Abstract
There is growing evidence highlighting the importance of monoubiquitination as part of the histone code. Monoubiquitination, the covalent attachment of a single ubiquitin molecule at specific lysines of histone tails, has been associated with transcriptional elongation and the DNA damage response. Sites function as scaffolds or docking platforms for proteins involved in transcription or DNA repair; however, not all sites are equal, with some sites resulting in actively transcribed chromatin and others associated with gene silencing. All events are written by E3 ubiquitin ligases, predominantly of the RING (really interesting new gene) finger type. One of the most well-studied events is monoubiquitination of histone H2B at lysine 120 (H2Bub1), written predominantly by the RING finger complex RNF20-RNF40 and generally associated with active transcription. Monoubiquitination of histone H2A at lysine 119 (H2AK119ub1) is also well-studied, its E3 ubiquitin ligase constituting part of the Polycomb Repressor Complex 1 (PRC1), RING1B-BMI1, associated with transcriptional silencing. Both modifications are activated as part of the DNA damage response. Histone monoubiquitination is a key epigenomic event shaping the chromatin landscape of malignancy and influencing how cells respond to DNA damage. This review discusses a number of these sites and the E3 RING finger ubiquitin ligases that write them.
Keywords:
E3 ligase; RING finger; monoubiquitination; histone; chromatin; RNF20; RNF40; polycomb repressor complex 1; RING1B; BRCA1 1. Introduction
Histones are basic proteins, with histone H2A, H2B, H3 and H4 constituting the core structure of the nucleosome as an octamer complex around which 147 bp of DNA wrap. An H1 linker protein is associated via DNA entry and exit sites [1,2,3]. N-terminal tails of histones in the nucleosome protrude from this structure and undergo a range of covalent modifications including acetylation, methylation, SUMOylation, phosphorylation, protein isomerisation and ubiquitination [3,4,5]. These structures comprise chromatin and are important for condensing or packaging DNA into higher order chromatin structures. Histone post-translational modifications (PTMs) act as docking sites or scaffolds for proteins that work together to direct cellular transcriptional programmes and the response to DNA damage [6]. Emphasising the importance of these fundamental roles, histones are encoded by multiple genes, including as parts of gene clusters [7,8].
The PTM of ubiquitination can take the form of mono-, i.e., covalent attachment of a single ubiquitin to specific lysines for purposes such as gene regulation, or polyubiquitination, where chains of end-to-end ubiquitin molecules form at lysines to mark the protein for degradation via the 26S proteasome requiring ATP hydrolysis [9]. Monoubiquitination is a major cellular event, in yeast shown to account for over half of all conjugated ubiquitin [10]. The most abundantly monoubiquitinated proteins within the nucleus of mammalian cells are histones [11,12]. At 8.5 kDa (76 residues), monoubiquitination of specific lysines on histone tails represents one of the larger histone PTMs relative to modifications such as methylation and acetylation [3,13]. As for polyubiquitination, the ubiquitin enzyme cascade required for monoubiquitination requires three key enzymes [14,15]. In an ATP-dependent reaction, the ubiquitin activating enzyme (E1) forms a thiol-ester bond between its active cysteine site and the carboxy-terminal glycine of ubiquitin. Ubiquitin is then transferred to a cysteine residue of a ubiquitin-conjugating enzyme (E2). Ubiquitin ligases (E3) are critical for substrate recognition, recruiting the E2 ubiquitin conjugate and catalysing substrate ubiquitination [15,16,17,18]. E2 enzymes can interact with multiple E3 enzymes, and, conversely, E3s can interact with different E2s, leading to different ubiquitin linkages and functional outcomes [19,20].
Humans have very few E1s, around 40 E2s and as many as 600–1000 E3 enzymes important for ensuring substrate recognition [14,15,21,22]. E3 ligases are classified as either HECT (homologous to E6-AP carboxy terminus) domain or RING (really interesting new gene) domain ligases, the latter being by far the more prevalent [18]. The RING domain is zinc finger domain-like, with 40–60 residues of a motif resembling Cys-X2-Cys-X9-39-Cys-X1-3-His-X2-3-Cys-X2-Cys-X4-48-Cys-X2-Cys (X represents any residue and His and Cys can be exchanged) [18,22,23]. It is this domain that provides a docking surface for the E2-ubiquitin conjugate [22].
RING finger ubiquitin protein ligases have roles in both the maintenance of the healthy cell and the oncogenic transformation, including genomic integrity and DNA damage and repair responses [15]. Numerous well-known proteins associated with malignancy function as RING finger E3 ligases, including tumour suppressors BRCA1 [24,25] and the FANC core complex [26], as well as the oncogene MDM2 [27,28]. RING finger E3 ligases also have a specific role in writing histone monoubiquitination, in this way having major influence on the chromatin landscape.
Sites of monoubiquitination of specific lysines on core histone tails will be discussed in this review. These include lysines 34 (K34) [29], 120 (K120, H2Bub1) and 125 (K125) on histone H2B [30,31,32,33,34]; lysines 13 (K13), 15 (K15), 119 (K119, H2AK119ub1), 127 (K127) and 129 (K129) on histone H2A [11,35,36,37,38] and lysine 31 (K31) on histone H4 [39,40] (Figure 1). Given the high levels of monoubiquitination of histone H2A and H2B in the cell, respectively 5–15% and 1-1.5/2%, the discussion of H2AK119ub1 and H2Bub1 will form a larger part of this review [12,41,42]. Core histone H3, the linker histone H1 and H2AX have also been reported to be modified by ubiquitin (reviewed in [42]). While little is known about the effect of some of these ubiquitination events, others have been shown to have opposing effects in the context of their association with either active or silent chromatin, impacting on gene transcription [31,43,44]. This fascinating dichotomy possibly relates to positioning within the nucleosome structure. These ubiquitinated histone sites and the RING finger E3 ligases that write them, including Ring Finger Protein 20 (RNF20), RNF40, RING 1A, RING 1B, MSL2, RNF8, RNF168 and Cullin 4A (CUL4A), are the focus of this review (Table 1).
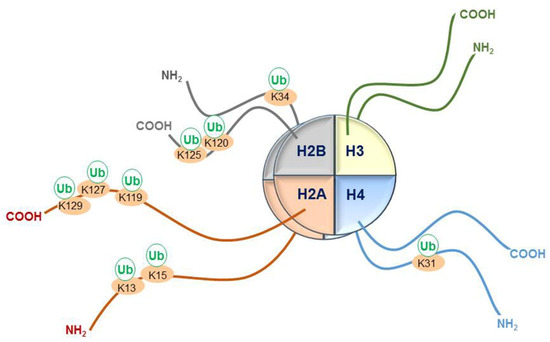
Figure 1.
Schematic representation of sites of monoubiquitination at lysines (K) on histone tails protruding from the core octamer structure of nucleosomes. Monoubiquitination at K34 and K120 on histone H2B, as well as at K31 on histone H4, has been associated with transcriptional elongation. Monoubiquitination of K119 on histone H2A is written by an E3 ligase in polycomb repressor complex 1 and is associated with inactive chromatin. K13 and K15 on the histone H2A tail are written by E3 ligases associated with DNA damage. K127 and K129 on histone tail H2A are ubiquitinated by E3 ligase activity of the Breast Cancer 1 (BRCA1)- BRCA1-Associated RING Domain Protein 1 (BARD1) complex. It is unclear whether K125 is monoubiquitinated in vivo. Monoubiquitination at K31 on histone H4, as well as at K34 and K120 on histone H2A, participates in histone crosstalk to promote the methylation of K4 and K79 on histone H3. Ub: Ubiquitin.

