BCL2L11 Induction Mediates Sensitivity to Src and MEK1/2 Inhibition in Thyroid Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cell Culture
2.3. Cell Viability Assay
2.4. Micorarray Gene Expression Profiling and RNA Sequencing
2.5. Reverse Phase Protein Array
2.6. Immunoblotting
2.7. siRNA Experiments
2.8. Doxycycline Inducible pTREX Expression Vectors
2.9. Myristoylated AKT
2.10. Apoptosis Assays
2.11. Statistical Analysis
3. Results
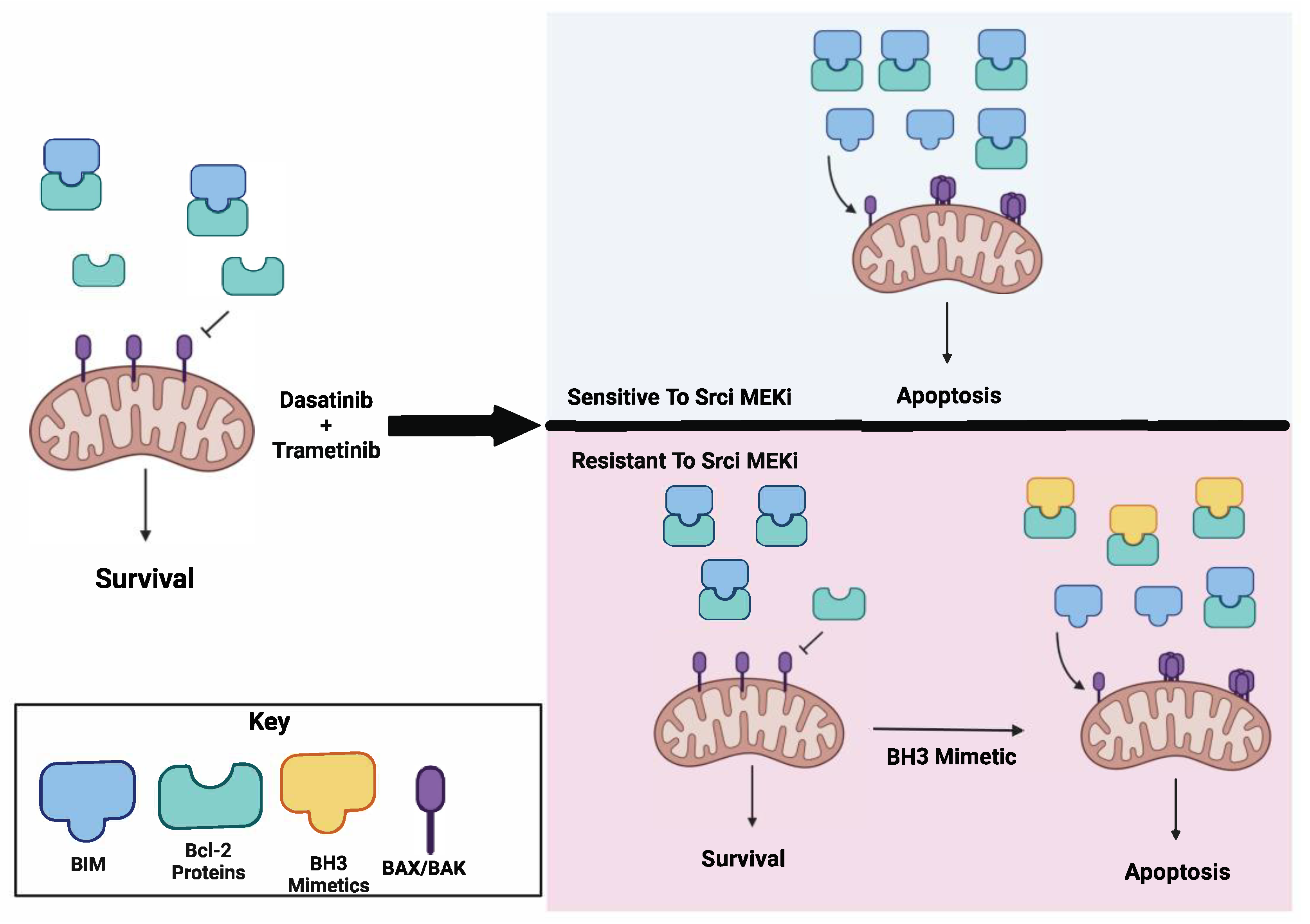
3.1. Combined Treatment with Dasatinib and Trametinib Identifies BIM as a Mediator of Sensitivity
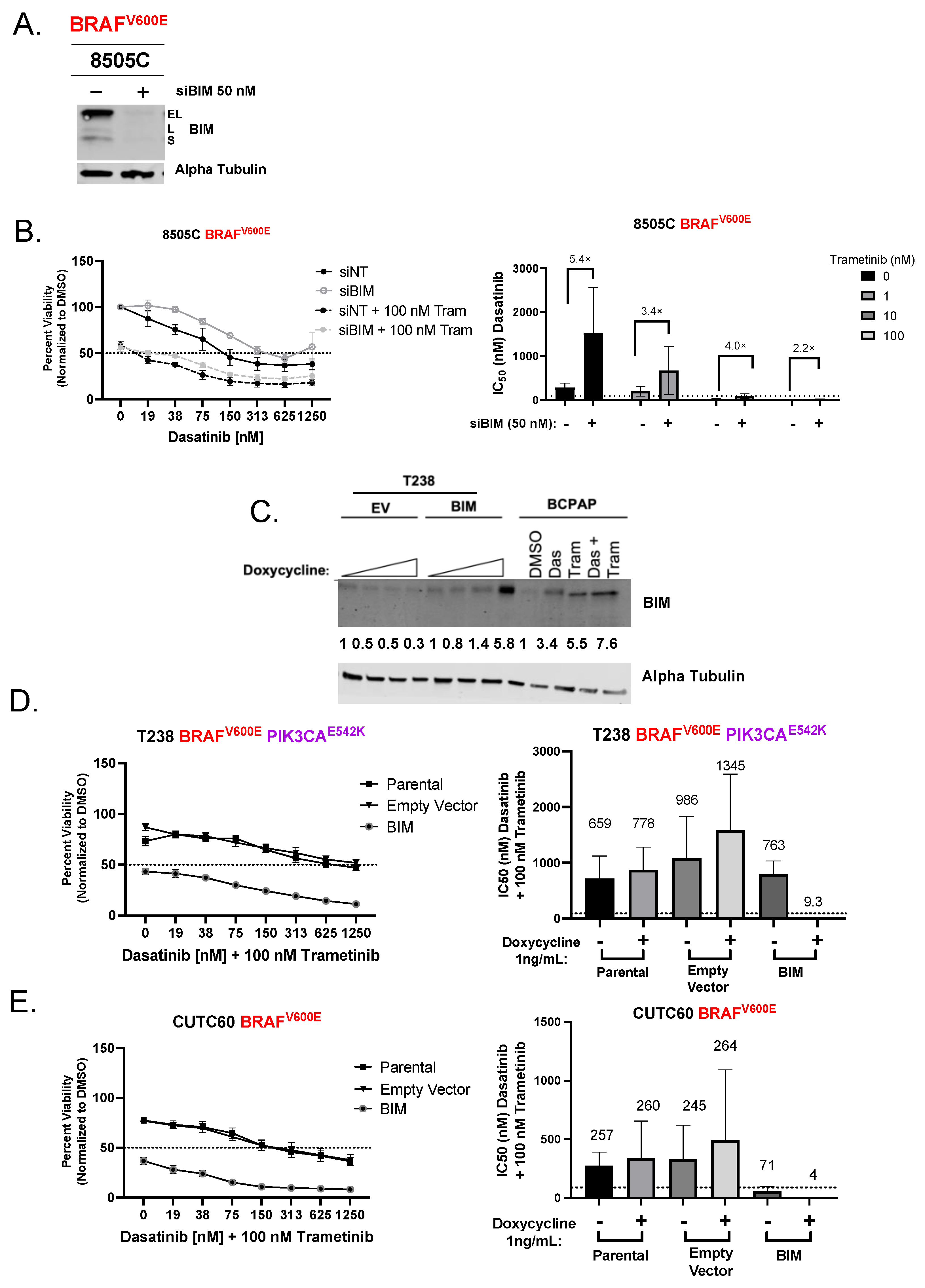
3.2. Induction of BIM Is Required for Growth Inhibition and Apoptosis Induction by Combined Dasatinib and Trametinib
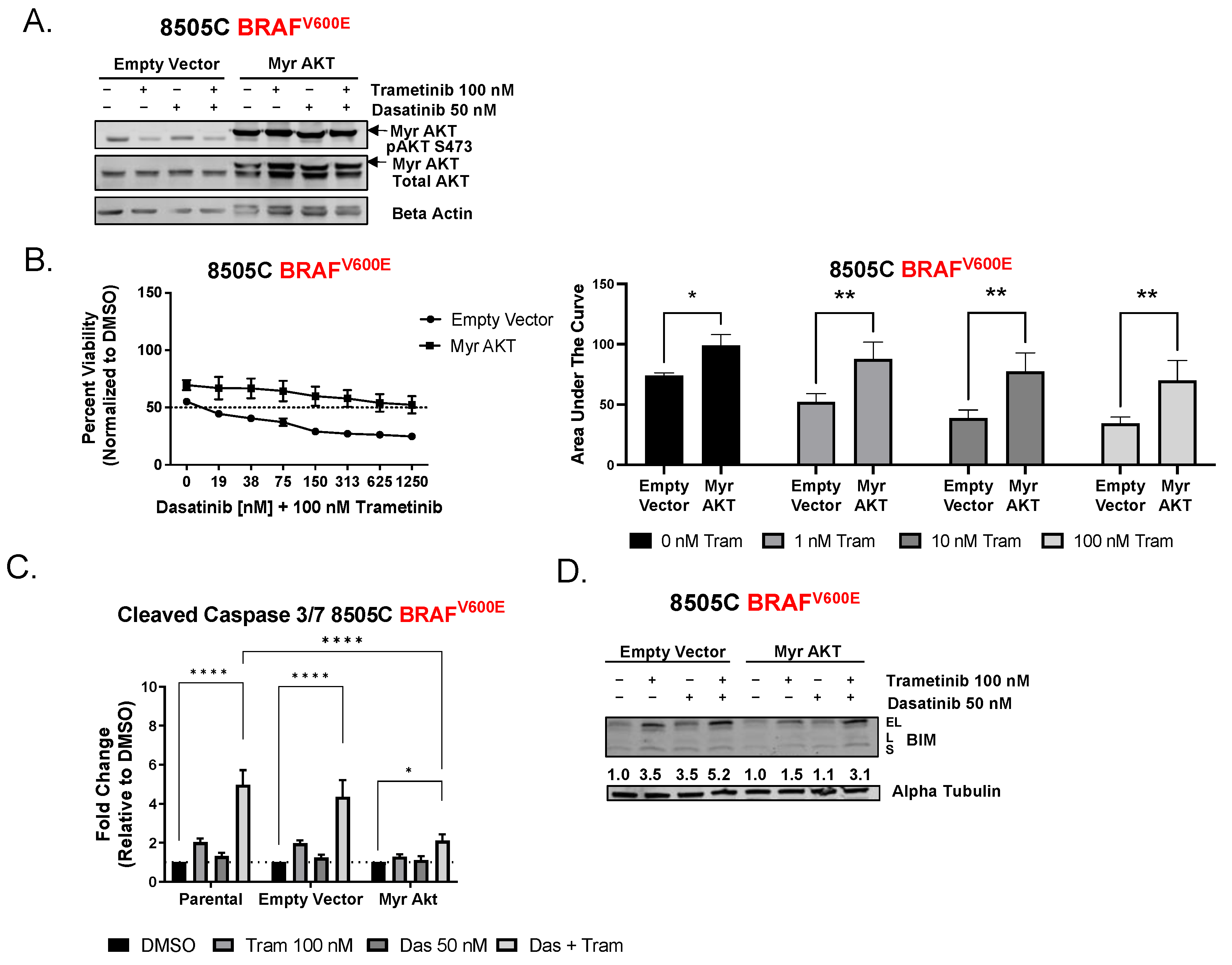
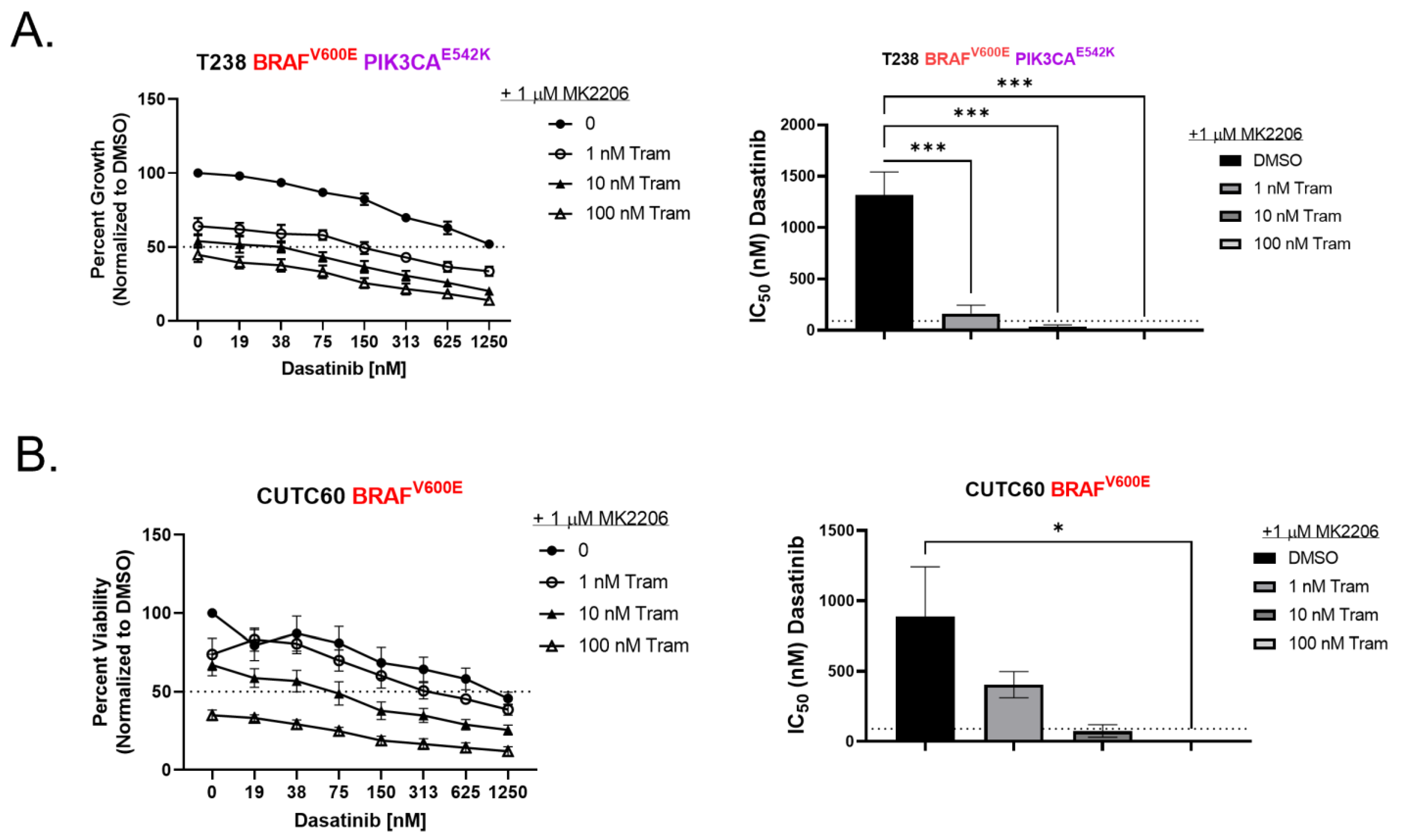
3.3. Inhibition of AKT Is Necessary for Sensitivity to Combined Dasatinib and Trametinib
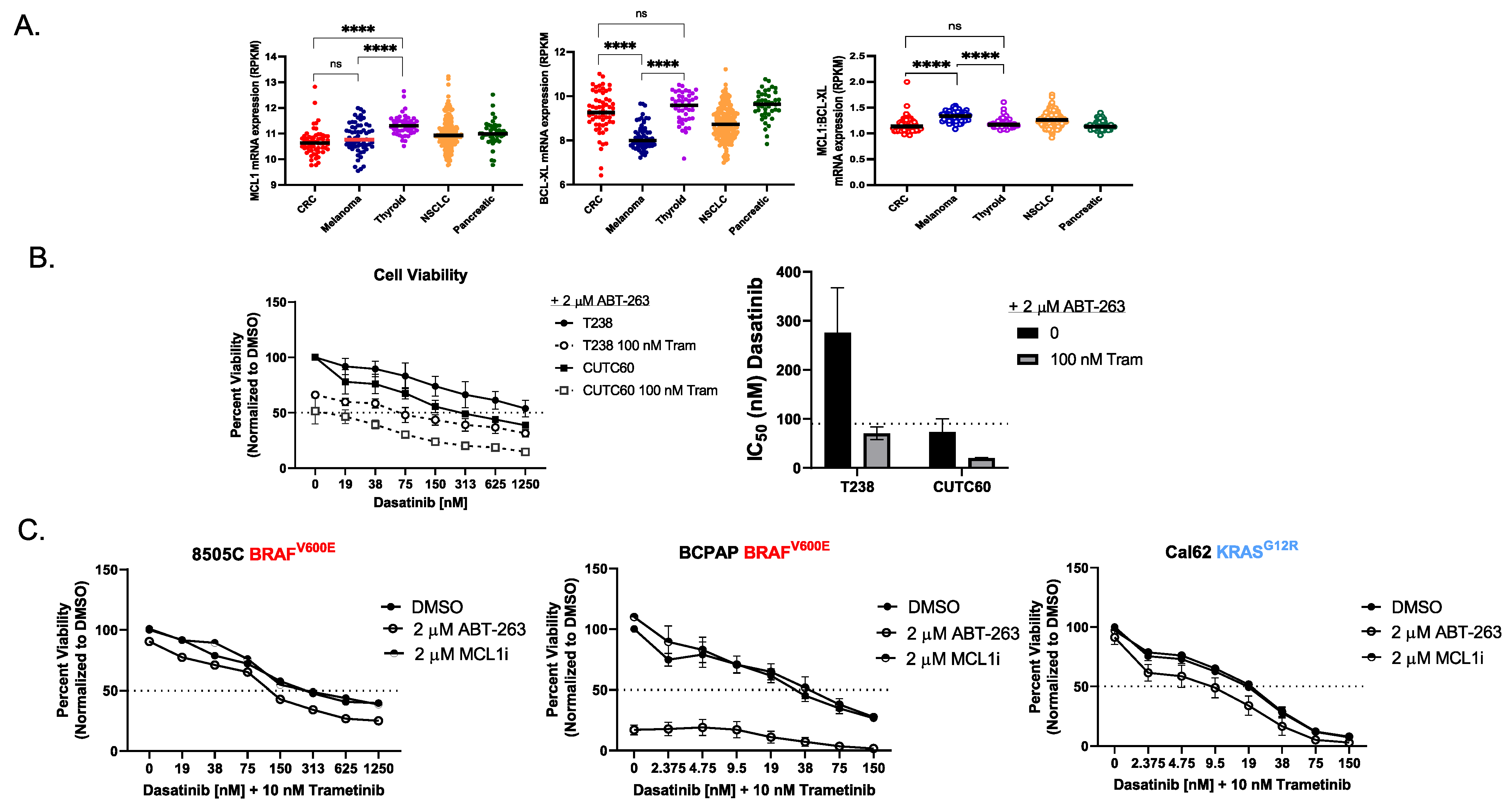
3.4. The RNA Ratio of MCL1:BCL-XL Predicts Sensitivity to the BH3 Mimetic ABT-263
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rao, S.N.; Cabanillas, M.E. Navigating Systemic Therapy in Advanced Thyroid Carcinoma: From Standard of Care to Personalized Therapy and Beyond. J. Endocr. Soc. 2018, 2, 1109–1130. [Google Scholar] [CrossRef] [PubMed]
- Prete, A.; Borges de Souza, P.; Censi, S.; Muzza, M.; Nucci, N.; Sponziello, M. Update on Fundamental Mechanisms of Thyroid Cancer. Front. Endocrinol. 2020, 11, 102. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, V.; Kreitman, R.J.; Wainberg, Z.A.; Cho, J.Y.; Schellens, J.H.M.; Soria, J.C.; Wen, P.Y.; Zielinski, C.C.; Cabanillas, M.E.; Boran, A.; et al. Dabrafenib plus trametinib in patients with <em>BRAF</em> V600E-mutant anaplastic thyroid cancer: Updated analysis from the phase II ROAR basket study. Ann. Oncol. 2022, 33, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Skoulidis, F.; Li, B.T.; Dy, G.K.; Price, T.J.; Falchook, G.S.; Wolf, J.; Italiano, A.; Schuler, M.; Borghaei, H.; Barlesi, F.; et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N. Engl. J. Med. 2021, 384, 2371–2381. [Google Scholar] [CrossRef] [PubMed]
