. However, the present investigation has limitations that necessitate further independent studies.
One significant limitation is the low number of markers and the inconsistencies between markers in the investigation and replication samples. This resulted in a heterogeneous representation of the same molecular pathways across the two genetic databases. Replication was possible because different genes covered in the various genetic databases were found to be enriched and clustered within the GO:0007417 pathway. The lack of comprehensive coverage and the differing SNPs included in the two arrays may have contributed to a decrease in the power of the current analysis.
The linkage disequilibrium (LD) structure of the variations under analysis was considered in the quality assessment of the results (using QQ-plots and lambda values), but the impact of LD cannot be entirely ruled out. The samples analyzed are considered medium to large in the context of GWAS, which may also have influenced the power of this investigation. We attempted to mitigate the risk of false-positive findings by including a replication sample and conducting permutation analyses. Nevertheless, further independent analyses are essential before confirming our results.
Finally, our results strengthen the idea that further research is needed focused on the genes and molecular cascades that characterize the GO:0007417 pathway. These genetic insights could inform the development of targeted therapies for autism.
5. Dataset
5.1. Investigation Sample
Genetic data were available from the NIMH Center for Collaborative Genetic Studies.
NIMH Study 65, also known as AGP or TASC (PI: Gallagher), deposited genotype data in four sets, cleaned and raw, stage1 and stage2. NIMH then combined the cleaned stage1 and stage2 data into one dataset named Autism Dataset 4 in the PLINK file format. The n = 2893 records in Dataset 4 occur in n = 964 families (99% trios), of which n = 935 families have one or more probands with a diagnosis, ranging from strict to broad autism; see the documentation for definitions.
The AGP Simplex Collection (TASC) was funded by an award from Autism Speaks and by funding support to the repository development by the NIMH. The principal investigator and co-investigators in this study were Louise Gallagher, Trinity College Dublin; Astrid Vicente, Instituto Gulbenkian de Ciencia, Oeira; Joseph Buxbaum, Mount Sinai School of Medicine; Peter Szatmari, McMaster University; William McMahon, University of Utah; Michael Cuccaro, University of Miami; James Sutcliffe, Vanderbilt University; Christine Freitag, Klinikum der Johann-Wolfgang Goethe-Universität, Frankfurt/Main; Sabine Klauck, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg; Veronica Vieland (DCC Director), Research Institute at Nationwide Children’s Hospital, Ohio; Dan Geschwind, AGRE/UCLA; John Nurnberger, University of Indiana; Ed Cook, University of Illinois at Chicago; Raphael Bernier, University of Washington/CPEA.
5.2. Replication Sample
Genotype data were generated by Dan Arking and Aravinda Chakravarti using 1264 samples that were obtained from the NIMH Center for Collaborative Genetic Studies on Mental Disorders. This study was supported by grants from the NIMH (R01 MH060007) to Aravinda Chakravarti and Dan Arking.
Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Jr., Chakravarti A. A Common Genetic Variant in the Neurexin Superfamily Member CNTNAP2 Increases Familial Risk of Autism. Am J Hum Genet 2008; 82:160–164.
5.3. Computational Power
A computer cluster was used to undertake the analyses. Computation power was required for the permutation analysis. The GenomeDK HPC cluster at Aarhus University comprises 190 nodes (3384 compute cores) connected with 10GigE/Infiniband. Each node has from 16 to 32 cores and either 64 GB, 128 GB, 256 GB, 512 GB, or 1 TB of RAM. The cluster has been designed specifically for bioinformatic workloads and has a storage capacity of 3.5 PB.
Author Contributions
Conceptualization, A.D. and C.C.; methodology, A.D. and M.C.; software, A.D. and M.C.; validation, A.D.; formal analysis, A.D. and M.C.; investigation, A.D. and M.C.; resources, A.D.; data curation, A.D.; writing—original draft preparation, A.D. and M.C.; writing—review and editing, A.D. and C.C.; visualization, M.C.; supervision, A.D. and C.C.; project administration, A.D. All authors have read and agreed to the published version of the manuscript.
