Abstract
Combined hepatocellular carcinoma–cholangiocarcinoma (cHCC-CCA) is a challenging primary liver cancer subtype with limited treatment options and a devastating prognosis. Recent studies have underscored the context-dependent roles of SOX9 in liver cancer formation in a preventive manner. Here, we revealed that liver-specific developmental Sox9 elimination using Alb-Cre;Sox9(flox/flox) (LKO) and CRISPR/Cas9-based tumor-specific acute Sox9 elimination (CKO) in SB-HDTVI-based Akt-YAP1 (AY) and Akt-NRAS (AN) cHCC-CCA models showed contrasting responses. LKO abrogates the AY CCA region while stimulating poorly differentiated HCC proliferation, whereas CKO prevents AY and AN cHCC-CCA development irrespective of tumor cell fate. Additionally, AN, but not AY, tumor formation partially depends on the Sox9-Dnmt1 cascade. SOX9 is dispensable for AY-mediated, HC-derived, LPC-like immature CCA formation but is required for their maintenance and transformation into mature CCA. Therapeutic Sox9 elimination using the OPN-CreERT2 strain combined with inducible Sox9 iKO specifically reduces AY but not AN cHCC-CCA tumors. This necessitates the careful consideration of genetic liver cancer studies using developmental Cre and somatic mutants, particularly for genes involved in liver development. Our findings suggest that SOX9 elimination may hold promise as a therapeutic approach for a subset of cHCC-CCA and highlight the need for further investigation to translate these preclinical insights into personalized clinical applications.
1. Introduction
Combined hepatocellular carcinoma–cholangiocarcinoma (cHCC-CCA) represents a rare and intriguing entity in the spectrum of primary liver cancers, characterized by the coexistence of hepatocellular and cholangiocellular differentiation within the same tumor [,,]. This dual histological phenotype poses significant diagnostic, prognostic, and therapeutic challenges, reflecting the complex interplay between hepatocytic and biliary lineages in liver tumorigenesis [,]. Especially noteworthy is the distinct response of this tumor type to broad-spectrum therapeutic and immune therapies, making the therapeutic strategy largely reliant on basic histological observations, such as measuring the ratio of respective tumor types []. Despite its clinical and pathological significance, the underlying molecular mechanisms driving the development and progression of cHCC-CCA remain poorly understood.
Traditionally, HCC and CCA are thought to originate from hepatocytes (HCs) and cholangiocytes (biliary epithelial cells; BECs), respectively, as evidenced by typical cellular morphology, unique structures, and the expression of cell type-specific markers. However, the frequent detection of human CCA and cHCC-CCA in the pericentral area of the liver lobule, a region anatomically different from native biliary structures, as well as the documented occurrence of HC-to-BEC differentiation in various chronic cholestasis models, has led to speculation that hepatocytes may also be the origin of a subset of human CCA and cHCC-CCA [,,].
Indeed, numerous studies have provided evidence supporting this theory by inducing HC-derived cHCC-CCA using the HC-specific co-expression of proto-oncogenes and biliary lineage commitment genes, such as myristoylated Akt (Akt) and constitutive-active YAP1 or NRAS [,,,]. The overexpression of these oncogenes successfully produces separate regions of HNF4α+;panCK+;CK19− poorly differentiated HCC and HNF4α−;CK19+ CCA, resembling the clinical cHCC-CCA tumor pathology [,,].
Recently, Liu et al. reported that the biliary-specific transcription factor SOX9 determines the fate of YAP1 alone-dependent murine liver cancer; the chronic deletion of SOX9 in HC suppresses YAP1-mediated CCA-like tumor formation while promoting aggressive HCC tumor development, suggesting SOX9 as a major commitment of YAP1-mediated liver cancer lineage []. However, whether SOX9 is required to maintain the biliary fate of CCA within fully developed advanced cHCC-CCA remains elusive. Additionally, it is unclear whether SOX9 removal directly converts CCA lesions into HCC fate or simply eliminates CCA tumors during tumor formation, thereby allowing HCC to remain in YAP1-independent cHCC-CCA settings. Furthermore, given the discrepancy between chronic and acute gene deletion in diseased liver [], the effects of chronic versus acute SOX9 elimination on the development of HC-derived cHCC-CCA remain to be thoroughly investigated.
Herein, we reveal that unlike chronic developmental Sox9 deletion, the acute and therapeutic elimination of Sox9 reduces overall cHCC-CCA tumor burden in Akt-YAP1 but not the Akt-NRAS model. Additionally, we found that transcriptional repressor DNA methyltransferase1 (DNMT1) is partially involved in the SOX9-dependent maintenance of Akt-NRAS cHCC-CCA. These findings underscore the context- and stage-dependent distinct roles of SOX9 in liver cancer, with potential biological and therapeutic implications.
2. Materials and Methods
2.1. Mouse Husbandry and Breeding
All animal care and experiments were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) at the University of Pittsburgh. Sox9(flox/flox) and Albumin-Cre mice were purchased from Jackson Laboratories for breeding. All transgenic and KO mouse lines were maintained on the immunocompetent C57BL/6 genetic background. All animals ranged from 6 to 12 weeks in age for analysis and were from either gender.
2.2. Patient Data
Study approval for all human tissue samples was obtained from the University of Pittsburgh (IRB# STUDY19070068). All samples were provided by the Pittsburgh Liver Research Center’s (PLRC’s) Clinical Biospecimen Repository and Processing Core (CBPRC), supported by P30DK120531.
TMAs were constructed from archival formalin-fixed paraffin-embedded tissue blocks from 108 cholangiocarcinoma patients seen at the University of Pittsburgh Medical Center and were also obtained from PLRC’s CBPRC. All tumor hematoxylin and eosin (H&E)-stained slides were reviewed, and representative areas were carefully selected for tissue microarray construction. Two random 1.0 mm sized cores were punched from each patient’s tumor and harvested into recipient blocks. The demographics and additional information about these cases are included in Supplementary Table S1. The TMAs were stained manually using an antibody against SOX9 (EMD Millipore, Burlington, MA, USA, 01803), YAP (Cell Signaling, Danvers, MA, USA, 01923), as described in the IHC sections. Whole-slide image capturing of the tissue microarray was acquired using the Aperio XT slide scanner (Leica Biosystems, Deer Park, IL, USA, 60010). The staining was evaluated and scored by an anatomic pathologist (A.S.). Staining for SOX9 and YAP was scored either as 0 (negative), 1+ (mostly cytoplasmic staining or very weak staining in CC tumor cells), or 2+ (strong positive nuclear staining in CC tumor cells). For SOX9 and YAP1, the scores for different tissue sections from each patient were averaged to obtain a single score per patient (Supplementary Table S1). Mean scores greater than or equal to 1.5 were considered “HIGH”, and mean scores less than 1.5 were considered “LOW/NEGATIVE”.