- Pohorelic, B.; Singh, R.; Parkin, S.; Koro, K.; Yang, A.D.; Egan, C.; Magliocco, A. Role of Src in breast cancer cell migration and invasion in a breast cell/bone-derived cell microenvironment. Breast Cancer Res. Treat. 2012, 133, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, F.; Wang, C.; Wang, C.; Tang, Y.; Jiang, Z. Src Promotes Metastasis of Human Non-Small Cell Lung Cancer Cells through Fn14-Mediated NF-κB Signaling. Med. Sci. Monit. 2018, 24, 1282–1294. [Google Scholar] [CrossRef] [PubMed]
- Jin, W. Regulation of Src Family Kinases during Colorectal Cancer Development and Its Clinical Implications. Cancers (Basel) 2020, 12, 1339. [Google Scholar] [CrossRef]
- Chan, C.M.; Jing, X.; Pike, L.A.; Zhou, Q.; Lim, D.J.; Sams, S.B.; Lund, G.S.; Sharma, V.; Haugen, B.R.; Schweppe, R.E. Targeted inhibition of Src kinase with dasatinib blocks thyroid cancer growth and metastasis. Clin. Cancer Res. 2012, 18, 3580–3591. [Google Scholar] [CrossRef]
- Liu, Z.; Falola, J.; Zhu, X.; Gu, Y.; Kim, L.T.; Sarosi, G.A.; Anthony, T.; Nwariaku, F.E. Antiproliferative effects of Src inhibition on medullary thyroid cancer. J. Clin. Endocrinol. Metab. 2004, 89, 3503–3509. [Google Scholar] [CrossRef]
- Schweppe, R.E.; Kerege, A.A.; French, J.D.; Sharma, V.; Grzywa, R.L.; Haugen, B.R. Inhibition of Src with AZD0530 reveals the Src-Focal Adhesion kinase complex as a novel therapeutic target in papillary and anaplastic thyroid cancer. J. Clin. Endocrinol. Metab. 2009, 94, 2199–2203. [Google Scholar] [CrossRef]
- Beadnell, T.C.; Mishall, K.M.; Zhou, Q.; Riffert, S.M.; Wuensch, K.E.; Kessler, B.E.; Corpuz, M.L.; Jing, X.; Kim, J.; Wang, G.; et al. The Mitogen-Activated Protein Kinase Pathway Facilitates Resistance to the Src Inhibitor Dasatinib in Thyroid Cancer. Mol. Cancer Ther. 2016, 15, 1952–1963. [Google Scholar] [CrossRef]