Table 1.
Sites of histone ubiquitination and the E3 ligases responsible for writing them.
2. Histone H2B Monoubiquitination and the E3 RING Finger Ligase Complex RNF20-RNF40
Histone H2B monoubiquitination was first discovered in yeast, occurring on lysine 123 and carried out by the E3 RING finger ubiquitin ligase Bre1 [45,46]. In 1980, histone H2B was shown to be ubiquitinated in mouse and man [12]. In both yeast and humans, RAD6A is the E2-conjugating enzyme, also referred to as the ubiquitin-conjugating enzyme 2A (UBE2A) [47]. A second E2, UBE2E1 (also known as UBCH6), for H2Bub1 has been reported [48,49]. In humans, H2Bub1 occurs at lysine 120 of histone H2B and functions to alter chromatin compaction, leading to open fibres that are more readily accessible to transcription factors and DNA repair proteins [50]. Lysine 120 of histone H2B resides at the interface of adjacent nucleosomes, with the addition of the bulky ubiquitin molecule likely interfering with nucleosome stacking and therefore chromatin configuration. This effect, however, is more than purely positional as the addition of the even bulkier modification of SUMO to this site does not behave in the same manner [50,51].
A number of different E3 ubiquitin ligases have been implicated as being able to monoubiquitinate histone H2B. These include MDM2 [34], the BRCA1-BARD1 complex [52,53] and BAF250B [54]. It is generally accepted, however, that the RNF20-RNF40, an orthologue of yeast Bre1, is the main RING finger E3 ligase complex capable of writing H2Bub1 in humans [30,32,33,47,48,55]. RNF20 and RNF40 form a heterotetrameric complex with two copies of each polypeptide [48]. While the key functional unit of this complex has been reported as RNF20 [56], it would appear that both subunits are likely important to the stability and function of this complex given that downregulation of either RNF20 or RNF40 leads to depletion of the other as well as to depletion of H2Bub1 [55,57,58]. Furthermore, overexpression of RNF20 has been shown to increase levels of H2Bub1 and subsequently histone methylation at lysines 4 and 79 on histone H3 as well as to stimulate the expression of HOX genes, a group of homeotic genes that are master controllers of embryonic development [48,56].
The importance of histone crosstalk involving this modification was reported in yeast, whereby monoubiquitination of yeast histone H2B K123 was shown to be required for methylation of histone H3K4 [59,60]. In humans, H2Bub1 is a central modification, able to recruit methyltransferase complexes such as COMPASS and DOTL1, leading to H3K4 di- and tri-methylation, as well as H3K79 tri-methylation (reviewed in [30]). This influence on histone methyltransferase events is not unique to H2Bub1, as discussed later in this review. It is important to note the dynamic nature of monoubiquitination, and while they will not be discussed further in this review, numerous deubiquitinases (DUBs), including USP7 and USP44, have been reported to erase H2Bub1 (reviewed in [30,32,33]).
2.1. RNF20-RNF40 and H2Bub1 in Transcriptional Elongation and DNA Damage
The interaction between RNF20-RNF40 and the PAF1 (RNA polymerase II-associated factor 1) complex is thought to be responsible for the increase in H2Bub1 at the coding regions of transcriptionally active genes [47]. The PAF1 complex is comprised of multiple subunits, a number of which, including the tumour suppressor CDC73, have been shown to interact with RNF20-RNF40 [30,57,61]. When the cell receives a signal to activate gene expression, cyclin-dependent kinase 9 (CDK9) phosphorylates both the H2Bub1 E2-conjugating enzyme UBE2A and Ser2 of the carboxy-terminal domain of RNA polymerase II. This creates a binding pocket for WAC (WW domain containing adaptor with coiled-coil) that associates with RNF20-RNF40 to enable H2Bub1 [62]. The RNF20-RNF40 association with the PAF1 complex that couples with RNA polymerase II at chromatin promotes transcriptional elongation (reviewed in [30]). The chromatin remodelling factor FACT is recruited, enabling removal of the H2B-H2A core histone dimer from the nucleosome structure, eliminating the physical block to RNA polymerase II and allowing transcriptional elongation to proceed (reviewed in [30]) [49,63]. Perhaps curiously, however, down-regulation of RNF20 or RNF40 does not affect the transcription of the majority of genes [64,65]. This introduces the concept that these E3 ligases may function as ‘selective’ tumour suppressors. It is possible that the location of genes throughout the genome plays a part in this apparent anomaly. One key gene that RNF20 depletion does reduce is the tumour suppressor p53 [64].
In response to DNA damage, the ATM (ataxia telangiectasia mutated) kinase phosphorylates RNF20 and RNF40 that are then directed to sites of double strand breaks (DSBs) where they function to monoubiquitinate histone H2B at lysine 120, facilitating open chromatin accessible by DNA repair proteins [55]. As part of this process, repair proteins, including those involved in homologous recombination (RAD51, BRCA1 and BRCA2) as well as those involved in non-homologous recombination (XRCC4 and Ku80), are recruited [55,66,67]. RNF20 is key in these processes, recruiting the chromatin remodelling factor SNF2h and establishing the importance of the RNF20 RING finger E3 ligase in shaping the chromatin landscape [67]. Furthermore, RNF20 has been shown to interact with p53, the guardian of the genome, associating with p53 at the promoters of p53 target genes [56]. Interestingly, a recent study has reported that the known apoptotic mark serine 14 on histone H2B is phosphorylated (H2BS14p) in response to DSBs induced by ionising radiation in an ATM-dependent manner, marking transcriptionally silent nucleolar chromatin [68].
2.2. RNF20, RNF40 and H2Bub1 in Cancer
Immunohistochemical assessment of overall, or global, levels of H2Bub1 has shown loss of H2Bub1 in a range of primary tumours, including breast, colon, lung, ovarian, parathyroid, gastric and testicular cancer [58,65,69,70,71,72,73,74,75]. Retention of H2Bub1 in non-cancer cells has been shown for a number of tissue types, including normal colonic mucosa and normal breast tissue adjacent to tumours [65,73]. In some tumour types, such as breast, loss of global levels of H2Bub1 has been associated with advancing tumour progression; however, this is not true for all malignancies, with H2Bub1 loss observed in both early and late stages of high-grade serous ovarian cancer [58,65]. Global loss of H2Bub1 has also been correlated with poor patient survival in colorectal cancer [73]. Loss of H2Bub1 has been associated with mutation of the tumour suppressor CDC73, a member of the PAF1 complex, in parathyroid tumours [57]. The clear mechanism underpinning this loss across multiple tumour types remains to be elucidated.
A small number of studies have assessed RNF20 in primary tumours. In a study of 424 high-grade serous ovarian cancers, loss of RNF20 was only seen in ~6% of tumours and, perhaps surprisingly, did not correlate with loss of H2Bub1 [58]. In apparent contrast, RNF20 and RNF40 mRNA levels were reduced in colons from patients with ulcerative colitis [71]. Further in this study, RNF20 and RNF40 levels were inversely correlated with mRNA levels of IL6 and IL8 inflammatory cytokines, causing the authors to speculate that constitutively reduced levels of RNF20 and RNF40 leading to reduced levels of H2Bub1 may increase the risk of developing certain chronic inflammatory diseases, at least those associated with the colon [71]. It would be interesting to see whether decreases at the gene expression level are recapitulated at the protein level for RNF20 and RNF40 in colorectal tissue associated with inflammatory conditions. Lastly, in a study of clear cell renal cell carcinoma, loss of RNF20 detected in primary tumours by immunohistochemistry was a marker of poor prognosis [76].
