Abstract
Interleukin 6 (IL-6) is an immunoregulatory cytokine involved in various inflammatory and immune responses. To investigate the effects of single nucleotide polymorphisms (SNPs) and haplotypes of IL-6 on resistance to Eimeria tenella (E. tenella), SNPs in the 5′ regulatory region of IL-6 were detected with direct sequencing, and the effects of SNPs and haplotypes on resistance to E. tenella were analyzed by the least square model in Jinghai yellow chickens. Nineteen SNPs were identified in the 5′ regulation region of IL-6, among which three SNPs were newly discovered. The SNP association analysis results showed that nine of the SNPs were significantly associated with E. tenella resistance indexes; the A-483G locus was significantly associated with the GSH-Px, IL-2, and IL-17 indexes (p < 0.05); the C-447G locus was significantly associated with the SOD, GSH-Px, IL-17, and IL-2 indexes (p < 0.05); and the G-357A locus had significant effects on the CAT and IL-16 indexes (p < 0.05). Haplotype analysis showed that H2H3 and H2H5 were favorable haplotype combinations with good coccidium resistance. Furthermore, we used qRT-PCR and observed that the expression of IL-6 in the infection group was higher than that in the control group in the liver, proventriculus, small intestine, thymus, kidney, and bursa of Fabricius and extremely significantly different than that in the cecum especially (p < 0.01). In summary, SNPs and haplotypes in the 5′ regulatory region of IL-6 have important effects on E. tenella resistance, and the results will provide a reference for molecular marker selection of E. tenella resistance in Jinghai yellow chickens.
1. Introduction
Coccidiosis is one of the intestinal parasitic diseases caused by the genus Eimeria, which seriously affects poultry production and animal welfare [1,2]. There are currently at least seven species of Eimeria found in chickens, among which the E. tenella parasite in the cecum is most common, mainly characterized by cecal epithelium hemorrhage and bloody stools [3,4]. So far, coccidiostats, live vaccines and strict hygiene control are the main measures to prevent coccidiosis in poultry production [5]. However, problems such as drug residues in food-producing animals, coccidiosis resistance, and vaccine safety still cannot be solved [6]. Therefore, studying the genes and SNPs associated with coccidia at the molecular level and breeding new strains resistant to coccidiosis are effective ways to solve this problem.
IL-6 is a pleiotropic cytokine that plays an important role in inflammation, the immune response, and hematopoiesis [7,8]. Produced mainly by macrophages, IL-6 performs protective functions in the healing of damaged tissues, participating in the acute phase immune response and coagulation in chickens [9]. Zhang and Zheng [10] reported that IL-6 triggers systemic inflammatory signals to initiate the host’s defense when the body is infected and damaged by pathogens. Rose-John et al. [11] discovered that the host’s defense against bacterial and fungal pathogen infection relied mainly on the classical IL-6 signaling pathway. By studying the effect of DNA plasmids carrying chicken IL-6 on infectious bursal disease virus (IBDV), Sun et al. [12] determined that injection with IL-6 plasmids resulted in a significantly increased protective effect in chickens. The RNA-seq technique was used to sequence the cecum tissue of an E. tenella-infected group and a noninfected group on the 7th day post-infection, and the results revealed that the differentially expressed genes (DEGs) included IL-6, IL-12β, and TGFB2, which were significantly enriched during coccidiosis infection [13]. In addition, Zhang et al. [14] showed that IL-1β, IL-6, and TNF-α play vital roles in the inflammatory response of chicken embryo fibroblasts infected with avian reovirus. However, there are relatively few studies on the association between single nucleotide polymorphisms (SNPs) of the chicken IL-6 gene and coccidium resistance. We previously performed a bioinformatics analysis of IL-6 gene SNPs in the 5′ promoter region of the Jinghai yellow chicken. In the present study, DNA direct sequencing technology was used to analyze the effect of SNPs and haplotypes in the 5′ regulatory region of IL-6 gene on E. tenella resistance indexes, and the results will provide basic data for molecular marker selection breeding of E. tenella resistance in the Jinghai yellow chicken.