- Beadnell, T.C.; Nassar, K.W.; Rose, M.M.; Clark, E.G.; Danysh, B.P.; Hofmann, M.C.; Pozdeyev, N.; Schweppe, R.E. Src-mediated regulation of the PI3K pathway in advanced papillary and anaplastic thyroid cancer. Oncogenesis 2018, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Ocana, A.; Gil-Martin, M.; Antolin, S.; Atienza, M.; Montano, A.; Ribelles, N.; Urruticoechea, A.; Falcon, A.; Pernas, S.; Orlando, J.; et al. Efficacy and safety of dasatinib with trastuzumab and paclitaxel in first line HER2-positive metastatic breast cancer: Results from the phase II GEICAM/2010-04 study. Breast Cancer Res. Treat. 2019, 174, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
- Landa, I.; Pozdeyev, N.; Korch, C.; Marlow, L.A.; Smallridge, R.C.; Copland, J.A.; Henderson, Y.C.; Lai, S.Y.; Clayman, G.L.; Onoda, N.; et al. Comprehensive Genetic Characterization of Human Thyroid Cancer Cell Lines: A Validated Panel for Preclinical Studies. Clin. Cancer Res. 2019, 25, 3141–3151. [Google Scholar] [CrossRef]
- Iadevaia, S.; Lu, Y.; Morales, F.C.; Mills, G.B.; Ram, P.T. Identification of optimal drug combinations targeting cellular networks: Integrating phospho-proteomics and computational network analysis. Cancer Res 2010, 70, 6704–6714. [Google Scholar] [CrossRef]
- Boehm, J.S.; Zhao, J.J.; Yao, J.; Kim, S.Y.; Firestein, R.; Dunn, I.F.; Sjostrom, S.K.; Garraway, L.A.; Weremowicz, S.; Richardson, A.L.; et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 2007, 129, 1065–1079. [Google Scholar] [CrossRef]
- Morgenstern, J.P.; Land, H. Advanced mammalian gene transfer: High titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990, 18, 3587–3596. [Google Scholar] [CrossRef]
- Weisberg, E.; Manley, P.W.; Cowan-Jacob, S.W.; Hochhaus, A.; Griffin, J.D. Second generation inhibitors of BCR-ABL for the treatment of imatinib-resistant chronic myeloid leukaemia. Nat. Rev. Cancer 2007, 7, 345–356. [Google Scholar] [CrossRef]
- Subbiah, V.; Kreitman, R.J.; Wainberg, Z.A.; Cho, J.Y.; Schellens, J.H.M.; Soria, J.C.; Wen, P.Y.; Zielinski, C.; Cabanillas, M.E.; Urbanowitz, G.; et al. Dabrafenib and Trametinib Treatment in Patients With Locally Advanced or Metastatic BRAF V600-Mutant Anaplastic Thyroid Cancer. J. Clin. Oncol. 2018, 36, 7–13. [Google Scholar] [CrossRef]