Funding
This paper received no funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available upon request at https://www.nimhgenetics.org/ (accessed on 15 July 2024).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Geschwind, D.H. Advances in autism. Annu. Rev. Med. 2009, 60, 367–380. [Google Scholar] [CrossRef]
- Lai, M.-C.; Lombardo, M.V.; Baron-Cohen, S. Autism. Lancet 2014, 383, 896–910. [Google Scholar] [CrossRef] [PubMed]
- Ozonoff, S.; Young, G.S.; Carter, A.; Messinger, D.; Yirmiya, N.; Zwaigenbaum, L.; Bryson, S.; Carver, L.J.; Constantino, J.N.; Dobkins, K.; et al. Recurrence risk for autism spectrum disorders: A Baby Siblings Research Consortium study. Pediatrics 2011, 128, e488–e495. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.; Le Couteur, A.; Gottesman, I.; Bolton, P.; Simonoff, E.; Yuzda, E.; Rutter, M. Autism as a strongly genetic disorder: Evidence from a British twin study. Psychol. Med. 1995, 25, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Cakir, J.; Frye, R.E.; Walker, S.J. The Lifetime Social Cost of Autism: 1990–2029. Res. Autism Spectr. Disord. 2020, 72, 101502. [Google Scholar] [CrossRef]
- Horecka-Lewitowicz, A.; Lewitowicz, W.; Wawszczak-Kasza, M.; Lim, H.; Lewitowicz, P. Autism Spectrum Disorder Pathogenesis-A Cross-Sectional Literature Review Emphasizing Molecular Aspects. Int. J. Mol. Sci. 2024, 25, 11283. [Google Scholar] [CrossRef]
- Autism Spectrum Disorders Working Group of the Psychiatric Genomics Consortium. Meta-Analysis of GWAS of over 16,000 Individuals with Autism Spectrum Disorder Highlights a Novel Locus at 10q24.32 and a Significant Overlap with Schizophrenia. Mol. Autism 2017, 8, 21. [Google Scholar] [CrossRef]
- Grove, J.; Ripke, S.; Als, T.D.; Mattheisen, M.; Walters, R.; Won, H.; Pallesen, J.; Agerbo, E.; Andreassen, O.A.; Anney, R.; et al. Common Risk Variants Identified in Autism Spectrum Disorder. bioRxiv 2017. [Google Scholar] [CrossRef]
- Choi, L.; An, J.-Y. Genetic Architecture of Autism Spectrum Disorder: Lessons from Large-Scale Genomic Studies. Neurosci. Biobehav. Rev. 2021, 128, 244–257. [Google Scholar] [CrossRef]
- Khoodoruth, M.A.S.; Khoodoruth, W.N.C.-K.; Uroos, M.; Al-Abdulla, M.; Khan, Y.S.; Mohammad, F. Diagnostic and Mechanistic Roles of MicroRNAs in Neurodevelopmental & Neurodegenerative Disorders. Neurobiol. Dis. 2024, 202, 106717. [Google Scholar] [CrossRef]
- Nóbrega, I.d.S.; Teles E Silva, A.L.; Yokota-Moreno, B.Y.; Sertié, A.L. The Importance of Large-Scale Genomic Studies to Unravel Genetic Risk Factors for Autism. Int. J. Mol. Sci. 2024, 25, 5816. [Google Scholar] [CrossRef] [PubMed]
- Vilela, J.; Rasga, C.; Santos, J.X.; Martiniano, H.; Marques, A.R.; Oliveira, G.; Vicente, A.M. Bridging Genetic Insights with Neuroimaging in Autism Spectrum Disorder—A Systematic Review. Int. J. Mol. Sci. 2024, 25, 4938. [Google Scholar] [CrossRef]
- Levy, D.; Ronemus, M.; Yamrom, B.; Lee, Y.; Leotta, A.; Kendall, J.; Marks, S.; Lakshmi, B.; Pai, D.; Ye, K.; et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron 2011, 70, 886–897. [Google Scholar] [CrossRef]