2. Materials and Methods
2.1. Animals
A total of 220 (110 ♂, 110 ♀) 1-day-old healthy and physiologically similar Jinghai yellow chickens were randomly selected from the Jinghai Yellow Chicken Resource Farm (Haimen, China), housed in a sterile animal room, and fed an antibiotic-free diet until 30 days old. All the chickens were free from parasitic infection according to fecal detection during the pre-test period. Each chicken in the infected group (110 birds—5M + 55F) was given an oral infection with 2.5 × 104 sporulated oocysts, and chickens in the uninfected group (110 birds—55M + 55F), receiving no oocysts, were used as uninfected controls. Parasite oocysts were collected from the Department of Parasitology, College of Veterinary Medicine, Yangzhou University [15]. The experiments were carried out in strict accordance with the regulation of the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, Yangzhou, China, revised in June 2012) and approved by experimental animal use permit No. SYXK (Su) 2012-0029.
2.2. Genomic DNA Extraction
Blood samples were obtained from the wing vein of each bird on the 7th day post-infection, and the blood was anticoagulated by heparin sodium. After centrifugation, blood cells and plasma were stored at −20 °C. Genomic DNA was extracted and purified using the conventional phenol-chloroform extraction method. The concentration of the extracted DNA samples was measured by an Eppendorf BioPhotometer (Eppendorf Scientific, Hamburg, Germany), and quality was based on gel electrophoresis.
2.3. Resistance Index Detection
The activity of CAT, SOD, and GSH-PX and the concentrations of NO, MDA, IL-2, IL-16, IL-17, IFN-γ, and β-carotene in plasma on the 7th day post-infection were determined by ELISA according to the manufacturer’s instructions (Nanjing Jiancheng Bioengineering Research Institute, Nanjing, China).
2.4. Primer Design and Polymerase Chain Reaction (PCR) Conditions
Based on the GenBank (https://www.ncbi.nlm.nih.gov/genbank/) chicken genomic DNA sequence (accession number: NC_006089.4), four pairs of primers were designed using Primer Premier 5.0 (PREMIER Biosoft, Palo Alto, CA, USA). The amplified fragments were located between −2200 bp and −1 bp upstream of the IL-6 gene. The primers were synthesized by Sangon Biotech Co. (Shanghai, China). Primer sequences can be seen in Table 1.

Table 1.
Primer sequences for PCR amplification of the chicken IL-6 gene.
PCR was carried out in a reaction containing 1 μL of DNA template, 10 μL of 2× Taq Master Mix, 1 μL each of the upstream and downstream primers, and by adding ddH2O to 20 μL. The PCR amplification procedures were preliminary denaturation at 95 °C for 5 min; 35 cycles of denaturation at 95 °C for 30 s, annealing for 30 s at the optimum primer annealing temperature, and elongation for 1 min at 72 °C; and a final extension at 72 °C for 7 min. The samples were stored at 4 °C.
2.5. SNP Identification in 5′ Regulatory Region of the IL-6 Gene
The purified PCR products were sent to Sangon Biotech Co. (Shanghai, China) and sequenced using an ABI3730XL DNA analyzer (Applied Biosystems, Foster City, CA, USA). We compared the sequences with the reference on NCBI-BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to discover potential SNPs of the chicken IL-6 gene by DNAMAN 5.2 (Lynnon Corporation, San Ramon, CA, USA) and MEGA6.06.
2.6. Haplotype Analysis and Estimation of Linkage Disequilibrium (LD)
SHEsis analysis software (http://analysis.bio-x.cn/SHEsisMain.htm) was used to estimate the extent of LD between the identified SNPs, and an imbalance coefficient of D’ > 0.8 with a correlation coefficient of r2 > 0.33 is considered a strong LD between the SNPs [16]. Furthermore, we used PHASE2.1 to calculate the type of haplotype and its frequency.
2.7. Statistical Analysis
Popgene 1.32 software was employed to analyze the population genetic diversity of identified SNPs. The associations of the SNPs and haplotype with the E. tenella resistance index in Jinghai yellow chickens were analyzed by the least square model:
where Y is the measured value of the resistance index for each bird; μ is the overall mean; Sex is the effect of sex; Group is the effect of the infected and uninfected groups; Genotype is the genotype or haplotype combination effect; Group × Genotype is the interaction effects between different infection states and genotypes; and e is the random error effect.
Y = μ + Sex + Group + Genotype + (Group × Genotype) + e,
All statistical analyses were performed using the univariate model of the general linear model (GLM) in SPSS 25.0 statistical software, and multiple comparisons were performed using the least significant difference (LSD) method.