- Menzies, A.M.; Long, G.V. Dabrafenib and trametinib, alone and in combination for BRAF-mutant metastatic melanoma. Clin. Cancer Res. 2014, 20, 2035–2043. [Google Scholar] [CrossRef] [PubMed]
- Gilmartin, A.G.; Bleam, M.R.; Groy, A.; Moss, K.G.; Minthorn, E.A.; Kulkarni, S.G.; Rominger, C.M.; Erskine, S.; Fisher, K.E.; Yang, J.; et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin. Cancer Res. 2011, 17, 989–1000. [Google Scholar] [CrossRef] [PubMed]
- Rix, U.; Hantschel, O.; Dürnberger, G.; Remsing Rix, L.L.; Planyavsky, M.; Fernbach, N.V.; Kaupe, I.; Bennett, K.L.; Valent, P.; Colinge, J.; et al. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood 2007, 110, 4055–4063. [Google Scholar] [CrossRef] [PubMed]
- Kessler, B.E.; Mishall, K.M.; Kellett, M.D.; Clark, E.G.; Pugazhenthi, U.; Pozdeyev, N.; Kim, J.; Tan, A.C.; Schweppe, R.E. Resistance to Src inhibition alters the BRAF-mutant tumor secretome to promote an invasive phenotype and therapeutic escape through a FAK>p130Cas>c-Jun signaling axis. Oncogene 2019, 38, 2565–2579. [Google Scholar] [CrossRef] [PubMed]
- Westin, S.N.; Sill, M.W.; Coleman, R.L.; Waggoner, S.; Moore, K.N.; Mathews, C.A.; Martin, L.P.; Modesitt, S.C.; Lee, S.; Ju, Z.; et al. Safety lead-in of the MEK inhibitor trametinib in combination with GSK2141795, an AKT inhibitor, in patients with recurrent endometrial cancer: An NRG Oncology/GOG study. Gynecol. Oncol. 2019, 155, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Tolcher, A.W.; Patnaik, A.; Papadopoulos, K.P.; Rasco, D.W.; Becerra, C.R.; Allred, A.J.; Orford, K.; Aktan, G.; Ferron-Brady, G.; Ibrahim, N.; et al. Phase I study of the MEK inhibitor trametinib in combination with the AKT inhibitor afuresertib in patients with solid tumors and multiple myeloma. Cancer Chemother. Pharm. 2015, 75, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Tolcher, A.W.; Kurzrock, R.; Valero, V.; Gonzalez, R.; Heist, R.S.; Tan, A.R.; Means-Powell, J.; Werner, T.L.; Becerra, C.; Wang, C.; et al. Phase I dose-escalation trial of the oral AKT inhibitor uprosertib in combination with the oral MEK1/MEK2 inhibitor trametinib in patients with solid tumors. Cancer Chemother. Pharm. 2020, 85, 673–683. [Google Scholar] [CrossRef]