2.3. RNF20, RNF40 and H2Bub1 Display Both Tumour-Suppressive and Oncogenic Functions
The RNF20 promoter has been reported to be hypermethylated in primary breast cancer cells [64,77] and mutated at low frequency in colorectal cancer [78,79]. RNF20 mRNA levels are also reduced in metastatic prostate cancer cells compared to benign disease and are lower in testicular germ cell cancer seminoma compared to normal testis [75,80]. Genomic loss of RNF20 has been reported in pre-invasive dysplastic airway lesions [81]. It will be interesting to observe whether these genetic and epigenetic changes impact upon the protein levels and function of RNF20 and RNF40. Furthermore, overexpression of RNF20 in renal cell cancer cell lines led to a decrease in proliferation, while suppression of RNF20 led to increased proliferation [76]. Another way that RNF20 may function as a tumour suppressor is by stopping recruitment of the transcription elongation factor TFIIS to chromatin. TFIIS works by releasing stalled RNA polymerase II, and its inhibition works to impede the expression of oncogenes such as MYC and FOS that normally reside in regions of compacted chromatin [77].
While most of the literature presented thus far would suggest that H2Bub1 and its E3 ligases function as tumour suppressors, there are a number of studies that suggest that higher levels and/or activity of these factors may actually promote tumorigenesis, i.e., have an oncogenic function (reviewed in [82]). Down-regulation of RNF20 has been shown to lead to the migration of MCF10A breast epithelial cells, as well as anchorage-independent growth of NIH3T3 cells [64]. In an opposite fashion, upregulation of RNF20 leading to increased Hox gene expression may also contribute to a malignant phenotype, and knockdown of RNF20 in the breast cancer cell line MCF7 led to reduced proliferation [48,83]. Furthermore, a recent study of different subtypes of breast cancer has shown that whether RNF20 and H2Bub1 inhibit or enhance cellular proliferation and migration is entirely dependent on the subtype. Here, silencing of RNF20 led to increased proliferation and migration in basal-like breast tumours, likely via upregulation of inflammatory cytokines, while silencing of RNF20 in luminal breast cancer cells decreased proliferation and migration, compromising transcription of the estrogen receptor [84]. In another apparent contrast, loss of RNF20 has been linked with an inflammatory phenotype in colorectal cancer [71], while loss of RNF40 would appear to have a protective effect against the development of an inflammatory phenotype in the same cancer [85], both involving NF-κB signalling. Fundamentally, whether RNF20, RNF40 and H2Bub1 predominantly drive or inhibit proliferative or inhibitory phenotypes via remodelling of the chromatin landscape might be influenced by models chosen for study and/or be cell-type- and disease-specific, likely underpinned by specific transcriptional activity. Furthermore, the effect of modulation of these proteins in the context of DNA damage and repair is currently unclear.
2.4. Non-Histone Substrates of RNF20 and RNF40
While there is a large and growing body of literature investigating histone H2B lysine 120 as a substrate of the RNF20-RNF40 complex, other substrates of these E3 ligases have also been reported. At present, it cannot be excluded that some tumour-suppressive or oncogenic functions of RNF20 and RNF40 might occur via non-histone substrates. RNF20-RNF40 is known to monoubiquitinate the motor protein Eg5 that has roles in spindle assembly during mitosis [86]. RNF20 has also been reported to polyubiquitinate the ErbB3 receptor binding protein Ebp1 [83]. Staring, the rat orthologue of RNF40, has been shown to polyubiquitinate Syntaxin 1, which is part of the neurotransmitter release machinery with links to learning and memory behaviour, facilitating its degradation via the ubiquitin proteasome [87]. It is currently unclear whether RNF20 and RNF40 may partner with different E2 enzymes for the purpose of ubiquitinating these non-histone substrates.
3. Histone H2A Monoubiquitination
The first protein identified over 40 years ago to be ubiquitinated was histone H2A [88]. Along with histone H2B, histone H2A is one of the most abundantly ubiquitinated nuclear proteins, with 5–15% of histone H2A being monoubiquitinated in human cells [11]. There is more than one site that undergoes monoubiquitination on histone H2A. Unlike H2Bub1, H2AK119ub1 does not impact on fibre compaction, likely related to its nucleosomal position [89]. Contrasting with H2Bub1, H2AK119ub1 is associated with gene silencing [43,44]. The mechanism underpinning the repression of gene expression is the prevention of FACT recruitment that otherwise relieves nucleosome barriers to RNA polymerase II-related transcriptional elongation [49,90]. Furthermore, H2AK119ub1 inhibits the transcriptionally active methylation marks H3K4me2 and me3 [91]. High levels of expression of H2AK119ub1 have been associated with a worse prognosis in some tumours [92].
3.1. H2AK119ub1 is Written by the Polycomb Repressive Complex 1
H2AK119ub1 is predominantly written by RING finger E3 ligases as part of the polycomb group (PcG) proteins, specifically polycomb repressive complex 1 (PRC1) [35,36,93,94,95]. Polycomb repressive complex 2 (PRC2) is comprised of four main components—EZH1/2, SUZ12, EED and RbAp46/48 (also referred to as RBBP7/4). It is through the methyltransferase activity of EZH1 and EZH2 (enhancer of zeste 1/2 polycomb repressive complex 2 subunit) that histone H3 at K27 is di- and tri-methylated (H3K27me2/3) [36]. PcG proteins work with their antagonists in the trithorax family during embryonic development and in adult cells to maintain the expression of key developmental genes important for the regulation of cell fate [96].
The RING domain ligases constituting PRC1 include RING1B (also known as Ring2/Rnf2), RING1A and BMI1 [44,94,95]. RING1B appears to be the dominant RING finger E3 ligase [44]. Through this mechanism of monoubiquitination, the PRC1 is involved in X chromosome inactivation [97], Hox gene silencing [95] and polycomb target gene silencing [94]. Loss of RING1A, RING1B or BMI1 leads to a global reduction of H2AK119ub1, cementing the importance of these PRC1 subunits as major E3 ligases for this histone modification [42].
RING1B associates with other repressive complexes, including E2F-6.com-1, which is involved in the silencing of E2F and MYC responsive genes in quiescent cells and the FBXL10-BcoR complex that likely also monoubiquitinates histone H2A at lysine 119 (reviewed in [44]). Monoubiquitination of the histone variant H2A.Z at K120 or K121 also involves RING1B as the E3 ligase [98]. Like many E3 ligases, RING1B has more than one substrate and has been shown to ubiquitinate the tumour suppressor p53, marking it for degradation [99]. The Cullin4B-E3 ligase complex, CRLB4, is physically associated with PRC2 and has been shown to catalyse H2AK119ub1 [100,101]. The RING finger domain E3 ligase TRIM37 has also been shown to catalyse H2AK119ub1, again interacting with the PRC2 complex [102]. Interestingly, the E3 ligase MDM2 interacts with the PRC2 complex, enhancing H2AK119ub1 [103]. Also able to monoubiquitinate histone H2A is the HECT domain E3 ligase LASU1 [104].