- Marshall, C.R.; Noor, A.; Vincent, J.B.; Lionel, A.C.; Feuk, L.; Skaug, J.; Shago, M.; Moessner, R.; Pinto, D.; Ren, Y.; et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008, 82, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Sebat, J.; Lakshmi, B.; Malhotra, D.; Troge, J.; Lese-Martin, C.; Walsh, T.; Yamrom, B.; Yoon, S.; Krasnitz, A.; Kendall, J.; et al. Strong association of de novo copy number mutations with autism. Science 2007, 316, 445–449. [Google Scholar] [CrossRef]
- Utami, K.H. The implications of de novo coding mutations in simplex autism families. Clin. Genet. 2015, 87, 428–429. [Google Scholar] [CrossRef] [PubMed]
- Gaugler, T.; Klei, L.; Sanders, S.J.; Bodea, C.A.; Goldberg, A.P.; Lee, A.B.; Mahajan, M.; Manaa, D.; Pawitan, Y.; Reichert, J.; et al. Most genetic risk for autism resides with common variation. Nat. Genet. 2014, 46, 881–885. [Google Scholar] [CrossRef] [PubMed]
- Veltman, M.W.M.; Craig, E.E.; Bolton, P.F. Autism spectrum disorders in Prader-Willi and Angelman syndromes: A systematic review. Psychiatr. Genet. 2005, 15, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Klei, L.; Sanders, S.J.; Murtha, M.T.; Hus, V.; Lowe, J.K.; Willsey, A.J.; Moreno-De-Luca, D.; Yu, T.W.; Fombonne, E.; Geschwind, D.; et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol. Autism 2012, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Aryal, S.; Bonanno, K.; Song, B.; Mani, D.R.; Keshishian, H.; Carr, S.A.; Sheng, M.; Dejanovic, B. Deep Proteomics Identifies Shared Molecular Pathway Alterations in Synapses of Patients with Schizophrenia and Bipolar Disorder and Mouse Model. Cell Rep. 2023, 42, 112497. [Google Scholar] [CrossRef] [PubMed]
- Calabró, M.; Briuglia, S.; Crisafulli, C.; Drago, A. Body Weight Changes and Bipolar Disorder: A Molecular Pathway Analysis. Pharmacogenet. Genom. 2022, 32, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Gong, M.; Li, J.; Qin, Z.; Machado Bressan Wilke, M.V.; Liu, Y.; Li, Q.; Liu, H.; Liang, C.; Morales-Rosado, J.A.; Cohen, A.S.A.; et al. MARK2 Variants Cause Autism Spectrum Disorder via the Downregulation of WNT/β-Catenin Signaling Pathway. Am. J. Hum. Genet. 2024, 111, 2392–2410. [Google Scholar] [CrossRef]
- Müller-Myhsok, B. The Usefulness of Single Nucleotide Polymorphisms (SNPs) for Genetic Epidemiological Investigation of Complex Psychiatric Diseases. Prog. Neuropsychopharmacol. Biol. Psychiatry 2005, 29, 1017–1020. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Croes, D.; van Helden, J. Prediction of metabolic pathways from genome-scale metabolic networks. Biosystems 2011, 105, 109–121. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- R Foundation for Statistical Computing. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and Visualization of LD and Haplotype Maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating High-Throughput Genomic Analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef]
- Yu, G.; He, Q.-Y. ReactomePA: An R/Bioconductor Package for Reactome Pathway Analysis and Visualization. Mol. Biosyst. 2016, 12, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Croft, D.; Mundo, A.F.; Haw, R.; Milacic, M.; Weiser, J.; Wu, G.; Caudy, M.; Garapati, P.; Gillespie, M.; Kamdar, M.R.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2014, 42, D472–D477. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Hodges, H.; Fealko, C.; Soares, N. Autism Spectrum Disorder: Definition, Epidemiology, Causes, and Clinical Evaluation. Transl. Pediatr. 2020, 9, S55–S65. [Google Scholar] [CrossRef]