3. Results
3.1. Forced Expression of Myristoylated Akt and YAP1 in HC Yields cHCC-CCA, While the Chronic Elimination of Sox9 Induces Molecular Phenotype Switch to an Aggressive HCC
Previously, it has been reported that the sleeping beauty transposon/transposase-hydrodynamic tail vein injection (SB-HDTVI) delivery of myristoylated Akt (Akt) and YAP1 S127A (YAP1) induced HC-derived cHCC-CCA in a Notch-dependent manner [,]. Tumor-specific Notch2 deletion switched the tumor type in the Akt-YAP1 model from cHCC-CCA to a benign hepatocellular adenoma-like tumor at the expense of CCA []. Moreover, HC-specific Sox9 deletion prevents YAP1 alone-mediated HC transformation into BEC-like HCC, while it provokes pure but aggressive HCC, indicating the critical role of Sox9 in YAP1-driven HC plasticity into biliary lineage []. Given that Sox9 is a well-established direct target of NOTCH2 [,], we aimed to investigate the effects of cell-autonomous Sox9 deletion on lineage commitment in Akt-YAP1-driven cHCC-CCA tumorigenesis. Akt and YAP1 plasmids were co-delivered by SB-HDTVI into Alb-Cre;Sox9flox/flox (Sox9 LKO) or Sox9flox/flox (LWT) in which Sox9 was deleted HC and BEC initiated at around day E15 [], resulting in chronic developmental deletion prior to tumor formation (Figure 1A). Notably, Akt-YAP1 in Sox9 LKO led to significantly decreased survival and a much greater and more lethal tumor burden, as seen by significantly greater LW/BW and macroscopically, as compared to the Akt-YAP1 in LWT mice (Figure 1B–D). Microscopically, in a representative tiled image of a lobe, the Sox9 LWT livers in the Akt-YAP1 model showed more intensely CK19-positive CCA nodules scattered throughout (Figure 1E). At higher magnification, the tumors contained separated clusters and showed CCA components which were positive for SOX9, YAP1, and panCK and an HCC component with nuclear HNF4α (Figure 2B). However, the entire Akt-YAP1 Sox9 LKO livers were full of circumscribed tumor foci which were negative or very weak for CK19 (Figure 1E). At higher magnification, the tumors revealed poorly differentiated HCCs that lacked SOX9, expressed HNF4α, and had small scattered subsets of YAP1 and panCK-positive cells with liver progenitor cell (LPC) characteristics (Figure 2B). The quantification of CK19 staining in the tiled image (Figure 1E) verified a significant decrease in the CK19-positive area in Sox9 LKO as compared to the LWT (Figure 1F). Further, tumors in the Sox9 LKO Akt-YAP1 model revealed the significantly lower expression of biliary markers, such as Krt19 and Epcam, along with the expected loss of Sox9. They also had increased expression levels of HC-specific markers, such as Hnf4α and Tryptophan 2,3-Dioxygenase (Tdo2), compared to LWT, suggesting a shift from CCA to HCC (Figure 2C,D). Together, these data support the role of Sox9 in commitment to the CCA phenotype in Akt-YAP1-driven cHCC-CCA tumorigenesis. Next, we examined cHCC-CCA (n = 32) in the available patient TMA for SOX9 and YAP1 localization. While the majority of the mixed tumors were concurrently positive for both markers (28/32 or 87.5%), a small subset was positive for YAP1 but negative or low for SOX9 (4/32 or 12.5%) (Figure 2E). While the numbers are too low to determine the impact on overall tumor behavior or prognosis in these cases, this observation does lend clinical credibility to our preclinical observation.
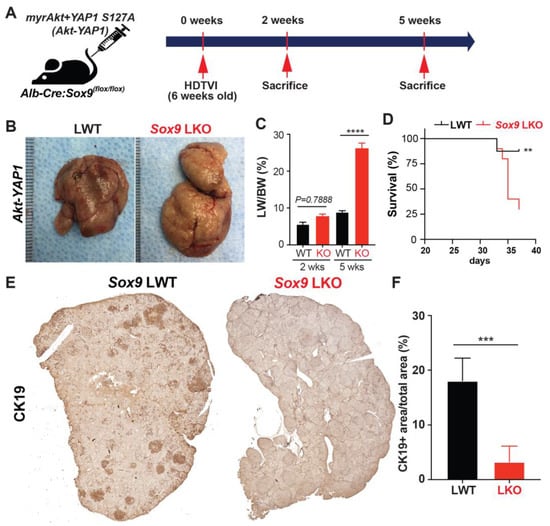
Figure 1.
Chronic developmental deletion of Sox9 switches the fate of Akt-YAP1-driven cHCC-CCA to aggressive HCC at the expense of CCA. (A) Experimental design illustrating plasmids used for HDTVI, mice used in study, and time-points analyzed. (B) Representative gross images from Akt-YAP1-injected Sox9-floxed mice (LWT) and Akt-YAP1-injected Alb-Cre;Sox9(f/f) liver-specific Sox9 knockout or Sox9 LKO mice showing tumor-laden enlarged livers in both cases. (C) LW/BW ratio depicts a significantly lower tumor burden in Akt-YAP1 Sox9 LKO as compared to LWT at 5 weeks but not earlier than the 2-week time-point. (D) Kaplan–Meier survival curve showing a significant decrease in the survival of Akt-YAP1 mice that were Sox9 LKO as compared to LWT. (E) Representative tiled image of tumor-bearing livers at 5 weeks in Akt-YAP1 LWT stained for CK19 IHC showing the CCA component of the positive cHCC-CCA staining. Sox9 LKO livers were full of circumscribed tumors that were negative for CK19 at the same time-point. (F) Quantification of CK19 IHC verifies significantly reduced staining in Akt-YAP1 Sox9 LKO as compared to Akt-YAP1 LWT at 5 weeks, as shown in E. Error bar: standard error of the mean; ** p < 0.01; *** p < 0.05; **** p < 0.0001.
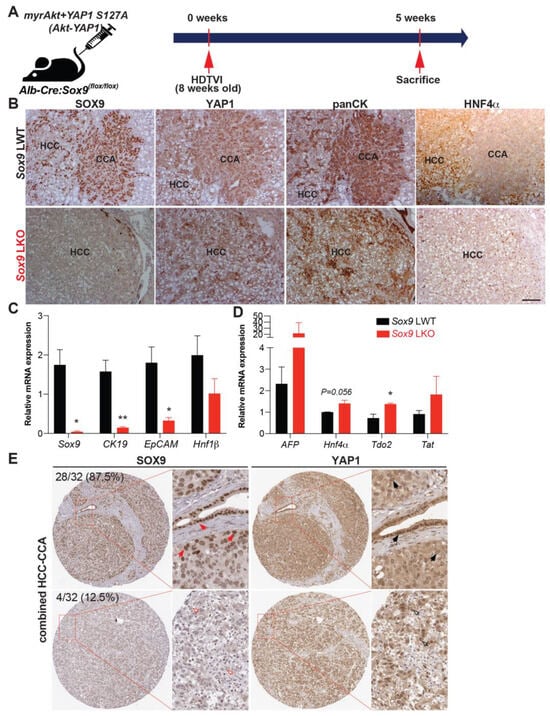
Figure 2.
Absence of Sox9 induces Akt-YAP1-mediated HC-derived panCK and HNF4α-positive HCC associated with liver progenitor cell characteristics. (A) Experimental design illustrating plasmids used for HDTVI, mice used in study, and time-points analyzed. (B) Representative serial sections of IHC images of 5-week Akt-YAP1 LWT show CCA component to be positive for SOX9, YAP1, and panCK and HCC to be strongly positive for HNF4α, while in Akt-YAP1 Sox9 LKO, no CCA was seen and HCC was negative for SOX9 but positive for HNF4α with panCK- and YAP1-positive cells interspersed in the tumor parenchyma. (C,D) qPCR data showing significantly decreased mRNA expression of Sox9, CK19, and EpCAM and significantly increased expression of Tdo2 when comparing tumor-bearing livers in Akt-YAP1 LWT and Sox9 LKO models at 5 weeks. (E) Representative IHC staining for SOX9 and YAP1 depicting SOX9-low and nuclear YAP1-high or SOX9-high and YAP1-high cHCC-CCA from TMA (32 patients). TMA sections were enlarged for better view of nuclear expression of SOX9 and YAP1. Red arrows point to nuclear SOX9-high cells; black arrows point to nuclear YAP1-high cells; red empty arrows point to nuclear SOX9-negative cells; and black empty arrows point to nuclear YAP1-high cells. Percentage of patients positive for each combination are indicated. Scale bars: 100 µm; Error bar: standard error of the mean; * p < 0.05; ** p < 0.01.
3.2. Transcriptomic Analysis of HCC in Akt-YAP1 Model in Sox9-LKO Reveals Significant Similarity to a Subset of Human HCCs
To directly address the clinical relevance of the HCCs observed in the Akt-YAP1 model in the absence of Sox9, we performed RNA-Seq analysis (GEO accession ID: GSE200472). When comparing the LWT and Akt-YAP1 Sox9-LKO livers, 525 genes were upregulated and 199 genes were downregulated by FDR = 5% and absolute log2 fold change of 1 (Figure 3A). To interpret the biological functions of these 724 DEGs, pathway enrichment analysis was performed by Ingenuity Pathway Analysis. Fifty-three pathways were significantly enriched in the Akt-YAP1 Sox9 LKO livers. To determine if the mouse model mimics a subtype of HCCs in patients, the LIHC cohort of the TCGA database was analyzed using a similar pipeline []. When comparing 50 normal or normal adjacent control livers and 374 HCCs, the DEGs were enriched in 59 pathways. Ten pathways were commonly altered in mouse and human tumors (Figure 3B). To directly compare mouse and human studies, the 724 DEGs from the Akt-YAP1 Sox9 LKO livers were converted to human homologous genes by the Mouse Genome Database []. Human expression data had 546 of the 724 DEGs, and using abs(log2FC) > 1 and FDR < 0.05, 118 of the 546 DEGs were referred to as the Akt-YAP1 Sox9 LKO (null) or AY signature and applied to the TCGA HCC (Figure 3C). These genes could clearly separate the human normal (orange bar) and HCC (light green bar) groups very well. Lastly, NTP analysis was performed using the Akt-YAP1 Sox9 LKO signature []. In the TCGA cohort, the AY signature captured 12% of HCC. This subgroup of patients is enriched in the S1 class [] (28/46 vs. 82/328 in rest of patients, p = 0.0015) and has a CCA-like signature [], (22/46 vs. 102/328, p = 0.03) (Figure 3D). Altogether, the HCC in the Akt-YAP1 Sox9 LKO model recapitulates a subset of human HCC.