3.2. RING1B in Malignancy
RING1B is highly expressed in a number of human malignancies including prostate cancer, pancreatic ductal adenocarcinoma, ovarian cancer and urothelial bladder carcinoma [92,105,106,107]. Overexpression of RING1B has been associated with a poorer prognosis for women with ovarian cancer [105] and shorter survival times for patients with urothelial bladder carcinoma [106].
3.3. H2AK119ub1 is Specifically Written by 2A-HUB to a Subset of Chemokine Genes
2A-HUB/hRUL138 is an alternative RING domain E3 ligase recruited by the N-CoR/HDAC1/3 complex to the promoters of some chemokine genes to monoubiquitinate histone H2A at lysine 119 and repress expression of these chemokines [90]. Unlike the case of PRC1, this does not appear to be a global effect; rather, it is specific to this subset of genes that have specific roles in ensuring an immune response to inflammatory stimuli [90].
3.4. Histones H2A, H2AX and the Response to DNA Damage
As for H2Bub1, histone H2A is monoubiquitinated at sites of DNA damage. RING1B-BMI1 is recruited to sites of DSBs where it catalyses H2A and H2AX monoubiquitination at lysine 119 [42,108,109]. These modifications are thought to be important for the recruitment of the ATM kinase to sites of DSBs, ATM being one of the earliest responders to DNA damage [41,110]. Furthermore, the DNA damage-responsive E3 ubiquitin ligases RNF8 and RNF168 monoubiquitinate histone H2A and H2AX at lysines 13 and 15 [37,111,112,113,114]. Modified sites then act as significant scaffolds for the recruitment of key proteins engaged in the DNA damage response, such as BRCA1 and 53BP1 [42].
As well as being able to function as an E3 ligase RING finger complex for H2Bub1, BRCA1-BARD1 co-operates with the E2 UbcH5c to monoubiquitinate H2A and H2AX [42,52,115]. BRCA1-deficient cells display loss of ubiquitinated histone H2A at pericentromeric tandemly repeated satellite DNA [116]. The BRCA1-BARD1 complex ubiquitinates histone H2A at lysines 127 and 129 [38,114].
4. Other Known Lysine Sites of Histone Tail Monoubiquitination and Their E3 RING Finger Ligases
Other sites of histone monoubiquitination have been reported in human cells but remain to be extensively investigated. K125 on the histone H2B tail is monoubiquitinated when K120 is mutated in vitro; however, there is no evidence to date that H2BK125ub1 occurs in vivo [34]. Monoubiquitination of histone H2B on lysine 34 (H2BK34ub1) is written by the RING finger E3 ligase MSL2 that is part of the MOF-MSL complex [29]. This complex consists of four subunits (MOF-MSL1-MSL2-MSL3) [117], with MSL1-MSL2 being required for optimal E3 ubiquitin ligase activity. Like H2Bub1, H2BK34ub1 facilitates transcriptional elongation by RNA polymerase II [118].
Interestingly, MOF (also known as KAT8 or MYST Histone Acetyltransferase 1) functions as part of the MOF-MSL complex to acetylate histone H4 at lysine 16, giving this complex a dual role of ubiquitination and acetylation [119,120]. Providing early evidence of a complex network of histone E3 RING finger ligases, investigators have shown that MSL2 is involved in the recruitment of the H2Bub1 E3 ubiquitin ligases RNF20 and RNF40 to chromatin [29]. Fitting the paradigm of E3 RING finger ligases having multiple substrates, MSL2 is known to ubiquitinate the tumour suppressor p53, but does so at different sites to the better-known p53 E3 ligase MDM2 [121,122]. As is H2Bub1, H2BK34ub1 is involved in histone cross-talk, regulating H3K4me3 and H3K79me2 [29].
Monoubiquitination of histone H4 at lysine 31 (H4K31ub1), like H2Bub1, is associated with a more open chromatin configuration conducive to the promotion of transcription [39,40]. The cullin 4A (CUL4A) RING finger E3 ubiquitin ligase has been associated with these ubiquitination events, albeit at much lower levels than H2Bub1 and H2AK119ub1 [40,123]. Furthermore, CUL4A-mediated ubiquitination of H4K31 is also associated with accumulation of the active methylation marks H3K4me3 and H3K79me2 [40].
5. Therapeutic Targeting of E3 Ligases that Write Histone Monoubiquitination
Given the significant role of E3 ubiquitin ligases in substrate recognition, their value as therapeutic targets for diseases such as cancer are being considered [124,125]. Although no drugs have yet been developed with the goal of impacting specifically upon E3s that write histones, knowledge gleaned from developments to target non-histone E3 writers should be considered. While the E3 RING finger ubiquitin ligase MDM2 is reported to write H2Bub1 in vivo [34], it is primarily known as the enzyme that polyubiquitinates the guardian of the genome tumour suppressor p53, marking it for degradation via the 26S proteasome [126,127]. Small molecule inhibitors that disrupt the interaction of MDM2 and p53, including the cis-imidazoline compounds nutlin and the nutlin derivative RG7388 (idasanutlin, RG), are being developed for the treatment of a number of cancers [128,129,130]. In preventing polyubiquitination of p53 and thus its degradation, wild-type p53 levels are increased, in turn promoting apoptosis.
Other RING finger E3 ligases being considered as therapeutic targets for a range of diseases include cereblon, XIAP, IAP, VHL complex and Parkin [125]. An important step that necessarily underpins drug design targeting all of the E3 ligases that monoubiquitinate histones will be a sound understanding of their non-histone substrates [16,125]. In this vein, it is interesting to speculate that disruption of E3 ubiquitin ligases that function as complexes, for example the RNF20-RNF40 interaction, may also have merit when considering strategies for therapeutic targeting.
A deeper understanding of the fact that single sites of histone monoubiquitination seem to be written by multiple E3 ubiquitin ligases will be important in order to maximise therapeutic targeting opportunities. Currently, the reasons for this apparent redundancy are poorly understood but would seem to highlight the importance of these ubiquitination events to decisions regarding cellular maintenance and fate, including those in response to DNA damage. It is also likely that this function of some of these E3 ligases may be specific to cell type, disease state and/or developmental stage.
6. Conclusions
The role of multiple E3 RING finger ubiquitin ligases in writing monoubiquitination at specific lysine residues of histone tails is clearly fundamental to RNA polymerase II-based transcriptional elongation and the DNA damage response. Cancer is frequently described as a disease of aberrant transcription, highlighting the potential for the development of new therapeutic strategies by targeting these key enzymes functioning as part of the transcriptional machinery. There is a high likelihood that additional sites of histone monoubiquitination and their writer enzymes will be identified. Whether these will affect global levels of histone monoubiquitination or offer new levels of complexity by being directed towards modifying the expression of only certain functional classes of genes remains to be determined.