2.8. Quantitative Real-Time PCR (qRT-PCR)
We randomly selected ten chickens for qRT-PCR from each group of the infected and control. qRT-PCR primers for IL-6 were designed using Primer-BLAST on the NCBI website (F: TGGTGATAAATCCCGATGAAG, R: GGCACTGAAACTCCTGGTCT). GAPDH and β-actin were used as reference genes [17]. qRT-PCR was performed using an ABI Prism 7500 sequence-detection system (Applied Biosystems, Foster City, CA, USA) with a SYBR Green PCR Master Mix (TaKaRa, Dalian, China) according to the manufacturers’ instructions. The procedure was as follows: preliminary denaturation at 94 °C for 30 s, followed by 40 cycles of denaturation for 5 s at 95 °C and annealing for 34 s at 60 °C. The individual measurements were performed in triplicate, and the relative gene expression was calculated using the 2−ΔΔCt method.
3. Results
3.1. SNP Identification in IL-6
By sequencing and sequence alignment, a total of nineteen SNP sites were identified in the 5′ regulation region of IL-6 and three genotypes were detected in each site, among which three SNPs were newly discovered (C-447G, G-357A, A-663G) [18]. Based on the polymorphism information analysis, the G-865A, A-663G and A-400G loci were lowly polymorphic (PIC < 0.25), and sixteen SNP sites were moderately polymorphic (0.25 < PIC < 0.5). The χ2-test indicated that fourteen SNP sites were consistent with the Hardy–Weinberg equilibrium. The chromosome position and variation of all the identified SNPs are summarized in Table 2.

Table 2.
Information on single nucleotide polymorphisms (SNPs) identified in the IL-6 gene.
3.2. Association between SNPs and the E. tenella Resistance Index
Associations between SNPs of the IL-6 gene and E. tenella resistance indexes are presented in Table 3. We found that these nine SNPs were significantly associated with at least one coccidiosis resistance index. At the T-1534C locus, the MDA concentration of the CT genotype was significantly higher than that of the CC genotype (p < 0.01) and significantly higher than that of the TT genotype (p < 0.05), but there was no significant difference among the three genotypes for other resistance indexes (p > 0.05). The MDA concentration of CG genotypes at the C-939G locus was significantly higher than that of the CC genotype (p < 0.01) and significantly higher than that of the GG genotype (p < 0.05). At the G-889A locus, the IL-2 level of the GA genotype was significantly higher than that of the AA genotype (p < 0.01), and those of the GA genotype were also higher than those of the other two genotypes for the NO, CAT, IL-17, IFN-γ, and weight gain indexes, but the difference was not significant (p > 0.05).The IL-2 indexes of AG genotypes at A-634G locus were significantly higher than that of AA and GG genotypes (p < 0.05) but significantly lower than those of GG genotypes for GSH-Px indexes. At the C-511T locus, the NO concentration of the TT genotype was significantly higher than that of the CC genotype (p < 0.05). At the A-483G locus, the GSH-Px, IL-17, and IL-2 indexes of the GG genotype were significantly higher than those of the GA and AA genotype (p < 0.05), and those of the GG genotype were also higher than those of the other two genotypes for the NO, CAT, SOD, IL-16, and weight gain indexes, but the difference was not significant (p > 0.05). The IL-2 level of the GT genotype at T-458G locus was significantly higher than that of the TT genotype (p < 0.05) and significantly higher than that of the GG genotype (p < 0.01). The SOD, IL-17, GSH-Px, and IL-2 indexes of CC genotypes at the C-447G locus were significantly different from those of the other two genotypes (p < 0.05). At the G-357A locus, the CAT and IL-16 indexes of the AA genotype were significantly higher than those of the GG and GA genotypes (p < 0.05) but significantly lower than that of the GG and GA genotypes (p < 0.01) for the IL-17 level.

Table 3.
Associations of nine SNPs in the IL-6 gene with the E. tenella resistance indexes in chickens.
3.3. Association between Haplotypes and the E. tenella Resistance Indexes
SHEsis software LD analysis was employed to estimate the extent of LD between the nine identified SNPs; the results are presented in Figure 1. The deeper the red color is in the figure, the stronger the LD. D’ greater than 0.8 shows a distinct red color, and the correlation coefficient r2 is greater than 0.33, which indicates a strong LD. The results showed that there was a strong LD between the G-889A and T-458G loci and between the A-634G and A-483G loci, and the A-634G, A-483G and C-447G loci were also highly linked.