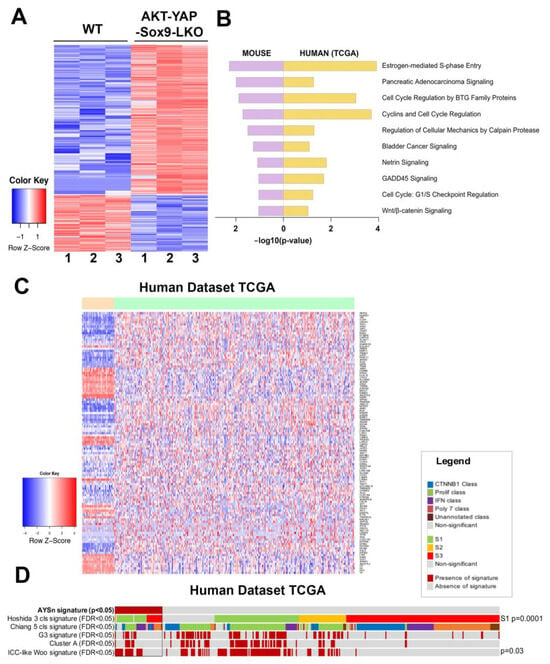
Figure 3.
RNA-seq analysis of mouse models and comparison with human liver cancer studies. (A) Heatmap for the differentially expressed genes comparing LWT and the Akt-YAP1 Sox9 LKO model. (B) Common top enriched pathways between mouse (Akt-YAP1 Sox9 LKO) and human (TCGA, LIHC) study. (C) Heatmap of gene signatures in the TCGA LIHC that are selected by the mouse model (LWT vs. Akt-YAP1 Sox9 LKO). (D) Nearest Template Prediction (NTP) analysis of the TCGA LIHC whole-tumor gene expression dataset using the Akt-YAP1 Sox9 LKO signature (AYSn signature) captured 12% of HCC. This subgroup of patients is enriched in the S1 class [] (p = 0.0015) and has an ICC-like signature [] (p = 0.03).
3.3. SOX9 Is Dispensable for Akt-YAP1-Mediated LPC-like Immature CCA Nodule Formation but Required for Their Transformation into Mature CCA
To explore the stage-specific roles of SOX9 in Akt-YAP1-driven HC-to-CCA lineage reprogramming, we carefully examined the liver histology in both LWT and Akt-YAP1 Sox9 LKO mice. We performed serial section immunostaining to detect HC and BEC markers, including HA-tag (AKT), YAP1, SOX9, HNF4α, panCK, and CK19, at 2 weeks and 5 weeks post-HDTVI, which correspond to the stages before the clonal expansion of transduced cells and after their transformation into mature CCA, respectively (Figure 4B,C) []. Importantly, LPC-like immature CCA nodules with Akt-YAP1 transduction (HA-tag+; nuclear YAP1+) were observed in both LWT and Sox9-LKO livers. These nodules retained an intermediate LPC morphology and co-expressed the HC marker HNF4α and the BEC/LPC marker panCK, while the presence or absence of SOX9 differed between the respective livers (Figure 4B, black dashed lines). Notably, none of these nodules in Sox9-LKO livers were positive for the mature BEC marker CK19, whereas a subset of panCK+ CCA nodules in LWT livers were CK19+, indicating further reprogramming into the biliary lineage with SOX9 (Figure 4C). At 5 weeks post-HDTVI, these nodules further transformed into CK19+ CCA nodules only in LWT livers. In contrast, panCK+;CK19+ CCA nodules were not detected in any Sox9-LKO livers at the same stage (Figure 4C). These data suggest that while SOX9 is not necessary for YAP1-driven HC dedifferentiation into LPCs or the initiation of biliary reprogramming, it is crucial for the maintenance and maturation of these CCA-like tumor nodules into a mature CCA lineage. This observation highlights the potential stage-dependent and distinct roles of SOX9 in fully developed Akt-YAP1-CCA at advanced stages, particularly from a therapeutic perspective.
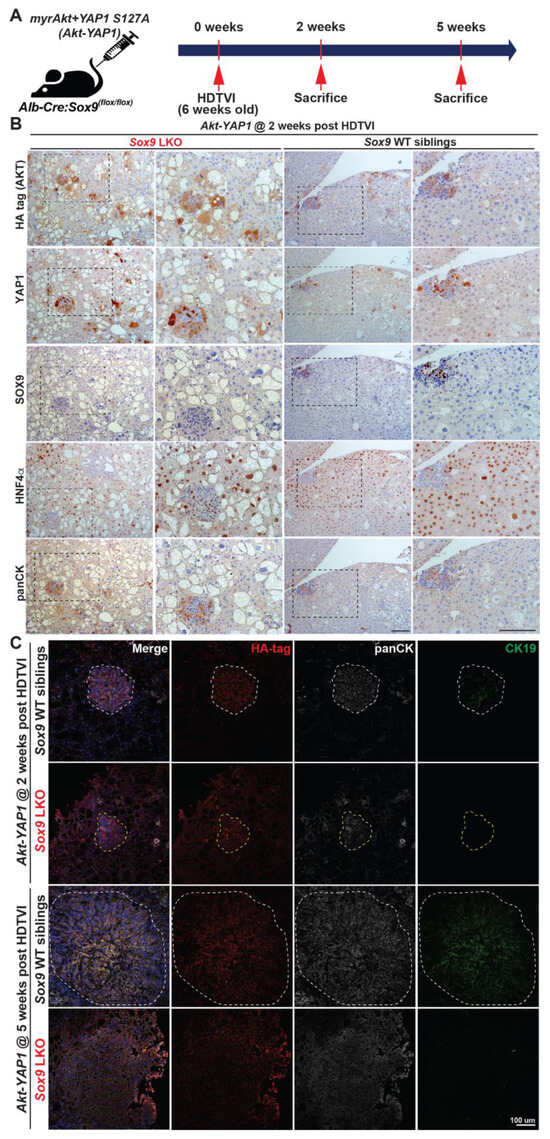
Figure 4.
SOX9 is required for Akt-YAP1-mediated HC reprogramming into CK19+ mature CCA. (A) Experimental design illustrating plasmids used for HDTVI, mice used in study, and time-points analyzed. (B) Representative serial section IHC images of both Akt-YAP1 LWT and Akt-YAP1 Sox9 LKO show CCA-like components to be positive for SOX9, YAP1, panCK, and weak positive HNF4α with liver progenitor cell morphology (black dash lined) and HCC to be strongly positive for HNF4α at 2 weeks post-HDTVI. (C) Confocal images of immunofluorescence staining of LWT or Sox9-LKO livers at 2 and 5 weeks verify the essential roles for Sox9 in CCA maturation. White dashed line points to panCK+;SOX9+;CK19+ CCA cells and yellow dashed line to panCK+;SOX9−;CK19− LPC-like immature CCA cells. Scale bars: 100 µm.
3.4. Distinct Roles SOX9 in Akt-YAP1-Driven HCC Tumor in Regulating Proliferation
Since Sox9 deletion decreases survival in Akt-YAP1 mice with a larger tumor burden, we next sought to evaluate tumor cell death and proliferation through histologic observation. To investigate, we analyzed LWT and Akt-YAP1 Sox9 LKO liver tissue at 5 weeks post-HDTVI using immunofluorescence for Ki-67 to assess proliferation and IHC for TUNEL to measure cell viability (Figure 5A). Interestingly, HCC tumors lacking SOX9 showed a significant increase in the number of Ki-67+;HA-tag+ tumor cells compared to WT (Figure 5B,C), whereas there was no significant difference in cell death between the absence of SOX9 and WT (Figure 5D,E). These findings suggest that Sox9 deletion exacerbates Akt-YAP1-mediated CCA-like tumors and promotes the proliferation of HCC, leading to a significantly larger tumor burden and reduced survival. This implies a distinct role for SOX9 in liver cancer lineage.