Author Contributions
D.J.M. conceptualised and wrote the manuscript. K.-A.D. assisted with reviewing the literature, as well as reviewing and editing the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Hergeth, S.P.; Schneider, R. The H1 linker histones: Multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015, 16, 1439–1453. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Kouzarides, T.; Huntly, B.J. Targeting epigenetic readers in cancer. N. Engl. J. Med. 2012, 367, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Santos-Rosa, H.; Caldas, C. Chromatin modifier enzymes, the histone code and cancer. Eur. J. Cancer 2005, 41, 2381–2402. [Google Scholar] [CrossRef] [PubMed]
- Albig, W.; Trappe, R.; Kardalinou, E.; Eick, S.; Doenecke, D. The human H2A and H2B histone gene complement. Biol. Chem. 1999, 380, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Sierra, F.; Lichtler, A.; Marashi, F.; Rickles, R.; Van Dyke, T.; Clark, S.; Wells, J.; Stein, G.; Stein, J. Organization of human histone genes. Proc. Natl. Acad. Sci. USA 1982, 79, 1795–1799. [Google Scholar] [CrossRef] [PubMed]
- Braun, S.; Madhani, H.D. Shaping the landscape: Mechanistic consequences of ubiquitin modification of chromatin. EMBO Rep. 2012, 13, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Ziv, I.; Matiuhin, Y.; Kirkpatrick, D.S.; Erpapazoglou, Z.; Leon, S.; Pantazopoulou, M.; Kim, W.; Gygi, S.P.; Haguenauer-Tsapis, R.; Reis, N.; et al. A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef] [PubMed]
- Clague, M.J.; Coulson, J.M.; Urbe, S. Deciphering histone 2A deubiquitination. Genome Biol. 2008, 9, 202. [Google Scholar] [CrossRef] [PubMed]
- West, M.H.; Bonner, W.M. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 1980, 8, 4671–4680. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, H.N.; Ye, Y. Cellular strategies for making monoubiquitin signals. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Lipkowitz, S.; Weissman, A.M. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer 2011, 11, 629–643. [Google Scholar] [CrossRef]
- Iconomou, M.; Saunders, D.N. Systematic approaches to identify E3 ligase substrates. Biochem. J. 2016, 473, 4083–4101. [Google Scholar] [CrossRef]
- Fang, S.; Weissman, A.M. A field guide to ubiquitylation. Cell. Mol. Life Sci. 2004, 61, 1546–1561. [Google Scholar] [CrossRef]
- Budhidarmo, R.; Nakatani, Y.; Day, C.L. RINGs hold the key to ubiquitin transfer. Trends Biochem. Sci. 2012, 37, 58–65. [Google Scholar] [CrossRef]
- Stewart, M.D.; Ritterhoff, T.; Klevit, R.E.; Brzovic, P.S. E2 enzymes: More than just middle men. Cell Res. 2016, 26, 423–440. [Google Scholar] [CrossRef]
- Metzger, M.B.; Pruneda, J.N.; Klevit, R.E.; Weissman, A.M. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 2014, 1843, 47–60. [Google Scholar] [CrossRef]
- Ye, Y.; Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009, 10, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Freemont, P.S.; Hanson, I.M.; Trowsdale, J. A novel cysteine-rich sequence motif. Cell 1991, 64, 483–484. [Google Scholar] [CrossRef]
- Stewart, M.D.; Duncan, E.D.; Coronado, E.; DaRosa, P.A.; Pruneda, J.N.; Brzovic, P.S.; Klevit, R.E. Tuning BRCA1 and BARD1 activity to investigate RING ubiquitin ligase mechanisms. Protein Sci. 2017, 26, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Densham, R.M.; Morris, J.R. The BRCA1 Ubiquitin ligase function sets a new trend for remodelling in DNA repair. Nucleus 2017, 8, 116–125. [Google Scholar] [CrossRef]
- Renaudin, X.; Koch Lerner, L.; Menck, C.F.; Rosselli, F. The ubiquitin family meets the Fanconi anemia proteins. Mutat. Res. Rev. Mutat. Res. 2016, 769, 36–46. [Google Scholar] [CrossRef]
- Wu, W.; Xu, C.; Ling, X.; Fan, C.; Buckley, B.P.; Chernov, M.V.; Ellis, L.; Li, F.; Munoz, I.G.; Wang, X. Targeting RING domains of Mdm2-MdmX E3 complex activates apoptotic arm of the p53 pathway in leukemia/lymphoma cells. Cell Death Dis. 2015, 6, e2035. [Google Scholar] [CrossRef]
- Fang, S.; Jensen, J.P.; Ludwig, R.L.; Vousden, K.H.; Weissman, A.M. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 2000, 275, 8945–8951. [Google Scholar] [CrossRef]
- Wu, L.; Zee, B.M.; Wang, Y.; Garcia, B.A.; Dou, Y. The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in crosstalk with H3 K4 and K79 methylation. Mol. Cell 2011, 43, 132–144. [Google Scholar] [CrossRef]
- Cole, A.J.; Clifton-Bligh, R.; Marsh, D.J. Histone H2B monoubiquitination: Roles to play in human malignancy. Endocr.-Relat. Cancer 2015, 22, T19–33. [Google Scholar] [CrossRef]
- Minsky, N.; Shema, E.; Field, Y.; Schuster, M.; Segal, E.; Oren, M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 2008, 10, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, S.A. The enigmatic role of H2Bub1 in cancer. FEBS Lett. 2012, 586, 1592–1601. [Google Scholar] [CrossRef]
- Fuchs, G.; Oren, M. Writing and reading H2B monoubiquitylation. Biochim. Biophys. Acta 2014, 1839, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Minsky, N.; Oren, M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol. Cell 2004, 16, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Marsh, D.J.; Shah, J.S.; Cole, A.J. Histones and their modifications in ovarian cancer—Drivers of disease and therapeutic targets. Front. Oncol. 2014, 4, 144. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Meas, R.; Mao, P. Histone ubiquitylation and its roles in transcription and DNA damage response. DNA Repair 2015, 36, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Kalb, R.; Mallery, D.L.; Larkin, C.; Huang, J.T.; Hiom, K. BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell Rep. 2014, 8, 999–1005. [Google Scholar] [CrossRef]
- Machida, S.; Sekine, S.; Nishiyama, Y.; Horikoshi, N.; Kurumizaka, H. Structural and biochemical analyses of monoubiquitinated human histones H2B and H4. Open Biol. 2016, 6. [Google Scholar] [CrossRef]
- Kim, K.; Lee, B.; Kim, J.; Choi, J.; Kim, J.M.; Xiong, Y.; Roeder, R.G.; An, W. Linker Histone H1.2 cooperates with Cul4A and PAF1 to drive H4K31 ubiquitylation-mediated transactivation. Cell Rep. 2013, 5, 1690–1703. [Google Scholar] [CrossRef]
- Pinder, J.B.; Attwood, K.M.; Dellaire, G. Reading, writing, and repair: The role of ubiquitin and the ubiquitin-like proteins in DNA damage signaling and repair. Front. Genet. 2013, 4, 45. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Yan, Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2012, 2, 26. [Google Scholar] [CrossRef] [PubMed]