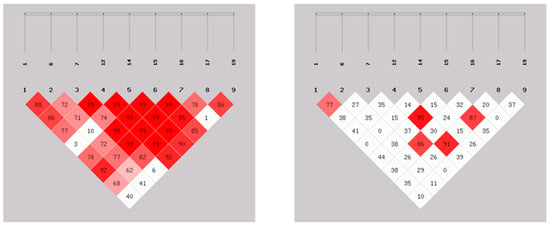
Figure 1.
D’ values (left) and r2 values (right) of pairwise LD analysis of the IL-6 gene. Note: the larger the value, the darker the color in boxes, the stronger the LD.
Moreover, haplotype analysis showed that ten haplotypes were formed by the nine SNPs, consisting of H1, H2, H3, H4, H5, etc. and that the frequency of the haplotypes was greater than 1% (Table 4). Haplotype-based association analysis showed that the resistance indexes for IL-17, GSH-Px, MDA, IL-2, IL-16, and weight gain of the H2H3 and H2H5 haplotype combinations were significantly (p < 0.05) or extremely significantly (p < 0.01) higher than those of the other haplotype combinations. IFN-γ and SOD indexes of the H2H3 and H2H5 genotypes were also higher than those of the other haplotypes, but the difference was not significant (p > 0.05). The information is presented in Table 5.

Table 4.
Haplotype analysis for the 5′ untranslated region (UTR).

Table 5.
Associations of haplotype with the E. tenella resistance indexes in chickens.
3.4. Expression of the IL-6 Gene in Nine Tissues
By using qRT-PCR, a comparison of IL-6 gene expression among different tissues was performed, and the results are presented in Figure 2. We found that the IL-6 gene was expressed in all nine tissues. The expression of IL-6 in the infection groups was higher than that in the control groups in the liver, proventriculus, small intestine, thymus, kidney, and bursa of Fabricius of Jinghai yellow chickens, and the expression level in the cecum was significantly higher in the infected group than in the control group (p < 0.01).
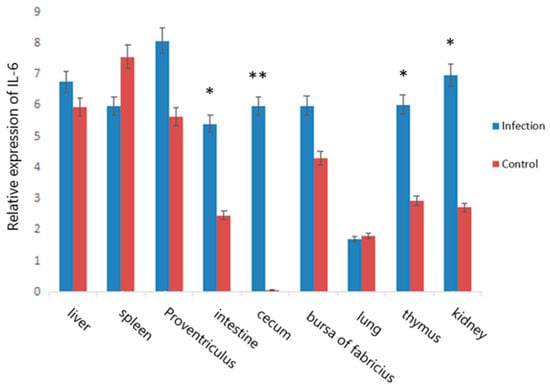
Figure 2.
Relative expression of the IL-6 gene in nine tissues of Jinghai yellow chickens. Note: “*” indicates a significant difference (p < 0.05), “**” indicates that the difference is extremely significant (p < 0.01).
4. Discussion
Research on the function and clinical application of the chicken IL-6 gene is still being conducted. Studies have shown that chicken IL-6 exhibits significant potential as an immune adjuvant, immune enhancer, and antibiotic substitute [19,20,21]. Rose-John et al. [11] discovered that the host’s defense against bacterial and fungal pathogen infection relied mainly on classical IL-6 signaling pathways. Swaggerty et al. [22] reported that high levels of IL-6, CXCLi2 (IL-8), and the chemokine CCLi2 expressed in broiler chickens resulted in significantly higher resistance than in those with low expression. These studies provide a basis for studying the relationship between IL-6 and coccidiosis infection. In this study, we used DNA direct sequencing technology to detect SNPs in the IL-6 gene 5′regulation region of the Jinghai yellow chicken and identified nineteen SNPs. Among them, three SNPs were newly discovered. All of the identified SNPs were lowly or moderately polymorphic, indicating that the genetic diversity among individuals within the population was good, which is conducive to subsequent breeding.