- Sale, M.J.; Minihane, E.; Monks, N.R.; Gilley, R.; Richards, F.M.; Schifferli, K.P.; Andersen, C.L.; Davies, E.J.; Vicente, M.A.; Ozono, E.; et al. Targeting melanoma’s MCL1 bias unleashes the apoptotic potential of BRAF and ERK1/2 pathway inhibitors. Nat. Commun. 2019, 10, 5167. [Google Scholar] [CrossRef]
- Schoenwaelder, S.M.; Jarman, K.E.; Gardiner, E.E.; Hua, M.; Qiao, J.; White, M.J.; Josefsson, E.C.; Alwis, I.; Ono, A.; Willcox, A.; et al. Bcl-xL–inhibitory BH3 mimetics can induce a transient thrombocytopathy that undermines the hemostatic function of platelets. Blood 2011, 118, 1663–1674. [Google Scholar] [CrossRef]
- Borre, P.V.; Gunda, V.; McFadden, D.G.; Sadow, P.M.; Varmeh, S.; Bernasconi, M.; Parangi, S. Combined BRAF V600E- and SRC-inhibition induces apoptosis, evokes an immune response and reduces tumor growth in an immunocompetent orthotopic mouse model of anaplastic thyroid cancer. Oncotarget 2014, 5, 3996–4010. [Google Scholar] [CrossRef]
- Koh, Y.W.; Shah, M.H.; Agarwal, K.; McCarty, S.K.; Koo, B.S.; Brendel, V.J.; Wang, C.; Porter, K.; Jarjoura, D.; Saji, M.; et al. Sorafenib and Mek inhibition is synergistic in medullary thyroid carcinoma in vitro. Endocr. Relat. Cancer 2012, 19, 29–38. [Google Scholar] [CrossRef]
- Yuan, M.; Xu, L.F.; Zhang, J.; Kong, S.Y.; Wu, M.; Lao, Y.Z.; Zhou, H.; Zhang, L.; Xu, H. SRC and MEK Co-inhibition Synergistically Enhances the Anti-tumor Effect in Both Non-small-cell Lung Cancer (NSCLC) and Erlotinib-Resistant NSCLC. Front. Oncol. 2019, 9, 586. [Google Scholar] [CrossRef]
- El Touny, L.H.; Vieira, A.; Mendoza, A.; Khanna, C.; Hoenerhoff, M.J.; Green, J.E. Combined SFK/MEK inhibition prevents metastatic outgrowth of dormant tumor cells. J. Clin. Investig. 2014, 124, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Simpkins, F.; Jang, K.; Yoon, H.; Hew, K.E.; Kim, M.; Azzam, D.J.; Sun, J.; Zhao, D.; Ince, T.A.; Liu, W.; et al. Dual Src and MEK Inhibition Decreases Ovarian Cancer Growth and Targets Tumor Initiating Stem-Like Cells. Clin. Cancer Res. 2018, 24, 4874–4886. [Google Scholar] [CrossRef] [PubMed]