- Osley, M.A.; Fleming, A.B.; Kao, C.F. Histone ubiquitylation and the regulation of transcription. Results Probl. Cell Differ. 2006, 41, 47–75. [Google Scholar] [PubMed]
- Weake, V.M.; Workman, J.L. Histone ubiquitination: Triggering gene activity. Mol. Cell 2008, 29, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.; Krogan, N.J.; Dover, J.; Schneider, J.; Heidt, J.; Boateng, M.A.; Dean, K.; Golshani, A.; Zhang, Y.; Greenblatt, J.F.; et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 2003, 11, 267–274. [Google Scholar] [CrossRef]
- Hwang, W.W.; Venkatasubrahmanyam, S.; Ianculescu, A.G.; Tong, A.; Boone, C.; Madhani, H.D. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 2003, 11, 261–266. [Google Scholar] [CrossRef]
- Kim, J.; Guermah, M.; McGinty, R.K.; Lee, J.S.; Tang, Z.; Milne, T.A.; Shilatifard, A.; Muir, T.W.; Roeder, R.G. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 2009, 137, 459–471. [Google Scholar] [CrossRef]
- Zhu, B.; Zheng, Y.; Pham, A.D.; Mandal, S.S.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol. Cell 2005, 20, 601–611. [Google Scholar] [CrossRef]
- Pavri, R.; Zhu, B.; Li, G.; Trojer, P.; Mandal, S.; Shilatifard, A.; Reinberg, D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 2006, 125, 703–717. [Google Scholar] [CrossRef]
- Fierz, B.; Chatterjee, C.; McGinty, R.K.; Bar-Dagan, M.; Raleigh, D.P.; Muir, T.W. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 2011, 7, 113–119. [Google Scholar] [CrossRef]
- Chandrasekharan, M.B.; Huang, F.; Sun, Z.W. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc. Natl. Acad. Sci. USA 2009, 106, 16686–16691. [Google Scholar] [CrossRef] [PubMed]
- Mallery, D.L.; Vandenberg, C.J.; Hiom, K. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 2002, 21, 6755–6762. [Google Scholar] [CrossRef] [PubMed]
- Thakar, A.; Parvin, J.; Zlatanova, J. BRCA1/BARD1 E3 ubiquitin ligase can modify histones H2A and H2B in the nucleosome particle. J. Biomol. Struct. Dyn. 2010, 27, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Li, X.S.; Trojer, P.; Matsumura, T.; Treisman, J.E.; Tanese, N. Mammalian SWI/SNF—A subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol. Cell. Biol. 2010, 30, 1673–1688. [Google Scholar] [CrossRef] [PubMed]
- Moyal, L.; Lerenthal, Y.; Gana-Weisz, M.; Mass, G.; So, S.; Wang, S.Y.; Eppink, B.; Chung, Y.M.; Shalev, G.; Shema, E.; et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell 2011, 41, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Hake, S.B.; Roeder, R.G. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell 2005, 20, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.A.; Dickson, K.A.; Jackson, S.; Clarkson, A.; Gill, A.J.; Marsh, D.J. The tumor suppressor CDC73 interacts with the ring finger proteins RNF20 and RNF40 and is required for the maintenance of histone 2B monoubiquitination. Hum. Mol. Genet. 2012, 21, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Dickson, K.A.; Cole, A.J.; Gill, A.J.; Clarkson, A.; Gard, G.B.; Chou, A.; Kennedy, C.J.; Henderson, B.R.; Fereday, S.; Traficante, N.; et al. The RING finger domain E3 ubiquitin ligases BRCA1 and the RNF20/RNF40 complex in global loss of the chromatin mark histone H2B monoubiquitination (H2Bub1) in cell line models and primary high-grade serous ovarian cancer. Hum. Mol. Genet. 2016, 25, 5460–5471. [Google Scholar] [CrossRef]
- Dover, J.; Schneider, J.; Tawiah-Boateng, M.A.; Wood, A.; Dean, K.; Johnston, M.; Shilatifard, A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002, 277, 28368–28371. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.W.; Allis, C.D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 2002, 418, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Jaehning, J.A. The Paf1 complex: Platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta 2010, 1799, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yu, X. WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol. Cell 2011, 41, 384–397. [Google Scholar] [CrossRef] [PubMed]
- Pirngruber, J.; Shchebet, A.; Schreiber, L.; Shema, E.; Minsky, N.; Chapman, R.D.; Eick, D.; Aylon, Y.; Oren, M.; Johnsen, S.A. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO Rep. 2009, 10, 894–900. [Google Scholar] [CrossRef] [PubMed]
- Shema, E.; Tirosh, I.; Aylon, Y.; Huang, J.; Ye, C.; Moskovits, N.; Raver-Shapira, N.; Minsky, N.; Pirngruber, J.; Tarcic, G.; et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008, 22, 2664–2676. [Google Scholar] [CrossRef] [PubMed]
- Prenzel, T.; Begus-Nahrmann, Y.; Kramer, F.; Hennion, M.; Hsu, C.; Gorsler, T.; Hintermair, C.; Eick, D.; Kremmer, E.; Simons, M.; et al. Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. Cancer Res. 2011, 71, 5739–5753. [Google Scholar] [CrossRef] [PubMed]
- Shiloh, Y.; Shema, E.; Moyal, L.; Oren, M. RNF20-RNF40: A ubiquitin-driven link between gene expression and the DNA damage response. FEBS Lett. 2011, 585, 2795–2802. [Google Scholar] [CrossRef]
- Nakamura, K.; Kato, A.; Kobayashi, J.; Yanagihara, H.; Sakamoto, S.; Oliveira, D.V.; Shimada, M.; Tauchi, H.; Suzuki, H.; Tashiro, S.; et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell 2011, 41, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Pefani, D.E.; Tognoli, M.L.; Pirincci Ercan, D.; Gorgoulis, V.; O’Neill, E. MST2 kinase suppresses rDNA transcription in response to DNA damage by phosphorylating nucleolar histone H2B. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, J.; Tong, T.R.; Wu, X.; Nelson, R.; Yuan, Y.C.; Reno, T.; Liu, Z.; Yun, X.; Kim, J.Y.; et al. Loss of H2B monoubiquitination is associated with poor-differentiation and enhanced malignancy of lung adenocarcinoma. Int. J. Cancer 2017, 141, 766–777. [Google Scholar] [CrossRef]
- Urasaki, Y.; Heath, L.; Xu, C.W. Coupling of glucose deprivation with impaired histone H2B monoubiquitination in tumors. PLoS ONE 2012, 7, e36775. [Google Scholar] [CrossRef]
- Tarcic, O.; Pateras, I.S.; Cooks, T.; Shema, E.; Kanterman, J.; Ashkenazi, H.; Boocholez, H.; Hubert, A.; Rotkopf, R.; Baniyash, M.; et al. RNF20 Links Histone H2B Ubiquitylation with Inflammation and Inflammation-Associated Cancer. Cell Rep. 2016, 14, 1462–1476. [Google Scholar] [CrossRef] [PubMed]