By bioinformatics analysis of the polymorphism of the promoter region of the IL-6 gene, Xin et al. [18] found that eleven SNPs are located in the core promoter region and may influence IL-6 gene expression by altering transcription factor binding or CpG methylation status. Pang et al. [23] analyzed the correlation between the SNPs in the IL-6 gene promoter region and the severity of enterovirus EV71 infection, indicating that IL-6 has a potential correlation with severe infection of human EV71. Zhang et al. [24] reported that SNPs in the MHC B-F and SPOCK1 genes are significantly associated with resistance to Salmonella pullorum. In this study, we analyzed the association between SNPs of the IL-6 gene in the regulation region and coccidium resistance indexes and found that nine SNPs were significantly associated with E. tenella resistance. The GG genotype of the A-483G locus was significantly higher than the GA and AA genotypes in the GSH-Px, IL-17, and IL-2 indexes. The NO, CAT, SOD, IL-16, and weight gain indexes of the GG genotype were also higher than those of the other two genotypes. Regarding the newly discovered SNPs, the CC genotype of the C-447G locus was significantly higher than the GG and CG genotypes in the SOD, GSH-Px, IL-17, and IL-2 indexes and higher than the GG and CG genotypes in the NO index. The AA genotype of the G-357A locus was significantly higher than the GG and GA genotypes in the CAT and IL-16 indexes and higher than the GG and GA genotypes in the SOD, MDA, β-carotene, and weight gain indexes. These resistance-related mutation sites may be involved in the regulation of IL-6 gene expression and affect its immune function during Eimeria infection.
SNPs on the same chromosome do not exist alone, and adjacent-allele SNPs may work simultaneously, indicating LD [25]. The use of haplotypes in association studies to identify common variants may be more effective than single allele studies [26,27]. Most traits are regulated by multiple genes or multiple sites of a certain gene, and it is impossible to accurately determine the true correlation between genes and traits by analyzing the polymorphism of a single locus in a certain gene. In this study, the haplotype combination association analysis results showed that the H2H3 and H2H5 haplotype combinations were significantly higher than other haplotype combinations in the resistance indexes for IL-17, GSH-Px, MDA, IL-2, IL-16, and weight gain. Thus, the H2H3 and H2H5 haplotype combinations are the dominant haplotype combinations of chickens against Eimeria infection.
Furthermore, we used qRT-PCR to reveal that the IL-6 gene was expressed in all nine tissues. The expression of IL-6 in the infection groups was higher than that in the control groups in the liver, proventriculus, small intestine, thymus, kidney, and bursa of Fabricius and significantly higher than that in the control group in the cecum. E. tenella is one of the most pathogenic species that exclusively occupies the cecum, causing significant swelling and bleeding of the cecum [28,29]. The Eimeria species infect cecum epithelial cells, with the potential to fully occupy the infection sites without appropriate control measurements [30]. Therefore, the study data suggest that IL-6 may play an important role in E. tenella resistance in Jinghai yellow chickens.
5. Conclusions
In this study, nineteen SNPs were identified in the 5′ regulation region of IL-6, among which nine of the SNPs were significantly associated with E. tenella resistance indexes, The A-483G locus were significantly associated with GSH-Px, IL-2, and IL-17 indexes; the C-447G locus were significantly associated with SOD, GSH-Px, IL-17, and IL-2 indexes; the G-357A locus had significant effects on the CAT and IL-16 indexes. Haplotype analysis showed that H2H3 and H2H5 were the favorable haplotype combinations with good coccidia resistance. Our results will provide a reference for molecular marker selection of E. tenella resistance in Jinghai Yellow Chicken.
Author Contributions
G.D.; Data curation, H.Y., G.D. and X.W.; Formal analysis, H.Y., G.D. and X.W.; Funding acquisition, G.D.; Investigation, H.Y., X.W., W.Z., S.X. and C.M.; Methodology, H.Y., G.D. and X.W.; Project administration, G.D.; Resources, C.Q.; Supervision, G.D., T.Z., G.Z. and K.X.; Validation, H.Y., W.Z., X.W. and S.X.; Visualization, H.Y., W.Z., S.X., X.W. and C.M.; Writing original draft, H.Y.; Writing, review & editing, H.Y., G.D., T.Z., G.Z., K.X. and J.W.
Funding
This research was supported by the Jiangsu Agricultural Industry Technology System (JATS[2018]303), the National Sci-Tech Support Plan (2014BAD13B02), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the China Agriculture Research System (CARS-41-G23).