- Karachaliou, N.; Codony-Servat, J.; Teixidó, C.; Pilotto, S.; Drozdowskyj, A.; Codony-Servat, C.; Giménez-Capitán, A.; Molina-Vila, M.A.; Bertrán-Alamillo, J.; Gervais, R.; et al. BIM and mTOR expression levels predict outcome to erlotinib in EGFR-mutant non-small-cell lung cancer. Sci. Rep. 2015, 5, 17499. [Google Scholar] [CrossRef] [PubMed]
- Faber, A.C.; Corcoran, R.B.; Ebi, H.; Sequist, L.V.; Waltman, B.A.; Chung, E.; Incio, J.; Digumarthy, S.R.; Pollack, S.F.; Song, Y.; et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011, 1, 352–365. [Google Scholar] [CrossRef]
- Dronca, R.S.; Liu, X.; Harrington, S.M.; Chen, L.; Cao, S.; Kottschade, L.A.; McWilliams, R.R.; Block, M.S.; Nevala, W.K.; Thompson, M.A.; et al. T cell Bim levels reflect responses to anti-PD-1 cancer therapy. JCI Insight 2016, 1, e86014. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.R.; Winter, P.S.; Lin, K.H.; Nussbaum, D.P.; Cakir, M.; Stein, E.M.; Soderquist, R.S.; Crawford, L.; Leeds, J.C.; Newcomb, R.; et al. A Landscape of Therapeutic Cooperativity in KRAS Mutant Cancers Reveals Principles for Controlling Tumor Evolution. Cell Rep. 2017, 20, 999–1015. [Google Scholar] [CrossRef]
- Luciano, F.; Jacquel, A.; Colosetti, P.; Herrant, M.; Cagnol, S.; Pages, G.; Auberger, P. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene 2003, 22, 6785–6793. [Google Scholar] [CrossRef]
- Chen, R.; Kim, O.; Yang, J.; Sato, K.; Eisenmann, K.M.; McCarthy, J.; Chen, H.; Qiu, Y. Regulation of Akt/PKB Activation by Tyrosine Phosphorylation. J. Biol. Chem. 2001, 276, 31858–31862. [Google Scholar] [CrossRef]
- Ahronian, L.G.; Corcoran, R.B. Effective MAPK Inhibition is critical for therapeutic responses in colorectal cancer with BRAF mutations. Mol. Cell. Oncol. 2016, 3, e1048405. [Google Scholar] [CrossRef]