- Bedi, U.; Scheel, A.H.; Hennion, M.; Begus-Nahrmann, Y.; Ruschoff, J.; Johnsen, S.A. SUPT6H controls estrogen receptor activity and cellular differentiation by multiple epigenomic mechanisms. Oncogene 2015, 34, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Melling, N.; Grimm, N.; Simon, R.; Stahl, P.; Bokemeyer, C.; Terracciano, L.; Sauter, G.; Izbicki, J.R.; Marx, A.H. Loss of H2Bub1 Expression is Linked to Poor Prognosis in Nodal Negative Colorectal Cancers. Pathol. Oncol. Res. 2016, 22, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.J.; Yang, J.L.; Wang, Y.P.; Lou, J.Y.; Chen, J.; Liu, C.; Guo, L.D. Decreased histone H2B monoubiquitination in malignant gastric carcinoma. World J. Gastroenterol. 2013, 19, 8099–8107. [Google Scholar] [CrossRef] [PubMed]
- Chernikova, S.B.; Razorenova, O.V.; Higgins, J.P.; Sishc, B.J.; Nicolau, M.; Dorth, J.A.; Chernikova, D.A.; Kwok, S.; Brooks, J.D.; Bailey, S.M.; et al. Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res. 2012, 72, 2111–2119. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Jeon, Y.G.; Lee, K.H.; Lee, H.W.; Park, J.; Jang, H.; Kang, M.; Lee, H.S.; Cho, H.J.; Nam, D.H.; et al. RNF20 Suppresses Tumorigenesis by Inhibiting SREBP1c-PTTG1 Axis in Kidney Cancer. Mol. Cell. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Shema, E.; Kim, J.; Roeder, R.G.; Oren, M. RNF20 inhibits TFIIS-facilitated transcriptional elongation to suppress pro-oncogenic gene expression. Mol. Cell 2011, 42, 477–488. [Google Scholar] [CrossRef]
- Barber, T.D.; McManus, K.; Yuen, K.W.; Reis, M.; Parmigiani, G.; Shen, D.; Barrett, I.; Nouhi, Y.; Spencer, F.; Markowitz, S.; et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc. Natl. Acad. Sci. USA 2008, 105, 3443–3448. [Google Scholar] [CrossRef]
- Tahara, T.; Yamamoto, E.; Madireddi, P.; Suzuki, H.; Maruyama, R.; Chung, W.; Garriga, J.; Jelinek, J.; Yamano, H.O.; Sugai, T.; et al. Colorectal carcinomas with CpG island methylator phenotype 1 frequently contain mutations in chromatin regulators. Gastroenterology 2014, 146, 530–538.e535. [Google Scholar] [CrossRef]
- Varambally, S.; Yu, J.; Laxman, B.; Rhodes, D.R.; Mehra, R.; Tomlins, S.A.; Shah, R.B.; Chandran, U.; Monzon, F.A.; Becich, M.J.; et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 2005, 8, 393–406. [Google Scholar] [CrossRef]
- Nakachi, I.; Rice, J.L.; Coldren, C.D.; Edwards, M.G.; Stearman, R.S.; Glidewell, S.C.; Varella-Garcia, M.; Franklin, W.A.; Keith, R.L.; Lewis, M.T.; et al. Application of SNP microarrays to the genome-wide analysis of chromosomal instability in premalignant airway lesions. Cancer Prev. Res. 2014, 7, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Wright, D.E.; Wang, C.Y.; Kao, C.F. Flickin’ the ubiquitin switch: The role of H2B ubiquitylation in development. Epigenetics 2011, 6, 1165–1175. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Oh, S.M.; Okada, M.; Liu, X.; Cheng, D.; Peng, J.; Brat, D.J.; Sun, S.Y.; Zhou, W.; Gu, W.; et al. Human BRE1 is an E3 ubiquitin ligase for Ebp1 tumor suppressor. Mol. Biol. Cell 2009, 20, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Tarcic, O.; Granit, R.Z.; Pateras, I.S.; Masury, H.; Maly, B.; Zwang, Y.; Yarden, Y.; Gorgoulis, V.G.; Pikarsky, E.; Ben-Porath, I.; et al. RNF20 and histone H2B ubiquitylation exert opposing effects in Basal-Like versus luminal breast cancer. Cell Death Differ. 2017, 24, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Kosinsky, R.L.; Chua, R.L.; Qui, M.; Saul, D.; Mehlich, D.; Strobel, P.; Schildhaus, H.U.; Wegwitz, F.; Faubion, W.A.; Johnsen, S.A. Loss of RNF40 decreases NF-kappaB activity in colorectal cancer cells and reduces colitis burden in mice. J. Crohn’s Colitis 2018. [Google Scholar] [CrossRef]
- Duan, Y.; Huo, D.; Gao, J.; Wu, H.; Ye, Z.; Liu, Z.; Zhang, K.; Shan, L.; Zhou, X.; Wang, Y.; et al. Ubiquitin ligase RNF20/40 facilitates spindle assembly and promotes breast carcinogenesis through stabilizing motor protein Eg5. Nat. Commun. 2016, 7, 12648. [Google Scholar] [CrossRef]
- Chin, L.S.; Vavalle, J.P.; Li, L. Staring, a novel E3 ubiquitin-protein ligase that targets syntaxin 1 for degradation. J. Biol. Chem. 2002, 277, 35071–35079. [Google Scholar] [CrossRef]
- Goldknopf, I.L.; Taylor, C.W.; Baum, R.M.; Yeoman, L.C.; Olson, M.O.; Prestayko, A.W.; Busch, H. Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. J. Biol. Chem. 1975, 250, 7182–7187. [Google Scholar] [PubMed]
- Jason, L.J.; Moore, S.C.; Ausio, J.; Lindsey, G. Magnesium-dependent association and folding of oligonucleosomes reconstituted with ubiquitinated H2A. J. Biol. Chem. 2001, 276, 14597–14601. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhu, P.; Wang, J.; Pascual, G.; Ohgi, K.A.; Lozach, J.; Glass, C.K.; Rosenfeld, M.G. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol. Cell 2008, 29, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Kajitani, T.; Togo, S.; Masuko, N.; Ohdan, H.; Hishikawa, Y.; Koji, T.; Matsuyama, T.; Ikura, T.; Muramatsu, M.; et al. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008, 22, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, J.; Zhan, Q.; Zhu, Y.; Chen, H.; Deng, X.; Hou, Z.; Shen, B.; Chen, Y.; Peng, C. H2AK119Ub1 and H3K27Me3 in molecular staging for survival prediction of patients with pancreatic ductal adenocarcinoma. Oncotarget 2014, 5, 10421–10433. [Google Scholar] [CrossRef] [PubMed]
- Lanzuolo, C.; Orlando, V. Memories from the polycomb group proteins. Annu. Rev. Genet. 2012, 46, 561–589. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Tsukada, Y.; Zhang, Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 2005, 20, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Cavalli, G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 2009, 136, 3531–3542. [Google Scholar] [CrossRef] [PubMed]
- de Napoles, M.; Mermoud, J.E.; Wakao, R.; Tang, Y.A.; Endoh, M.; Appanah, R.; Nesterova, T.B.; Silva, J.; Otte, A.P.; Vidal, M.; et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 2004, 7, 663–676. [Google Scholar] [CrossRef]
- Sarcinella, E.; Zuzarte, P.C.; Lau, P.N.; Draker, R.; Cheung, P. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol. Cell. Biol. 2007, 27, 6457–6468. [Google Scholar] [CrossRef]
- Su, W.J.; Fang, J.S.; Cheng, F.; Liu, C.; Zhou, F.; Zhang, J. RNF2/Ring1b negatively regulates p53 expression in selective cancer cell types to promote tumor development. Proc. Natl. Acad. Sci. USA 2013, 110, 1720–1725. [Google Scholar] [CrossRef]