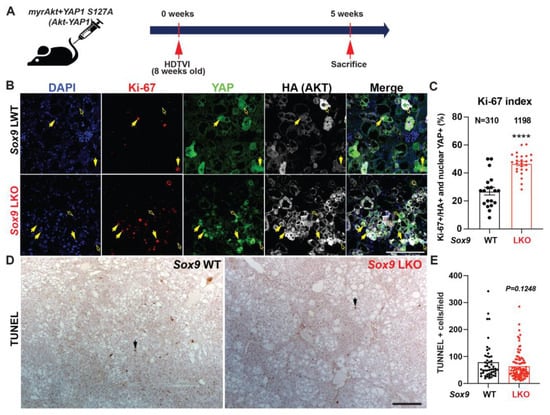
Figure 5.
Developmental removal of Sox9 promotes proliferation of Akt-YAP1-mediated liver cancer. (A) Experimental design illustrating plasmids used for HDTVI, mice used in study, and time-points analyzed. (B) Representative IF for Ki-67 (red), YAP1 (green), HA-tag (gray), and DAPI (blue) in liver sections from 5 weeks for Akt-YAP1 LWT or Sox9 LKO. Yellow arrows indicate Ki-67+;HA−tag+;YAP+ proliferating liver cancer cells and Yellow empty arrows point to Ki-67−;HA-tag+;YAP+ non-proliferating liver cancer cells. (C) The percentage of Ki-67-positive nuclei normalized to HA-tag-positive total tumor cell nuclei in representative images shown in B demonstrate significant increase in proliferation of transduced tumor cells in Sox9 LKO as compared to LWT. (D) IHC for TUNEL to detect non-viable tumor cells shows comparable cell death was evident between Sox9 LKO and LWT in Akt-YAP1 model at 5-week time-point. Black arrows indicate nuclear TUNEL-positive apoptotic cells. (E) The number of TUNEL-positive nuclei normalized to field in representative images shown in D demonstrates comparable cell death in Sox9 LKO as compared to WT. Scale bars: 100 µm; error bar: standard error of the mean; **** p < 0.0001.
3.5. Tumor-Specific Acute Sox9 Loss Represses YAP1 or NRAS-Dependent cHCC-CCA Development
Recently, there have been several publications demonstrating phenotypic differences between chronic developmental gene deletion in the liver using the Albumin (Alb)-Cre strain and acute gene deletion mediated by the AAV8-Tbg-Cre/CRISPR-Cas9 system [,], suggesting an adaptation of HC and BEC against developmental gene deletion. Notably, Sox9 deletion in HC and BEC from E15 days using Alb-Cre;Sox9(f/f) animals results in delayed bile duct formation [,], indicating a compensatory mechanism. This suggests a potential disparity between acute Sox9 elimination and the developmental adaptation observed in Alb-Cre;Sox9(f/f) animals [,]. These observations prompted us to investigate the effect of Sox9 deletion concurrently with oncogenic events in the malignant transformation of HCs. To address this, we utilized an SB-based CRISPR/Cas9-mediated inducible Sox9 deletion vector system applicable via HDTVI delivery (Figure 6B). We employed two validated HC-derived cHCC-CCA models driven by two different combinations of oncogenes. We delivered SB-STOP(flf)−Cas9-sg-Sox9 and Cre expression plasmids to specifically eliminate Sox9 in tumors along with Akt-YAP1 or Akt-NRAS to induce cHCC-CCA in the presence (Sox9 CWT) or absence of Sox9 (Sox9 CKO).
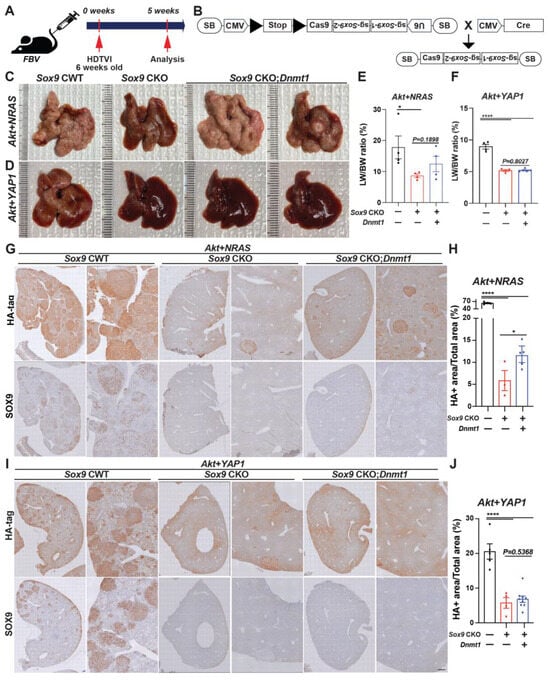
Figure 6.
Acute Sox9 deletion prevents the formation of combined HCC-CCA mediated by Akt-YAP1 or Akt-NRAS, although Akt-NRAS tumor exhibit partial dependence on Dnmt1. (A) Experimental design illustrating plasmids used for HDTVI, mice used in study, and time-points analyzed. (B) A model illustrating the experimental design utilizing sleeping beauty transposon/transposase-CRISPR/Cas9-based inducible Sox9 knockout plasmid. Representative gross images from Akt-NRAS (C) or Akt-YAP1 (D)-injected WT (CWT), acute Sox9-knockout (CKO), and Dnmt1-injected Sox9-CKO (Sox9 CKO-Dnmt1) livers showing tumor burden. (E) LW/BW ratio depicts significantly lower tumor burden in Akt-NRAS Sox9 CKO and Akt-NRAS Sox9 CKO-Dnmt1 mice as compared to CWT at 5 weeks. (F) LW/BW ratio depicts significantly lower tumor burden in Akt-YAP1 Sox9 CKO and Akt-YAP1 Sox9 CKO-Dnmt1 mice as compared to CWT at 5 weeks. (G,H) Representative IHC images of tumor-bearing livers at 5 weeks in Akt-NRAS CWT, Akt-NRAS Sox9 CKO, and Akt-NRAS Sox9 CKO-Dnmt1 liver stained for HA-tag and SOX9 showing cHCC-CCA component. HA-tag+ Akt-NRAS cHCC/CCA tumor burden was robustly abrogated in Sox9 CKO livers while being slightly but significantly restored in Dnmt1-injected Sox9-CKO livers. (I,J) Representative IHC images of tumor-bearing livers at 5 weeks in Akt-YAP1 CWT, Akt-YAP1 Sox9 CKO, and Akt-YAP1 Sox9 CKO-Dnmt1 liver stained for HA-tag and SOX9 showing cHCC-CCA component. HA-tag+ Akt-YAP1 cHCC/CCA tumor burden was robustly abrogated in both Sox9 CKO and Sox9 CKO-Dnmt1 livers. Each dot in the graphs represent an individual mouse. Scale bars: 100 µm. Error bar: standard error of the mean; * p < 0.05; **** p < 0.0001.