- Kopetz, S.; Desai, J.; Chan, E.; Hecht, J.R.; O’Dwyer, P.J.; Maru, D.; Morris, V.; Janku, F.; Dasari, A.; Chung, W.; et al. Phase II Pilot Study of Vemurafenib in Patients With Metastatic BRAF-Mutated Colorectal Cancer. J. Clin. Oncol. 2015, 33, 4032–4038. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.H.; Wei, L.; Wirth, L.J.; Daniels, G.A.; Souza, J.A.D.; Timmers, C.D.; Sexton, J.L.; Beshara, M.; Nichols, D.; Snyder, N.; et al. Results of randomized phase II trial of dabrafenib versus dabrafenib plus trametinib in BRAF-mutated papillary thyroid carcinoma. J. Clin. Oncol. 2017, 35, 6022. [Google Scholar] [CrossRef]
- Gao, L.; Liu, H.; Sun, X.; Gao, D.; Zhang, C.; Jia, B.; Zhu, Z.; Wang, F.; Liu, Z. Molecular Imaging of Post-Src Inhibition Tumor Signatures for Guiding Dasatinib Combination Therapy. J. Nucl. Med. 2016, 57, 321–326. [Google Scholar] [CrossRef]
- Bridgeman, V.L.; Wan, E.; Foo, S.; Nathan, M.R.; Welti, J.C.; Frentzas, S.; Vermeulen, P.B.; Preece, N.; Springer, C.J.; Powles, T.; et al. Preclinical Evidence That Trametinib Enhances the Response to Antiangiogenic Tyrosine Kinase Inhibitors in Renal Cell Carcinoma. Mol. Cancer Ther. 2016, 15, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Starodub, A.; Cushman, I.; Liu, Y.; Marshall, D.J.; Hurwitz, H.I.; Nixon, A.B. Dual inhibition of αV integrins and Src kinase activity as a combination therapy strategy for colorectal cancer. Anticancer Drugs 2013, 24, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Schlumberger, M.; Tahara, M.; Wirth, L.J.; Robinson, B.; Brose, M.S.; Elisei, R.; Habra, M.A.; Newbold, K.; Shah, M.H.; Hoff, A.O.; et al. Lenvatinib versus Placebo in Radioiodine-Refractory Thyroid Cancer. N. Engl. J. Med. 2015, 372, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Brose, M.S.; Robinson, B.; Sherman, S.I.; Krajewska, J.; Lin, C.C.; Vaisman, F.; Hoff, A.O.; Hitre, E.; Bowles, D.W.; Hernando, J.; et al. Cabozantinib for radioiodine-refractory differentiated thyroid cancer (COSMIC-311): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2021, 22, 1126–1138. [Google Scholar] [CrossRef]
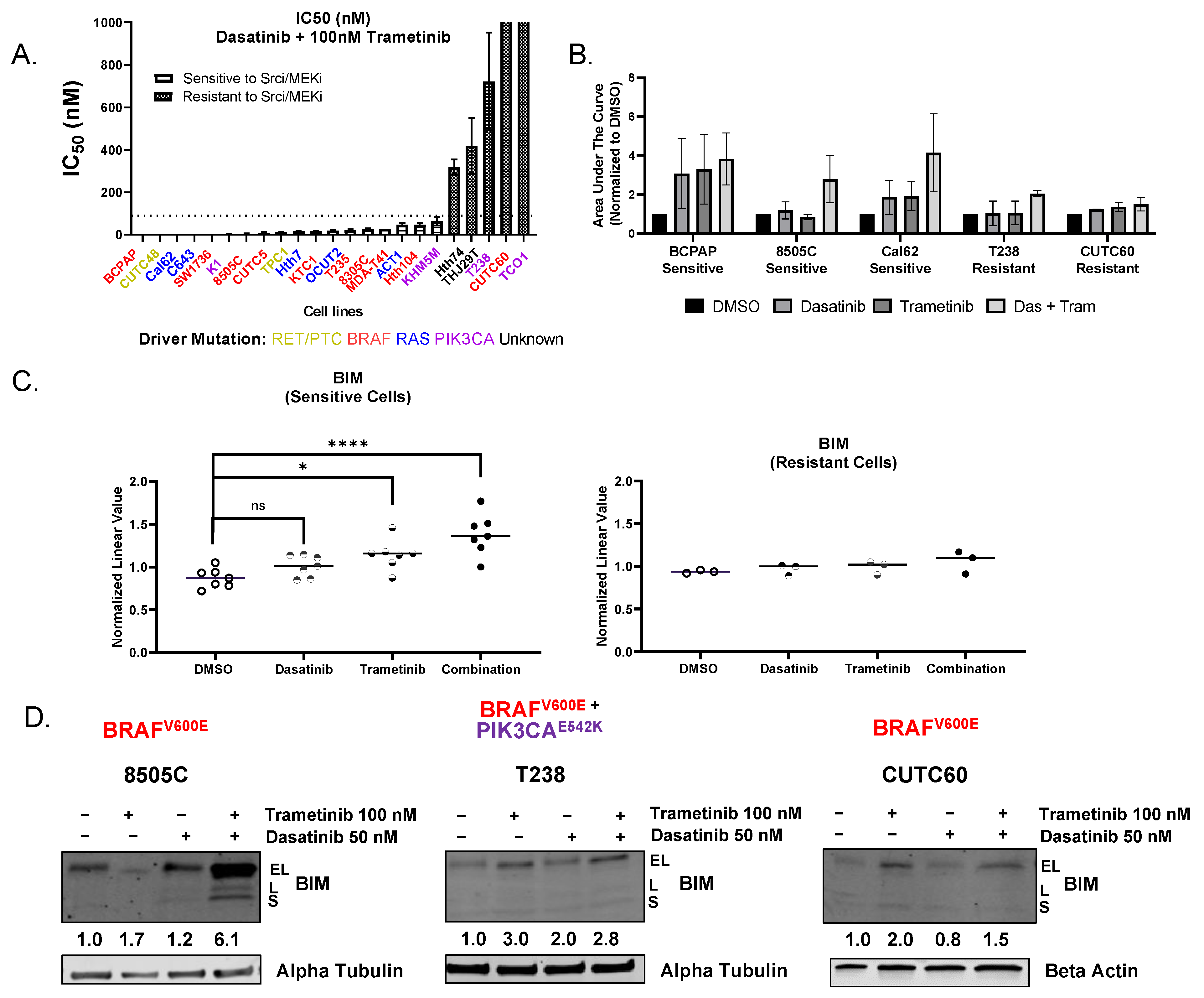
| Cell Line | Driver Oncoprotein | IC50 nM of Dasatinib + 100 nM Trametinib | Sensitive or Resistant |
|---|---|---|---|
| BCPAP *† | BRAF V600E | 0.03 | Sensitive |
| CUTC48 | RET/PTC1 | 0.08 | Sensitive |
| Cal62 *† | KRAS G12R | 0.24 | Sensitive |
| C643 * | HRAS G13R | 0.28 | Sensitive |