- Defo, J.; Awany, D.; Ramesar, R. From SNP to Pathway-Based GWAS Meta-Analysis: Do Current Meta-Analysis Approaches Resolve Power and Replication in Genetic Association Studies? Brief. Bioinform. 2023, 24, bbac600. [Google Scholar] [CrossRef]
- Schulmann, A.; Ryu, E.; Goncalves, V.; Rollins, B.; Christiansen, M.; Frye, M.A.; Biernacka, J.; Vawter, M.P. Novel Complex Interactions between Mitochondrial and Nuclear DNA in Schizophrenia and Bipolar Disorder. Mol. Neuropsychiatry 2019, 5, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Dhroso, A.; Johnson, N.; Korkin, D. The Variation Game: Cracking Complex Genetic Disorders with NGS and Omics Data. Methods 2015, 79–80, 18–31. [Google Scholar] [CrossRef]
- Andrews, S.V.; Sheppard, B.; Windham, G.C.; Schieve, L.A.; Schendel, D.E.; Croen, L.A.; Chopra, P.; Alisch, R.S.; Newschaffer, C.J.; Warren, S.T.; et al. Case-Control Meta-Analysis of Blood DNA Methylation and Autism Spectrum Disorder. Mol. Autism 2018, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Glessner, J.T.; Li, J.; Qi, X.; Hou, X.; Zhu, C.; Li, X.; March, M.E.; Yang, L.; Mentch, F.D.; et al. Integrative Analysis of Genome-Wide Association Studies Identifies Novel Loci Associated with Neuropsychiatric Disorders. Transl. Psychiatry 2021, 11, 69. [Google Scholar] [CrossRef]
- Lipska, B.K.; Mitkus, S.N.; Mathew, S.V.; Fatula, R.; Hyde, T.M.; Weinberger, D.R.; Kleinman, J.E. Functional Genomics in Postmortem Human Brain: Abnormalities in a DISC1 Molecular Pathway in Schizophrenia. Dialogues Clin. Neurosci. 2006, 8, 353–357. [Google Scholar] [CrossRef] [PubMed]
- O’Dushlaine, C.; Kenny, E.; Heron, E.; Donohoe, G.; Gill, M.; Morris, D.; International Schizophrenia Consortium; Corvin, A. Molecular Pathways Involved in Neuronal Cell Adhesion and Membrane Scaffolding Contribute to Schizophrenia and Bipolar Disorder Susceptibility. Mol. Psychiatry 2011, 16, 286–292. [Google Scholar] [CrossRef]
- Espregueira Themudo, G.; Leerschool, A.-R.; Rodriguez-Proano, C.; Christiansen, S.L.; Andersen, J.D.; Busch, J.R.; Christensen, M.R.; Banner, J.; Morling, N. Targeted Exon Sequencing in Deceased Schizophrenia Patients in Denmark. Int. J. Legal Med. 2020, 134, 135–147. [Google Scholar] [CrossRef]
- Bhattacharyya, U.; John, J.; Lencz, T.; Lam, M. Dissecting Schizophrenia Biology Using Pleiotropy with Cognitive Genomics. MedRxiv Prepr. Serv. Health Sci. 2024. [Google Scholar] [CrossRef]
- Wang, G.; Zhao, J.; Zhou, M.; Lu, H.; Mao, F. Unveiling Diabetic Nephropathy: A Novel Diagnostic Model through Single-Cell Sequencing and Co-Expression Analysis. Aging 2024, 16, 10972–10984. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Ou, Y.; Xiao, L.; Gu, R.; He, F.; Peng, J.; Shu, Y.; Li, T.; Hao, L. Identification and Construction of a Disulfidptosis-Mediated Diagnostic Model and Associated Immune Microenvironment of Osteoarthritis from the Perspective of PPPM. J. Inflamm. Res. 2024, 17, 3753–3770. [Google Scholar] [CrossRef]