Additionally, we previously reported the indispensable roles of DNMT1 in repressing the transcription of HC-specific transcription factors, which is crucial for the biliary fate commitment during Notch- or YAP1-mediated CCA cancer formation. We also showed that DNMT1 inhibition eliminates the Akt-YAP1-CCA region while leaving the HCC region intact, indicating that DNMT1 is required for AKT-YAP-mediated HC-to-CCA reprogramming but is dispensable for HC transformation into HCC. Since the effect of Sox9 deletion in the AY model is similar to that of DNMT1 inhibition, we investigated whether there is any crosstalk between SOX9 and DNMT1 in the Akt-YAP1 and Akt-NRAS models. To explore the involvement of DNMT1 in the Sox9-mediated reduction in AY/AN-cHCC-CCA, we delivered a plasmid expressing full-length Dnmt1 into Sox9 CKO mice to investigate its association with SOX9 during liver cancer development. We carefully evaluated tumor formation and examined tumor characteristics at 5 weeks post-HDTVI (Figure 6A). All CWT mice developed a significant burden of liver cancer, with liver LW/BW reaching 30 for Akt-NRAS (Figure 6C,E) and 8 for Akt-YAP1 (Figure 6D,F) animals, respectively. Remarkably, Cas9-mediated acute Sox9 disruption significantly suppressed both Akt-YAP1 and Akt-NRAS-driven cHCC-CCA development regardless of Dnmt1 delivery, as all Sox9 CKO mice remained healthy at 5 weeks when they were sacrificed for comparison to CWT mice. Grossly, Akt-YAP1 Sox9 CKO or Akt-NRAS Sox9 CKO mice showed only rare tumors and significantly lower LW/BW ratios compared to the widespread gross disease in CWT (Figure 6C–F). Microscopic observation also depicts the significant decrease in HA-tag+ Akt-NRAS and Akt-YAP1 cHCC/CCA tumors in Sox9 CKO livers, supporting gross and LW/BW observations (Figure 6G,I). However, Dnmt1 re-expression in Sox9 CKO livers slightly but significantly restored tumor formation driven by Akt-NRAS (Figure 6H), but not Akt-YAP1 (Figure 6J), implying the partial involvement of Dnmt1 in Akt-NRAS tumor development under the SOX9. Importantly, SOX9 IHC confirmed effective elimination both in Sox9 CKO and Sox9 CKO-Dnmt1 livers, indicating successful Sox9 deletion using our plasmid system. Together, in contrast to Alb-Cre strain-driven developmental Sox9 removal, tumor-specific acute Sox9 elimination robustly prevents Akt-YAP1- or Akt-NRAS-mediated cHCC/CCA formation, irrespective of the lineage of liver cancer, while Dnmt1 is partially responsible for Sox9 contribution in Akt-NRAS liver cancer development. These data may imply the existence of the adaptive compensation of HCs against liver-specific developmental deletion of Sox9, which induces a distinct response against Sox9 elimination, necessitating the examination of the therapeutic potential of Sox9 targeting at an advanced stage.
3.6. Therapeutic Deletion of Sox9 Significantly Reduces Advanced Akt-YAP1 Liver Cancer
To assess the therapeutic effect of Sox9 deletion in advanced Akt-YAP1 or Akt-NRAS cHCC/CCA, we employed Osteopontin (OPN)-CreERT2 strains, a well-validated BEC-specific Tamoxifen (TM)-inducible Cre expression system []. As previously confirmed by other groups, the administration of triple intraperitoneal (i.p.) injections of 100 mg/kg of TM effectively induces the Cre-mediated deletion of the Stop cassette, along with the floxed forms of Tdtomato reporter, as evidenced by the absence of RFP in the corresponding CCA and cholangiocyte (manuscript submitted elsewhere). Given the widespread expression of SOX9 in fully developed Akt-YAP1 or Akt-NRAS CCA regions as well as poorly differentiated HCC regions (Figure 2B), we delivered SB-STOP(flf)−Cas9-sg-Sox9 along with the Akt-YAP1 or Akt-NRAS plasmid into OPN-CreERT2 mice to induce liver cancer. We then injected 100 mg/kg of TM (i.p.) 3–4 weeks after HDTVI, for a total of three injections, to achieve the tumor-specific elimination of Sox9 when liver cancer was fully developed at an advanced stage (Sox9 iKO). This was followed by assessments at 6–8 weeks post-HDTVI (Figure 7A). As a control, we injected TM into the WT littermates or corn oil into the OPN-CreERT2 strains (Sox9 iWT) that were also injected with the same dose of Akt-YAP1 or Akt-NRAS along with the SB-STOP(flf)−Cas9-sg-Sox9 plasmid and sacrificed at the same stage as the experimental group with Sox9 iKO mice. Consistently, the Akt-YAP1 or Akt-NRAS iWT mice developed significant liver cancer, with LW/BW reaching 10 and 12, respectively (Figure 7B–E). Interestingly, therapeutic Sox9 iKO significantly decreased Akt-YAP1 tumor burden at 6–8 weeks post-HDTVI (Figure 7B,C), bringing the LW/BW ratios down to 4–5, which is comparable to mice without tumors, suggesting a strong therapeutic effect in Akt-YAP1-driven liver cancer. However, there was no significant tumor reduction in Akt-NRAS Sox9 iKO, with variable gross tumor presence and LW/BW ratios comparable to those of iWT mice (Figure 7D,E). Notably, a subset of CCA-like tumors in iWT liver displayed SOX9− nodules, suggesting evident leakiness of the floxed Stop cassette in our system. Despite this, the significant and robust reduction in HA-tag+ liver cancer specifically in Akt-YAP1 Sox9 iKO livers indicates the context- and oncogenic driver-dependent therapeutic potential of Sox9 elimination in advanced Akt-driven cHCC-CCA tumors (Figure 7F).
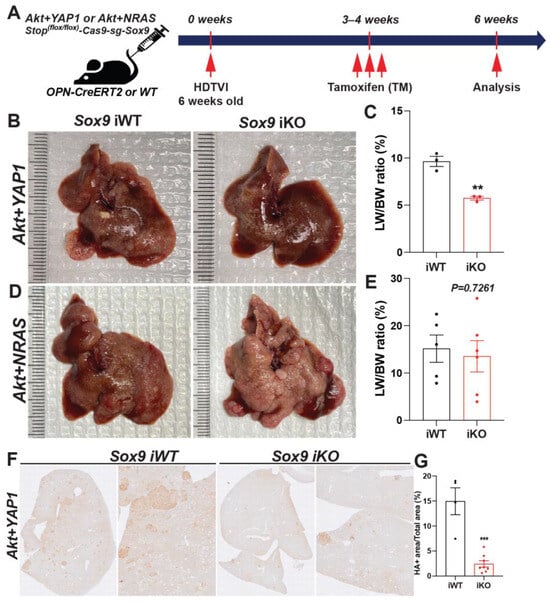
Figure 7.
Therapeutic Sox9 elimination reduces Akt-YAP1-dependent advanced combined HCC-CCA. (A) Experimental design illustrating plasmids used for HDTVI, Tamoxifen treatment, mouse strain used in study, and time-points analyzed. (B,D) Representative gross images from Akt-YAP1 (B) or Akt-NRAS (D)-injected OPN-CreERT2 mice along with Stop(f/f)−Cas9-sg-empty (Sox9-iWT) and Stop(f/f)-Cas9-sg-Sox9 (Sox9-iKO) treated with Tamoxifen (100 mg/kg triple) displaying gross tumor burden. (C,E) LW/BW ratio depicts significantly lower tumor burden in Akt-YAP1 (C) but not in Akt-NRAS (E) Sox9 iKO as compared to iWT at 6 weeks. (F) Representative IHC image of tumor-bearing livers at 6 weeks in Akt-YAP1 iWT stained for HA-tag IHC showing component of the cHCC-CCA staining. Sox9 iKO livers significantly reduced HA-tag+ tumor burden. (G) Quantification of HA-tag IHC verifies significantly reduced staining in Akt-YAP1 Sox9 iKO as compared to Akt-YAP1 Sox9 iWT at 6 weeks, as shown in (B). Each dot in the graphs represent an individual mouse. Error bar: standard error of the mean; ** p < 0.01; *** p < 0.05.
4. Discussion
Primary liver cancer, including HCC and CCA, arises from malignancies of hepatic parenchymal epithelial cells, HC and BEC, derived from the same parental cell, the hepatoblast, during development []. Interestingly, the liver exhibits a remarkable capacity for regeneration, characterized by the cellular plasticity of these two adult cell populations, involving diverse and complex molecular signaling pathways [,]. Particularly, crucial lineage-specific transcriptional regulatory components such as SOX9, HNF4α, HIPPO-YAP1, and HNF1α/β play important roles in forced cellular reprogramming both in vitro and in vivo [,,,]. Indeed, several groups have demonstrated the translation of this plasticity into cancer settings, especially in mixed HCC/iCCA and/or cHCC-CCA tumors [,,,,,]. Mixed HCC-iCCA and/or cHCC/CCA are also evident in clinical practice, and the cellular and molecular basis remains unknown []. The Akt-YAP1 but not the Akt-NICD model displayed such combined tumors, suggesting distinct roles of YAP1 and Notch signaling in cooperating with AKT activation in hepatobiliary tumorigenesis [,,]. The activation of YAP1 appears to drive a more hepatoblast/LPC-like cell fate which then evolves into either CCA or HCC [,,]. Importantly, YAP1 activation in conjunction with active-β-catenin yielded hepatoblastoma in the SB-HDTVI model []. Additionally, SOX9 was critical in directing cholangiocyte and eventually CCA cell fate in the Akt-YAP1 model since chronic Sox9 LKO drove the Akt-YAP1-reprogramed cell towards HC at the expense of biliary fate. Indeed, SOX9 has been shown to be essential for proper bile duct differentiation during hepatic development [,]. What was also unexpected was that the elimination of Sox9 in the Akt-YAP1 models not only prevented the development of the CCA component of the cHCC-CCA tumors, but it also led to a more aggressive HCC with higher proliferative index. This suggests that in the context of YAP1 activation, SOX9 may be restricting HC or HCC cell proliferation. This is a novel observation, and while SOX9 has been shown to be both upregulated [] or downregulated [] by YAP1 signaling and appears context-dependent, how SOX9 restricts YAP1 signaling and suppresses cell proliferation in transformed hepatocytes remains unknown. It should be noted that SOX9 upregulation and downregulation have both been observed in various tumors, and thus, the overall biological outcome may be tissue-dependent [,]. In our study, SOX9 has dual roles of not just regulating the biliary differentiation of HC to yield CCA, but also restricting YAP1-dependent HC proliferation, such that in its absence, the Akt-YAP1-driven HCC is more proliferative and aggressive.