| SW1736 * | BRAF V600E | 0.29 | Sensitive |
| K1 * | BRAF V600E PIK3CA E542K W11C | 3.08 | Sensitive |
| 8505C *† | BRAF V600E | 3.99 | Sensitive |
| CUTC5 | BRAF V600E | 8.83 | Sensitive |
| TPC1 | RET/PTC1 | 11.74 | Sensitive |
| Hth7 | NRAS Q61R | 15.46 | Sensitive |
| KTC1 | BRAF V600E | 16.55 | Sensitive |
| OCUT2 | BRAF V600E PIK3CA H1047R | 18.53 | Sensitive |
| T235 | BRAF V600E | 21.86 | Sensitive |
| 8305C | BRAF V600E | 25.77 | Sensitive |
| MDA-T41 * | BRAF V600E | 28.00 | Sensitive |
| ACT1 | NRAS Q61K | 46.53 | Sensitive |
| Hth104 | BRAF V600E | 46.64 | Sensitive |
| KHM5M | BRAF V600E PIK3CA M10431 | 62.90 | Sensitive |
| Hth74 | None | 318.07 | Resistant |
| THJ29T | None | 418.47 | Resistant |
| T238 *† | BRAF V600E PIK3CA E542K | 722.37 | Resistant |
| CUTC60 * | BRAF V600E | 3364.00 | Resistant |
| TCO1 *† | BRAF V600E PIK3CA N1044S | 22,637.00 | Resistant |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rose, M.M.; Espinoza, V.L.; Hoff, K.J.; Pike, L.A.; Sharma, V.; Hofmann, M.-C.; Tan, A.C.; Pozdeyev, N.; Schweppe, R.E. BCL2L11 Induction Mediates Sensitivity to Src and MEK1/2 Inhibition in Thyroid Cancer. Cancers 2023, 15, 378. https://doi.org/10.3390/cancers15020378
Rose MM, Espinoza VL, Hoff KJ, Pike LA, Sharma V, Hofmann M-C, Tan AC, Pozdeyev N, Schweppe RE. BCL2L11 Induction Mediates Sensitivity to Src and MEK1/2 Inhibition in Thyroid Cancer. Cancers. 2023; 15(2):378. https://doi.org/10.3390/cancers15020378
Chicago/Turabian StyleRose, Madison M., Veronica L. Espinoza, Katelyn J. Hoff, Laura A. Pike, Vibha Sharma, Marie-Claude Hofmann, Aik Choon Tan, Nikita Pozdeyev, and Rebecca E. Schweppe. 2023. "BCL2L11 Induction Mediates Sensitivity to Src and MEK1/2 Inhibition in Thyroid Cancer" Cancers 15, no. 2: 378. https://doi.org/10.3390/cancers15020378
APA StyleRose, M. M., Espinoza, V. L., Hoff, K. J., Pike, L. A., Sharma, V., Hofmann, M.-C., Tan, A. C., Pozdeyev, N., & Schweppe, R. E. (2023). BCL2L11 Induction Mediates Sensitivity to Src and MEK1/2 Inhibition in Thyroid Cancer. Cancers, 15(2), 378. https://doi.org/10.3390/cancers15020378