- Xiong, Z.; Fang, Y.; Lu, S.; Sun, Q.; Sun, Y.; Yang, P.; Huang, J. Exploring the Relevance of Disulfidptosis to the Pathophysiology of Ulcerative Colitis by Bioinformatics Analysis. J. Inflamm. Res. 2024, 17, 2757–2774. [Google Scholar] [CrossRef]
- Liu, H.; Tang, T. Pan-Cancer Genetic Analysis of Cuproptosis and Copper Metabolism-Related Gene Set. Front. Oncol. 2022, 12, 952290. [Google Scholar] [CrossRef]
- Liu, H.; Xie, R.; Dai, Q.; Fang, J.; Xu, Y.; Li, B. Exploring the Mechanism Underlying Hyperuricemia Using Comprehensive Research on Multi-Omics. Sci. Rep. 2023, 13, 7161. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Garg, P.K.; Wu, J. Associations between Daytime Napping, Sleep Duration, and Depression and 15 Cardiovascular Diseases: A Mendelian Randomization Study. Cardiovasc. Diagn. Ther. 2024, 14, 771–787. [Google Scholar] [CrossRef] [PubMed]
- Rahnama, M.; Tehrani, H.A.; Mirzaie, M. Vahid Ziaee Identification of Key Genes and Convergent Pathways Disrupted in Autism Spectrum Disorder via Comprehensive Bioinformatic Analysis. Inform. Med. Unlocked 2021, 24, 100589. [Google Scholar] [CrossRef]
- Huang, G.; Chen, S.; Chen, X.; Zheng, J.; Xu, Z.; Doostparast Torshizi, A.; Gong, S.; Chen, Q.; Ma, X.; Yu, J.; et al. Uncovering the Functional Link Between SHANK3 Deletions and Deficiency in Neurodevelopment Using iPSC-Derived Human Neurons. Front. Neuroanat. 2019, 13, 23. [Google Scholar] [CrossRef]
- Miozzo, F.; Murru, L.; Maiellano, G.; Zippo, A.; Moretto, E.; Avendano, A.Z.; Metodieva, V.D.; Riccardi, S.; D’Aliberti, D.; Spinelli, S.; et al. Disruption of Autism-Associated Pcdh9 Gene Leads to Transcriptional Alterations, Synapses Overgrowth and Aberrant Excitatory Transmission in the CA1. 2024. Available online: https://www.biorxiv.org/content/10.1101/2024.01.27.577576v1.abstract (accessed on 15 July 2024).
- Gupta, V.; Ben-Mahmoud, A.; Ku, B.; Velayutham, D.; Jan, Z.; Yousef Aden, A.; Kubbar, A.; Alshaban, F.; Stanton, L.W.; Jithesh, P.V.; et al. Identification of Two Novel Autism Genes, TRPC4 and SCFD2, in Qatar Simplex Families through Exome Sequencing. Front. Psychiatry 2023, 14, 1251884. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, A.; Chanez-Paredes, S.D.; Haest, X.; Turner, J.R. Paracellular Permeability and Tight Junction Regulation in Gut Health and Disease. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 417–432. [Google Scholar] [CrossRef]
- Peñagarikano, O. New Therapeutic Options for Autism Spectrum Disorder: Experimental Evidences. Exp. Neurobiol. 2015, 24, 301–311. [Google Scholar] [CrossRef]
- Kolevzon, A.; Bush, L.; Wang, A.T.; Halpern, D.; Frank, Y.; Grodberg, D.; Rapaport, R.; Tavassoli, T.; Chaplin, W.; Soorya, L.; et al. A pilot controlled trial of insulin-like growth factor-1 in children with Phelan-McDermid syndrome. Mol. Autism 2014, 5, 54. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Dobkin, C.; Sheikh, A.M.; Malik, M.; Brown, W.T.; Li, X. The therapeutic effect of memantine through the stimula-tion of synapse formation and dendritic spine maturation in autism and fragile X syndrome. PLoS ONE 2012, 7, e36981. [Google Scholar] [CrossRef]