Acknowledgments
The authors would like to thank all members of this work for their advice and technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shirley, M.; Lillehoj, H. The long view: A selective review of 40 years of coccidiosis research. Avian Pathol. 2012, 41, 111–121. [Google Scholar] [CrossRef]
- Li, C.; Yan, X.; Lillehoj, H.S.; Oh, S.; Liu, L.; Sun, Z.; Gu, C.; Lee, Y.; Xianyu, Z.; Zhao, H. Eimeria maxima-induced transcriptional changes in the cecal mucosa of broiler chickens. Parasites Vectors 2019, 12, 285. [Google Scholar] [CrossRef] [PubMed]
- Clark, E.L.; Macdonald, S.E.; Thenmozhi, V.; Kundu, K.; Garg, R.; Kumar, S.; Ayoade, S.; Fornace, K.M.; Jatau, I.D.; Moftah, A. Cryptic Eimeria genotypes are common across the southern but not northern hemisphere. Int. J. Parasitol. 2016, 46, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Noack, S.; Chapman, H.D.; Selzer, P.M. Anticoccidial drugs of the livestock industry. Parasitol. Res. 2019, 118, 2009–2026. [Google Scholar] [CrossRef] [PubMed]
- Chapman, H. A landmark contribution to poultry science—Prophylactic control of coccidiosis in poultry. Poult. Sci. 2009, 88, 813–815. [Google Scholar] [CrossRef]
- Wang, X.; Zou, W.; Yu, H.; Lin, Y.; Dai, G.; Zhang, T.; Zhang, G.; Xie, K.; Wang, J.; Shi, H. RNA sequencing analysis of chicken cecum tissues following Eimeria tenella infection in vivo. Genes 2019, 10, 420–432. [Google Scholar] [CrossRef]
- Neurath, M.F.; Finotto, S. IL-6 signaling in autoimmunity, chronic inflammation and inflammation-associated cancer. Cytokine Growth Factor Rev. 2011, 22, 83–89. [Google Scholar] [CrossRef]
- Nishimichi, N.; Kawashima, T.; Hojyo, S.; Horiuchi, H.; Furusawa, S.; Matsuda, H. Characterization and expression analysis of a chicken interleukin-6 receptor α. Dev. Comp. Immunol. 2006, 30, 419–429. [Google Scholar] [CrossRef]
- Schett, G.; Elewaut, D.; McInnes, I.B.; Dayer, J.M.; Neurath, M.F. How cytokine networks fuel inflammation: Toward a cytokine-based disease taxonomy. Nat. Med. 2013, 19, 822–824. [Google Scholar] [CrossRef]
- Zhang, W.J.; Zheng, H. Advances in IL-6-mediated immune inflammatory response and its relationship with disease. J. Cell. Mol. Immunol. 2017, 33, 699–703. [Google Scholar]
- Rose-John, S.; Winthrop, K.; Calabrese, L. The role of IL-6 in host defence against infections: Immunobiology and clinical implications. Nat. Rev. Rheumatol. 2017, 13, 399–409. [Google Scholar] [CrossRef]
- Sun, J.H.; Yan, Y.X.; Jiang, J.; Lu, P. DNA immunization against very virulent infectious bursal disease virus with VP2-4-3 gene and chicken IL-6 gene. J. Vet. Med. Ser. B 2005, 52, 1–7. [Google Scholar] [CrossRef]
- Lin, Y.X. RNA Sequencing Analysis of Chicken Cecum Tissues Following E. Tenella Infection and Coccidiosis Evaluation of Jinghai Yellow Chicken Cross-Breeding System Parents. Master’s Thesis, Yangzhou University, Yangzhou, China, 2015. [Google Scholar]
- Zhang, K.L.; Xie, Z.X.; Huang, L.; Xie, L.J.; Liu, J.B.; Deng, X.W.; Xie, Z.Q.; Fan, Q.; Luo, S.S. Dynamic changes of IL-1β, IL-6 and TNF-α mRNA levels in chicken embryo fibroblast infected with avian reovirus. Chin. Vet. Sci. 2014, 44, 805–810. [Google Scholar]