A recent study led by Dr. Yang’s group demonstrated two dominant roles for SOX9 in YAP1 alone-driven liver cancer: indispensable roles in lineage determinants for HC-to-iCCA formation and responsibility for the severity of YAP1-HCC using AAV8-Cre;Sox9(f/f)-mediated Sox9 ablation []. However, we previously reported that SOX9 is dispensable in NOTCH-driven HC-to-BEC/iCCA reprogramming, whereas it is involved in tumor cell viability and proliferation in Akt-NICD HC-derived iCCA models []. This indicates context-dependent roles for SOX9 in fate control and cell viability within the mammalian liver cancer. These observations also suggest that anticipating the roles of biliary factors in clinical liver cancer settings is extremely difficult and complex due to the complicated and heterogeneous molecular signature of human liver cancers. In the current study, we mainly used the SB-HDTVI-based Akt-YAP1 and Akt-NRAS HC-derived cHCC-CCA models to investigate the roles of Sox9 in the liver cancer setting, including cHCC-CCA. Importantly, some of our observations are similar to those of YAP1 alone-driven liver cancer studies, while a large part of our investigation demonstrates distinct responses, with multiple caveats. Consistent with observations in YAP1 alone-mediated liver cancer settings, chronic developmental deletion by Sox9 LKO prior to the HDTVI delivery of Akt-YAP1 induced a switch of tumor lineage from cHCC-CCA to aggressive and poorly differentiated HCC with LPC characteristics, genetically representing a subset of clinical HCC cases. However, in contrast to Sox9 LKO or YAP1 alone-mediated liver cancer studies, acute Sox9 disruption using the CRISPR/Cas9 system robustly repressed Akt-YAP1 cHCC-CCA formation, irrespective of tumor fate.
These discrepancies raise several caveats that need to be carefully considered in murine liver cancer models. In particular, the difference between Sox9 LKO and Sox9 CKO may underscore the importance of hepatic adaptation against the deletion of the genes involved in hepatic competence and specification during development, Sox9 in the current case, which may be responsible for the dependency cHCC-CCA formations. From this perspective, a comprehensive investigation of the adaptive roles for validated compensation genes will be pertinent studies to elucidate the mechanism behind these differences. Furthermore, the distinct tumor microenvironment between the HDTVI delivery of YAP1 and non-invasive TET-ON YAP1 expression in HC used for the YAP1-alone liver cancer model, along with the collaboration of constitutive active AKT, needs to be carefully compared to conclude these different observations.
Previously, we reported that the NOTCH-YAP1-DNMT1 axis plays indispensable and permissive roles in HC reprogramming into CCA, whereas DNMT1 is dispensable for the maintenance of CCA []. Given that Sox9 LKO specifically abrogates the Akt-YAP1 CCA region, similar to pharmacologic DNMT1 inhibition, we posited an association between SOX9 and DNMT1 in CCA fate commitment in our cHCC-CCA models. Interestingly, SOX9-DNMT1 is partially involved only in Akt-NRAS tumor formation but not in Akt-YAP1 tumor development. This may suggest that DNMT1 and SOX9 are independently regulated under YAP1-driven HC-originated CCA tumor region, while the NRAS-SOX9-DNMT1 signaling cascade may be active in Akt-NRAS CCA tumor cells. However, further functional validation, including examining the association of YAP1 or SOX9 with pharmacologic/genetic DNMT1 regulation on Akt-NRAS CCA, will be an interesting focus for future studies.
Importantly, we observed the successful formation of SOX9− LPC-like immature CCA nodules in Akt-YAP1 Sox9 LKO livers at an early stage at 2 weeks post-HDTVI, expressing equivalent markers to SOX9+ CCA-like nodules in Sox9 WT livers, which is similar with the case of Akt-NICD Sox9 KO livers forming SOX9− AKT-NICD CCA nodules. However, we revealed that SOX9 is specifically required for the maintenance of fully developed Akt-YAP1 but not Akt-NICD CCA tumors. This further supports our claim of overlapping and/or context/stage-dependent roles for SOX9 in liver cancer including CCA.
Lastly, considering the significant therapeutic effect of Sox9 ablation in advanced Akt-YAP1 cHCC-CCA, similar to our Sox9 CKO (acute deletion) data but contrasting with Sox9 LKO (developmental deletion) in a preventive manner, there is a need to revisit tumor studies conducted using developmental Cre strains-mediated gene KO systems, which may overlook adaptation. This suggests the necessity for future studies to reevaluate therapeutic potentials, anticipating distinct response impact information, as preclinical data translates into relevance for human cancer patients.
5. Conclusions
Our study underscores the complex, context-dependent roles of SOX9 in the development and progression of cHCC-CCA. The contrasting outcomes observed between developmental Sox9 elimination and tumor-specific acute Sox9 ablation highlight the nuanced functions of SOX9 in liver cancer biology. Our findings advocate the need for careful consideration when using developmental Cre models in liver cancer research. Moreover, our results point to the potential for precise, personalized therapeutic strategies targeting SOX9 in treating clinical cHCC-CCA. Further research is essential to translate these preclinical insights into effective clinical treatments.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/cells13171451/s1: File S1. Online Supplementary. Table S1. List of antibodies for IHC and IF in this study. Primary Antibodies. Table S2. Secondary Antibodies. Tabel S3. Nucleotide sequence of primers used in this. File S2. Clinical and demographic patient information for 108 cholangiocarcinoma patients from UPMC cohort [,,,,,,,,,,,,,,].
Author Contributions
Conceptualization, S.K.; methodology, S.H., Y.P., M.K., M.O., A.S., S.P.M. and S.L.; formal analysis, S.H., Y.P., M.K. and S.L.; investigation, S.H., Y.P., M.K., and M.O.; resources, S.K., A.S. and S.P.M.; data curation, S.H., Y.P., M.K. and M.O.; writing—original draft preparation, S.K.; writing—review and editing, S.K.; visualization, S.K. and Y.P. supervision, S.K.; project administration, S.K.; funding acquisition, S.K., S.L., S.P.M. and M.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NIH grants R01CA258449 to S.K. and 1P30DK120531-01 to Pittsburgh Liver Research Center (PLRC) and an Innovation in Cancer Informatics Discover grant (https://www.the-ici-fund.org, accessed on 1 January 2024) to S.K. and S.L., and partly by R01 DK130949 to M.O. This work was partially supported by NIH grants R01CA251155 and R01CA250227 to S.P.M.
Institutional Review Board Statement
All human tissue samples were provided by Pittsburgh Liver Research Center’s (PLRC’s) Clinical Biospecimen Repository and Processing Core supported by P30DK120531 under approved Institutional Review Board STUDY19070068.
Informed Consent Statement
Informed consent was waived under the Pittsburgh Liver Research Center’s Clinical Biospecimen Repository and Processing, as approved by the Institutional Review Board (IRB) under STUDY19070068.
Data Availability Statement
RNA-seq data have been submitted to the online database gene expression omnibus (GEO) accession ID: GSE200472.