- Hu, H.; Yang, Y.; Ji, Q.; Zhao, W.; Jiang, B.; Liu, R.; Yuan, J.; Liu, Q.; Li, X.; Zou, Y.; et al. CRL4B catalyzes H2AK119 monoubiquitination and coordinates with PRC2 to promote tumorigenesis. Cancer Cell 2012, 22, 781–795. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, R.; Qiu, R.; Zheng, Y.; Huang, W.; Hu, H.; Ji, Q.; He, H.; Shang, Y.; Gong, Y.; et al. CRL4B promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA methylation-based epigenetic silencing. Oncogene 2015, 34, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, S.; Gazin, C.; Chamberlain, L.; Ou, J.; Zhu, X.; Tushir, J.S.; Virbasius, C.M.; Lin, L.; Zhu, L.J.; Wajapeyee, N.; et al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature 2014, 516, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Wienken, M.; Dickmanns, A.; Nemajerova, A.; Kramer, D.; Najafova, Z.; Weiss, M.; Karpiuk, O.; Kassem, M.; Zhang, Y.; Lozano, G.; et al. MDM2 Associates with Polycomb Repressor Complex 2 and Enhances Stemness-Promoting Chromatin Modifications Independent of p53. Mol. Cell 2016, 61, 68–83. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Oughtred, R.; Wing, S.S. Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol. Cell. Biol. 2005, 25, 2819–2831. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Q.; He, W.-P.; Hou, W.-H.; Chen, J.-W.; Fan, R.-R.; Yuan, L.-J.; Yang, G.-P.; Cai, M.-Y.; Chen, L.; He, S.-Y.; et al. Overexpression of RNF2 is positively associated with ovarian carcinoma aggressiveness and indicative of poor patient survival. Oncotarget 2016. [Google Scholar] [CrossRef]
- Li, X.D.; Chen, S.L.; Dong, P.; Chen, J.W.; Wang, F.W.; Guo, S.J.; Jiang, L.J.; Zhou, F.J.; Xie, D.; Liu, Z.W. Overexpression of RNF2 Is an Independent Predictor of Outcome in Patients with Urothelial Carcinoma of the Bladder Undergoing Radical Cystectomy. Sci. Rep. 2016, 6, 20894. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Jiao, D.; Han, D.; Wu, J.; Wei, F.; Zheng, G.; Guo, Z.; Xi, W.; Yang, F.; Xie, P.; et al. Knockdown of RNF2 induces cell cycle arrest and apoptosis in prostate cancer cells through the upregulation of TXNIP. Oncotarget 2017, 8, 5323–5338. [Google Scholar] [CrossRef] [PubMed]
- Bergink, S.; Salomons, F.A.; Hoogstraten, D.; Groothuis, T.A.; de Waard, H.; Wu, J.; Yuan, L.; Citterio, E.; Houtsmuller, A.B.; Neefjes, J.; et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006, 20, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Kang, H.Y.; Yang, W.L.; Wu, J.; Jeong, Y.S.; Wang, J.; Chan, C.H.; Lee, S.W.; Zhang, X.; Lamothe, B.; et al. Critical role of monoubiquitination of histone H2AX protein in histone H2AX phosphorylation and DNA damage response. J. Biol. Chem. 2011, 286, 30806–30815. [Google Scholar] [CrossRef]
- Pan, M.R.; Peng, G.; Hung, W.C.; Lin, S.Y. Monoubiquitination of H2AX protein regulates DNA damage response signaling. J. Biol. Chem. 2011, 286, 28599–28607. [Google Scholar] [CrossRef]
- Gatti, M.; Pinato, S.; Maspero, E.; Soffientini, P.; Polo, S.; Penengo, L. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle 2012, 11, 2538–2544. [Google Scholar] [CrossRef] [PubMed]
- Bekker-Jensen, S.; Mailand, N. Ubiquitin and the DNA damage response: A new handle on histones. Cell Cycle 2012, 11, 3153. [Google Scholar] [CrossRef] [PubMed]
- Mattiroli, F.; Vissers, J.H.; van Dijk, W.J.; Ikpa, P.; Citterio, E.; Vermeulen, W.; Marteijn, J.A.; Sixma, T.K. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 2012, 150, 1182–1195. [Google Scholar] [CrossRef] [PubMed]
- Uckelmann, M.; Sixma, T.K. Histone ubiquitination in the DNA damage response. DNA Repair 2017, 56, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Kleiman, F.E.; Manley, J.L.; Ouchi, T.; Pan, Z.Q. Autoubiquitination of the BRCA1*BARD1 RING ubiquitin ligase. J. Biol. Chem. 2002, 277, 22085–22092. [Google Scholar] [CrossRef]
- Zhu, Q.; Pao, G.M.; Huynh, A.M.; Suh, H.; Tonnu, N.; Nederlof, P.M.; Gage, F.H.; Verma, I.M. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 2011, 477, 179–184. [Google Scholar] [CrossRef]
- Smith, E.R.; Cayrou, C.; Huang, R.; Lane, W.S.; Cote, J.; Lucchesi, J.C. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol. Cell. Biol. 2005, 25, 9175–9188. [Google Scholar] [CrossRef]
- Wu, L.; Li, L.; Zhou, B.; Qin, Z.; Dou, Y. H2B ubiquitylation promotes RNA Pol II processivity via PAF1 and pTEFb. Mol. Cell 2014, 54, 920–931. [Google Scholar] [CrossRef]
- Akhtar, A.; Becker, P.B. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 2000, 5, 367–375. [Google Scholar] [CrossRef]
- Taipale, M.; Rea, S.; Richter, K.; Vilar, A.; Lichter, P.; Imhof, A.; Akhtar, A. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol. Cell. Biol. 2005, 25, 6798–6810. [Google Scholar] [CrossRef]
- Mendoza, M.; Mandani, G.; Momand, J. The MDM2 gene family. Biomol. Concepts 2014, 5, 9–19. [Google Scholar] [CrossRef]
- Kruse, J.P.; Gu, W. MSL2 promotes Mdm2-independent cytoplasmic localization of p53. J. Biol. Chem. 2009, 284, 3250–3263. [Google Scholar] [CrossRef]
- Wang, H.; Zhai, L.; Xu, J.; Joo, H.Y.; Jackson, S.; Erdjument-Bromage, H.; Tempst, P.; Xiong, Y.; Zhang, Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 2006, 22, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Dixit, V.M. Drugging the undruggables: Exploring the ubiquitin system for drug development. Cell Res. 2016, 26, 484. [Google Scholar] [CrossRef] [PubMed]
- Galdeano, C. Drugging the undruggable: Targeting challenging E3 ligases for personalized medicine. Future Med. Chem. 2017, 9, 347–350. [Google Scholar] [CrossRef]
- Piette, J.; Neel, H.; Marechal, V. Mdm2: Keeping p53 under control. Oncogene 1997, 15, 1001–1010. [Google Scholar] [CrossRef]
- Khoo, K.H.; Verma, C.S.; Lane, D.P. Drugging the p53 pathway: Understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 2014, 13, 217–236. [Google Scholar] [CrossRef] [PubMed]
- Fischer, P.M.; Lane, D.P. Small-molecule inhibitors of the p53 suppressor HDM2: Have protein-protein interactions come of age as drug targets? Trends Pharmacol. Sci. 2004, 25, 343–346. [Google Scholar] [CrossRef]
- Ding, Q.; Zhang, Z.; Liu, J.J.; Jiang, N.; Zhang, J.; Ross, T.M.; Chu, X.J.; Bartkovitz, D.; Podlaski, F.; Janson, C.; et al. Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J. Med. Chem. 2013, 56, 5979–5983. [Google Scholar] [CrossRef]
- Zanjirband, M.; Edmondson, R.J.; Lunec, J. Pre-clinical efficacy and synergistic potential of the MDM2-p53 antagonists, Nutlin-3 and RG7388, as single agents and in combined treatment with cisplatin in ovarian cancer. Oncotarget 2016, 7, 40115–40134. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).