- Chez, M.G.; Burton, Q.; Dowling, T.; Chang, M.; Khanna, P.; Kramer, C. Memantine as adjunctive therapy in children diagnosed with autistic spectrum disorders: An observation of initial clinical response and maintenance tolerability. J. Child Neurol. 2007, 22, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Erickson, C.A.; Posey, D.J.; Stigler, K.A.; Mullett, J.; Katschke, A.R.; McDougle, C.J. A retrospective study of memantine in children and adolescents with pervasive developmental disorders. Psychopharmacology 2007, 191, 141–147. [Google Scholar] [CrossRef]
- Berry-Kravis, E.M.; Hessl, D.; Rathmell, B.; Zarevics, P.; Cherubini, M.; Walton-Bowen, K.; Mu, Y.; Nguyen, D.V.; Gonza-lez-Heydrich, J.; Wang, P.P.; et al. Effects of STX209 (arbaclofen) on neurobehavioral function in children and adults with fragile X syndrome: A randomized, controlled, phase 2 trial. Sci. Transl. Med. 2012, 4, 152ra127. [Google Scholar] [CrossRef]
- Erickson, C.A.; Veenstra-Vanderweele, J.M.; Melmed, R.D.; McCracken, J.T.; Ginsberg, L.D.; Sikich, L.; Scahill, L.; Cherubini, M.; Zarevics, P.; Walton-Bowen, K.; et al. King BHSTX209 (arbaclofen) for autism spectrum disorders: An 8-week open-label study. J. Autism Dev. Disord. 2014, 44, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, J.L.; Mishaal, R.A.; Nabetani, M.; Goldman, R.D. Oxytocin for the Treatment of Autism Spectrum Disorder in Children. Can. Fam. Physician Med. Fam. Can. 2022, 68, 103–105. [Google Scholar] [CrossRef] [PubMed]
- Yenkoyan, K.; Ounanian, Z.; Mirumyan, M.; Hayrapetyan, L.; Zakaryan, N.; Sahakyan, R.; Bjørklund, G. Advances in the Treatment of Autism Spectrum Disorder: Current and Promising Strategies. Curr. Med. Chem. 2024, 31, 1485–1511. [Google Scholar] [CrossRef]
- Kurup, D.; FitzPatrick, A.M.; Badura, A.; Serra, I. Bridging the Gap: Neurodevelopmental Disorder Risks in Inborn Errors of Immunity. Curr. Opin. Allergy Clin. Immunol. 2024, 24, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Courchesne, E. Brainstem, Cerebellar and Limbic Neuroanatomical Abnormalities in Autism. Curr. Opin. Neurobiol. 1997, 7, 269–278. [Google Scholar] [CrossRef]
- Catani, M.; Dell’acqua, F.; Thiebaut de Schotten, M. A Revised Limbic System Model for Memory, Emotion and Behaviour. Neurosci. Biobehav. Rev. 2013, 37, 1724–1737. [Google Scholar] [CrossRef] [PubMed]
- Borrell, V.; Götz, M. Role of radial glial cells in cerebral cortex folding. Curr. Opin. Neurobiol. 2014, 27, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Li, Q. The Role of Parvalbumin Interneurons in Autism Spectrum Disorder. J. Neurosci. Res. 2024, 102, e25391. [Google Scholar] [CrossRef] [PubMed]
- Casanova, M.F.; Buxhoeveden, D.P.; Switala, A.E.; Roy, E. Minicolumnar pathology in autism. Neurology 2002, 58, 428–432. [Google Scholar] [CrossRef]
- Kim, Y.S.; Choi, J.; Yoon, B.-E. Neuron-Glia Interactions in Neurodevelopmental Disorders. Cells 2020, 9, 2176. [Google Scholar] [CrossRef] [PubMed]
- Gzielo, K.; Nikiforuk, A. Astroglia in Autism Spectrum Disorder. Int. J. Mol. Sci. 2021, 22, 11544. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).