Acknowledgments
We extend special thanks to Daniela Sia at Icahn School of Medicine for her kind guidance in bioinformatic analysis.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gigante, E.; Paradis, V.; Ronot, M.; Cauchy, F.; Soubrane, O.; Ganne-Carrie, N.; Nault, J.C. New insights into the pathophysiology and clinical care of rare primary liver cancers. JHEP Rep. 2021, 3, 100174. [Google Scholar] [CrossRef] [PubMed]
- Moeini, A.; Sia, D.; Zhang, Z.; Camprecios, G.; Stueck, A.; Dong, H.; Montal, R.; Torrens, L.; Martinez-Quetglas, I.; Fiel, M.I.; et al. Mixed hepatocellular cholangiocarcinoma tumors: Cholangiolocellular carcinoma is a distinct molecular entity. J. Hepatol. 2017, 66, 952–961. [Google Scholar] [CrossRef] [PubMed]
- Rossner, F.; Sinn, B.V.; Horst, D. Pathology of Combined Hepatocellular Carcinoma-Cholangiocarcinoma: An Update. Cancers 2023, 15, 494. [Google Scholar] [CrossRef] [PubMed]
- Moeini, A.; Haber, P.K.; Sia, D. Cell of origin in biliary tract cancers and clinical implications. JHEP Rep. 2021, 3, 100226. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Ro, J.Y. Combined Hepatocellular-Cholangiocarcinoma: An Update on Pathology and Diagnostic Approach. Biomedicines 2022, 10, 1826. [Google Scholar] [CrossRef]
- Feng, M.; Pan, Y.; Kong, R.; Shu, S. Therapy of Primary Liver Cancer. Innovation 2020, 1, 100032. [Google Scholar] [CrossRef]
- Brindley, P.J.; Bachini, M.; Ilyas, S.I.; Khan, S.A.; Loukas, A.; Sirica, A.E.; Teh, B.T.; Wongkham, S.; Gores, G.J. Cholangiocarcinoma. Nat. Rev. Dis. Primers 2021, 7, 65. [Google Scholar] [CrossRef]
- Hsu, B.Y.; Driscoll, J.; Tateno, C.; Mattis, A.N.; Kelley, R.K.; Willenbring, H. Human Hepatocytes Can Give Rise to Intrahepatic Cholangiocarcinomas. Gastroenterology 2024, in press. [Google Scholar] [CrossRef]
- Wang, H.; Wang, J.; Zhang, S.; Jia, J.; Liu, X.; Zhang, J.; Wang, P.; Song, X.; Che, L.; Liu, K.; et al. Distinct and Overlapping Roles of Hippo Effectors YAP and TAZ During Human and Mouse Hepatocarcinogenesis. Cell Mol. Gastroenterol. Hepatol. 2021, 11, 1095–1117. [Google Scholar] [CrossRef]
- Wang, J.; Dong, M.; Xu, Z.; Song, X.; Zhang, S.; Qiao, Y.; Che, L.; Gordan, J.; Hu, K.; Liu, Y.; et al. Notch2 controls hepatocyte-derived cholangiocarcinoma formation in mice. Oncogene 2018, 37, 3229–3242. [Google Scholar] [CrossRef]
- Matter, M.S.; Marquardt, J.U.; Andersen, J.B.; Quintavalle, C.; Korokhov, N.; Stauffer, J.K.; Kaji, K.; Decaens, T.; Quagliata, L.; Elloumi, F.; et al. Oncogenic driver genes and the inflammatory microenvironment dictate liver tumor phenotype. Hepatology 2016, 63, 1888–1899. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.; Wang, C.; Mattu, S.; Destefanis, G.; Ladu, S.; Delogu, S.; Armbruster, J.; Fan, L.; Lee, S.A.; Jiang, L.; et al. AKT (v-akt murine thymoma viral oncogene homolog 1) and N-Ras (neuroblastoma ras viral oncogene homolog) coactivation in the mouse liver promotes rapid carcinogenesis by way of mTOR (mammalian target of rapamycin complex 1), FOXM1 (forkhead box M1)/SKP2, and c-Myc pathways. Hepatology 2012, 55, 833–845. [Google Scholar] [CrossRef]
- Hu, S.; Molina, L.; Tao, J.; Liu, S.; Hassan, M.; Singh, S.; Poddar, M.; Bell, A.; Sia, D.; Oertel, M.; et al. NOTCH-YAP1/TEAD-DNMT1 Axis Drives Hepatocyte Reprogramming Into Intrahepatic Cholangiocarcinoma. Gastroenterology 2022, 163, 449–465. [Google Scholar] [CrossRef]
- Liu, Y.; Zhuo, S.; Zhou, Y.; Ma, L.; Sun, Z.; Wu, X.; Wang, X.W.; Gao, B.; Yang, Y. Yap-Sox9 signaling determines hepatocyte plasticity and lineage-specific hepatocarcinogenesis. J. Hepatol. 2022, 76, 652–664. [Google Scholar] [CrossRef] [PubMed]
- Poncy, A.; Antoniou, A.; Cordi, S.; Pierreux, C.E.; Jacquemin, P.; Lemaigre, F.P. Transcription factors SOX4 and SOX9 cooperatively control development of bile ducts. Dev. Biol. 2015, 404, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Panikkar, A.; Xu, J.; Antoniou, A.; Raynaud, P.; Lemaigre, F.; Stanger, B.Z. Notch signaling controls liver development by regulating biliary differentiation. Development 2009, 136, 1727–1739. [Google Scholar] [CrossRef]
- Yin, C. Molecular mechanisms of Sox transcription factors during the development of liver, bile duct, and pancreas. Semin. Cell Dev. Biol. 2017, 63, 68–78. [Google Scholar] [CrossRef]
- Weisend, C.M.; Kundert, J.A.; Suvorova, E.S.; Prigge, J.R.; Schmidt, E.E. Cre activity in fetal albCre mouse hepatocytes: Utility for developmental studies. Genesis 2009, 47, 789–792. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341.e23. [Google Scholar] [CrossRef]
- Bult, C.J.; Blake, J.A.; Smith, C.L.; Kadin, J.A.; Richardson, J.E.; Mouse Genome Database Group. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 2019, 47, D801–D806. [Google Scholar] [CrossRef]
- Hoshida, Y. Nearest template prediction: A single-sample-based flexible class prediction with confidence assessment. PLoS ONE 2010, 5, e15543. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y.; Nijman, S.M.; Kobayashi, M.; Chan, J.A.; Brunet, J.P.; Chiang, D.Y.; Villanueva, A.; Newell, P.; Ikeda, K.; Hashimoto, M.; et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009, 69, 7385–7392. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.G.; Lee, J.H.; Yoon, J.H.; Kim, C.Y.; Lee, H.S.; Jang, J.J.; Yi, N.J.; Suh, K.S.; Lee, K.U.; Park, E.S.; et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 2010, 70, 3034–3041. [Google Scholar] [CrossRef] [PubMed]
- Pibiri, M.; Simbula, G. Role of the Hippo pathway in liver regeneration and repair: Recent advances. Inflamm. Regen. 2022, 42, 59. [Google Scholar] [CrossRef]
- Hu, S.; Cao, C.; Poddar, M.; Delgado, E.; Singh, S.; Singh-Varma, A.; Stolz, D.B.; Bell, A.; Monga, S.P. Hepatocyte β-catenin loss is compensated by Insulin-mTORC1 activation to promote liver regeneration. Hepatology 2023, 77, 1593–1611. [Google Scholar] [CrossRef]
- Antoniou, A.; Raynaud, P.; Cordi, S.; Zong, Y.; Tronche, F.; Stanger, B.Z.; Jacquemin, P.; Pierreux, C.E.; Clotman, F.; Lemaigre, F.P. Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology 2009, 136, 2325–2333. [Google Scholar] [CrossRef]
- Xu, W.P.; Cui, Y.L.; Chen, L.L.; Ding, K.; Ding, C.H.; Chen, F.; Zhang, X.; Xie, W.F. Deletion of Sox9 in the liver leads to hepatic cystogenesis in mice by transcriptionally downregulating Sec63. J. Pathol. 2021, 254, 57–69. [Google Scholar] [CrossRef]
- Lesaffer, B.; Verboven, E.; Van Huffel, L.; Moya, I.M.; van Grunsven, L.A.; Leclercq, I.A.; Lemaigre, F.P.; Halder, G. Comparison of the Opn-CreER and Ck19-CreER Drivers in Bile Ducts of Normal and Injured Mouse Livers. Cells 2019, 8, 380. [Google Scholar] [CrossRef]
- Ko, S.; Russell, J.O.; Molina, L.M.; Monga, S.P. Liver Progenitors and Adult Cell Plasticity in Hepatic Injury and Repair: Knowns and Unknowns. Annu. Rev. Pathol. 2020, 15, 23–50. [Google Scholar] [CrossRef]