- Jiao, J.; Yang, Y.; Liu, M.; Li, J.; Cui, Y.; Yin, S.; Tao, J. Artemisinin and Artemisia annua leaves alleviate Eimeria tenella infection by facilitating apoptosis of host cells and suppressing inflammatory response. Vet. Parasitol. 2018, 254, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Guryev, V.; Smits, B.M.; Van de Belt, J.; Verheul, M.; Hubner, N.; Cuppen, E. Haplotype block structure is conserved across mammals. PLoS Genet. 2006, 2, e121. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q. Cloning and Sequence Analysis of Avian Toll-Like Receptors Genes and the Differences of Innate Immunity in Different Chicken Lines Against Salmonella Infection. Master’s Thesis, Yangzhou University, Yangzhou, China, 2012. [Google Scholar]
- Xin, S.; Wang, X.; Dai, G.; Zhang, J.; An, T.; Zou, W.; Zhang, G.; Xie, K.; Wang, J. Bioinformatics analysis of SNPs in IL-6 gene promoter of Jinghai yellow chickens. Genes 2018, 9, 446. [Google Scholar] [CrossRef]
- Hui, G.; Hashimoto, C. Interleukin-6 has differential influence on the ability of adjuvant formulations to potentiate antibody reponses to a Plasmodium falciparum blood-stage vaccine. Vaccine 2007, 25, 6598–6603. [Google Scholar] [CrossRef]
- Zhao, N.; Li, Z.C.; Zhang, X.R.; Wu, Y.T. Research progress of chicken interleukin-6. Shanghai J. Anim. Husb. Vet. Med. 2013, 1, 16–17. [Google Scholar] [CrossRef]
- Su, B.; Wang, J.; Wang, X.; Jin, H.; Zhao, G.; Ding, Z.; Kang, Y.; Wang, B. The effects of IL-6 and TNF-α as molecular adjuvants on immune responses to FMDV and maturation of dendritic cells by DNA vaccination. Vaccine 2008, 26, 5111–5122. [Google Scholar] [CrossRef]
- Swaggerty, C.; Pevzner, I.; Kogut, M. Selection for pro-inflammatory mediators produces chickens more resistant to Eimeria tenella. Poult. Sci. 2015, 94, 37–42. [Google Scholar] [CrossRef]
- Pang, L.L.; Zhang, S.X.; Zhang, Q.; Gong, X.; Xiang-Yu, K.; Gao, W.J.; Li, X.L.; Jin, Y.; Duan, Z.J. Association of IL-6-572G/C polymorphism with severity of enterovirus 71 infection. Chin. J. Biol. Prod. 2016, 29, 620–622. [Google Scholar]
- Zhang, Z.T.; Gan, J.K.; Zhang, W.W.; Zhang, D.X.; Zhang, X.Q.; Luo, Q.B. The SNPs C.513A> T in the MHC BF gene and rs15001532 in the SPOCK1 gene are associated with Salmonella pullorum disease resistance in chickens. J. Integr. Agric. 2016, 15, 1856–1862. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, T.; Zhang, G.; Wang, J.; Fan, Q.; Chen, X.; Wei, Y.; Han, K.; Wang, Y. Eight SNPs of the Myf5 gene and diplotypes associated with growth and reproductive traits in Jinghai yellow chicken. Mol. Biol. Rep. 2014, 41, 6837–6844. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.C.L.; Esposito, L.; Barratt, B.J.; Smith, A.N.; Heward, J.; Di Genova, G.; Ueda, H.; Cordell, H.J.; Eaves, I.A.; Dudbridge, F. Haplotype tagging for the identification of common disease genes. Nat. Genet. 2001, 29, 233–237. [Google Scholar] [CrossRef]
- Bonnen, P.E.; Wang, P.J.; Kimmel, M.; Chakraborty, R.; Nelson, D.L. Haplotype and linkage disequilibrium architecture for human cancer-associated genes. Genome Res. 2002, 12, 1846–1853. [Google Scholar] [CrossRef]
- Hammond, D.M.; Sampson, J.R. Fine structural aspects of development of Eimeria alabamensis schizonts in cell cultures. J. Parasitol. 1972, 58, 311–322. [Google Scholar] [CrossRef]
- Lopez-Bernad, F.; Del Cacho, E.; Gallego, M.; Quílez, J.; Sánchez-Acedo, C. Immunohistochemical identification of the cells parasitized by second-generation schizonts of Eimeria tenella. Parasitol. Res. 1997, 84, 132–135. [Google Scholar] [CrossRef]
- Guo, A.J.; Cai, J.; Gong, W.; Yan, H.; Luo, X.; Tian, G.; Zhang, S.; Zhang, H.; Zhu, G.; Cai, X. Transcriptome analysis in chicken cecal epithelia upon infection by Eimeria tenella in vivo. PLoS ONE 2013, 8, e64236. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).