- Michalopoulos, G.K.; Bhushan, B. Liver regeneration: Biological and pathological mechanisms and implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 40–55. [Google Scholar] [CrossRef]
- Jiang, M.; Ren, J.; Belmonte, J.C.I.; Liu, G.H. Hepatocyte reprogramming in liver regeneration: Biological mechanisms and applications. FEBS J. 2023, 290, 5674–5688. [Google Scholar] [CrossRef] [PubMed]
- Romanazzo, S.; Lin, K.; Srivastava, P.; Kilian, K.A. Targeting cell plasticity for regeneration: From in vitro to in vivo reprogramming. Adv. Drug. Deliv. Rev. 2020, 161-162, 124–144. [Google Scholar] [CrossRef]
- Gadd, V.L.; Aleksieva, N.; Forbes, S.J. Epithelial Plasticity during Liver Injury and Regeneration. Cell Stem Cell 2020, 27, 557–573. [Google Scholar] [CrossRef]
- Li, W.; Li, L.; Hui, L. Cell Plasticity in Liver Regeneration. Trends Cell Biol. 2020, 30, 329–338. [Google Scholar] [CrossRef]
- Cigliano, A.; Zhang, S.; Ribback, S.; Steinmann, S.; Sini, M.; Ament, C.E.; Utpatel, K.; Song, X.; Wang, J.; Pilo, M.G.; et al. The Hippo pathway effector TAZ induces intrahepatic cholangiocarcinoma in mice and is ubiquitously activated in the human disease. J. Exp. Clin. Cancer Res. 2022, 41, 192. [Google Scholar] [CrossRef]
- Kim, M.; Delgado, E.; Ko, S. DNA methylation in cell plasticity and malignant transformation in liver diseases. Pharmacol. Ther. 2023, 241, 108334. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.; Kim, M.; Molina, L.; Sirica, A.E.; Monga, S.P. YAP1 activation and Hippo pathway signaling in the pathogenesis and treatment of intrahepatic cholangiocarcinoma. Adv. Cancer Res. 2022, 156, 283–317. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, H.; Cui, G.; Liang, B.; Chen, X.; Ko, S.; Affo, S.; Song, X.; Liao, Y.; Feng, J.; et al. beta-Catenin Sustains and Is Required for YES-associated Protein Oncogenic Activity in Cholangiocarcinoma. Gastroenterology 2022, 163, 481–494. [Google Scholar] [CrossRef]
- Razumilava, N.; Gores, G.J. Cholangiocarcinoma. Lancet 2014, 383, 2168–2179. [Google Scholar] [CrossRef]
- Driskill, J.H.; Pan, D. The Hippo Pathway in Liver Homeostasis and Pathophysiology. Annu. Rev. Pathol. 2021, 16, 299–322. [Google Scholar] [CrossRef]
- Yamamoto, M.; Xin, B.; Watanabe, K.; Ooshio, T.; Fujii, K.; Chen, X.; Okada, Y.; Abe, H.; Taguchi, Y.; Miyokawa, N.; et al. Oncogenic Determination of a Broad Spectrum of Phenotypes of Hepatocyte-Derived Mouse Liver Tumors. Am. J. Pathol. 2017, 187, 2711–2725. [Google Scholar] [CrossRef]
- Russell, J.O.; Camargo, F.D. Hippo signalling in the liver: Role in development, regeneration and disease. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 297–312. [Google Scholar] [CrossRef]
- Zhang, S.; Song, X.; Cao, D.; Xu, Z.; Fan, B.; Che, L.; Hu, J.; Chen, B.; Dong, M.; Pilo, M.G.; et al. Pan-mTOR inhibitor MLN0128 is effective against intrahepatic cholangiocarcinoma in mice. J. Hepatol. 2017, 67, 1194–1203. [Google Scholar] [CrossRef]
- Tao, J.; Calvisi, D.F.; Ranganathan, S.; Cigliano, A.; Zhou, L.; Singh, S.; Jiang, L.; Fan, B.; Terracciano, L.; Armeanu-Ebinger, S.; et al. Activation of beta-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology 2014, 147, 690–701. [Google Scholar] [CrossRef]
- Song, S.; Ajani, J.A.; Honjo, S.; Maru, D.M.; Chen, Q.; Scott, A.W.; Heallen, T.R.; Xiao, L.; Hofstetter, W.L.; Weston, B.; et al. Hippo coactivator YAP1 upregulates SOX9 and endows esophageal cancer cells with stem-like properties. Cancer Res. 2014, 74, 4170–4182. [Google Scholar] [CrossRef]
- Goto, H.; Nishio, M.; To, Y.; Oishi, T.; Miyachi, Y.; Maehama, T.; Nishina, H.; Akiyama, H.; Mak, T.W.; Makii, Y.; et al. Loss of Mob1a/b in mice results in chondrodysplasia due to YAP1/TAZ-TEAD-dependent repression of SOX9. Development 2018, 145, dev159244. [Google Scholar] [CrossRef]
- Aguilar-Medina, M.; Avendano-Felix, M.; Lizarraga-Verdugo, E.; Bermudez, M.; Romero-Quintana, J.G.; Ramos-Payan, R.; Ruiz-Garcia, E.; Lopez-Camarillo, C. SOX9 Stem-Cell Factor: Clinical and Functional Relevance in Cancer. J. Oncol. 2019, 2019, 6754040. [Google Scholar] [CrossRef]
- Tripathi, S.K.; Sahoo, R.K.; Biswal, B.K. SOX9 as an emerging target for anticancer drugs and a prognostic biomarker for cancer drug resistance. Drug Discov. Today 2022, 27, 2541–2550. [Google Scholar] [CrossRef]
- Tao, J.; Zhang, R.; Singh, S.; Poddar, M.; Xu, E.; Oertel, M.; Chen, X.; Ganesh, S.; Abrams, M.; Monga, S.P. Targeting beta-catenin in hepatocellular cancers induced by coexpression of mutant beta-catenin and K-Ras in mice. Hepatology 2017, 65, 1581–1599. [Google Scholar] [CrossRef]
- Fan, B.; Malato, Y.; Calvisi, D.F.; Naqvi, S.; Razumilava, N.; Ribback, S.; Gores, G.J.; Dombrowski, F.; Evert, M.; Chen, X.; et al. Cholangiocarcinomas can originate from hepatocytes in mice. J. Clin. Investig. 2012, 122, 2911–2915. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Yu, Y.P.; Tao, J.; Liu, S.; Tseng, G.; Nalesnik, M.; Hamilton, R.; Bhargava, R.; Nelson, J.B.; Pennathur, A.; et al. MAN2A1-FER Fusion Gene Is Expressed by Human Liver and Other Tumor Types and Has Oncogenic Activity in Mice. Gastroenterology 2017, 153, 1120–1132.e15. [Google Scholar] [CrossRef]
- Chen, Z.H.; Yu, Y.P.; Zuo, Z.H.; Nelson, J.B.; Michalopoulos, G.K.; Monga, S.; Liu, S.; Tseng, G.; Luo, J.H. Targeting genomic rearrangements in tumor cells through Cas9-mediated insertion of a suicide gene. Nat. Biotechnol. 2017, 35, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szczesniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC—A Quality Control Tool for High throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 26 June 2024).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Smyth, G.K. Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2005; pp. 397–420. [Google Scholar]
- Reich, M.; Liefeld, T.; Gould, J.; Lerner, J.; Tamayo, P.; Mesirov, J.P. GenePattern 2.0. Nat. Genet. 2006, 38, 500–501. [Google Scholar] [CrossRef]
- Sia, D.; Hoshida, Y.; Villanueva, A.; Roayaie, S.; Ferrer, J.; Tabak, B.; Peix, J.; Sole, M.; Tovar, V.; Alsinet, C.; et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology 2013, 144, 829–840. [Google Scholar] [CrossRef]
- Villanueva, A.; Alsinet, C.; Yanger, K.; Hoshida, Y.; Zong, Y.; Toffanin, S.; Rodriguez-Carunchio, L.; Sole, M.; Thung, S.; Stanger, B.Z.; et al. Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology 2012, 143, 1660–1669.e1667. [Google Scholar] [CrossRef]
- Oishi, N.; Kumar, M.R.; Roessler, S.; Ji, J.; Forgues, M.; Budhu, A.; Zhao, X.; Andersen, J.B.; Ye, Q.H.; Jia, H.L.; et al. Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of miR-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology 2012, 56, 1792–1803. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).