Genomic, Probiotic, and Functional Properties of Bacteroides dorei RX2020 Isolated from Gut Microbiota
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial and Animals
2.2. 16S rRNA Analysis
2.3. Genome Sequencing and Annotation
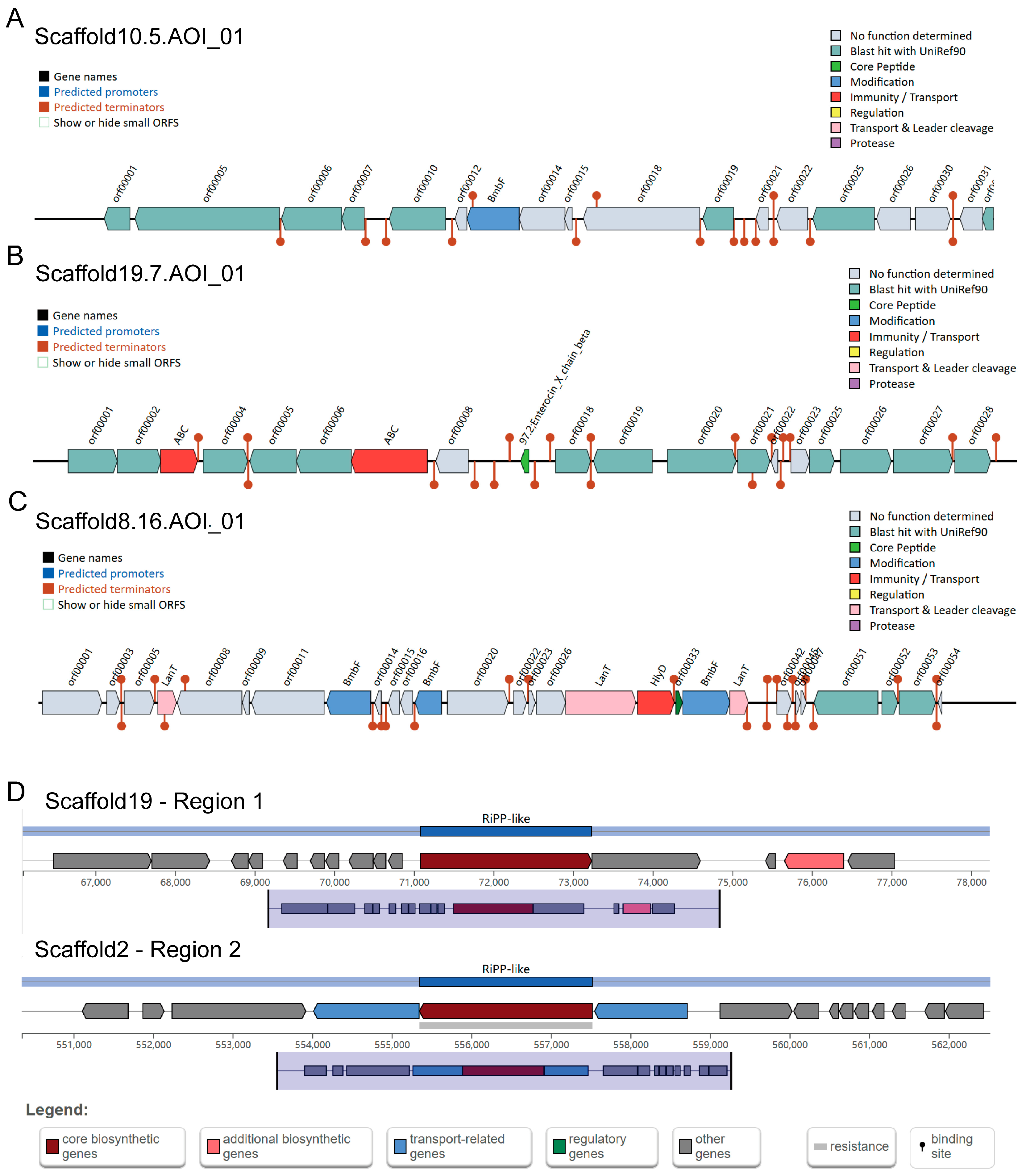
2.4. Bacteriocin and Secondary Metabolite
2.5. Tolerance to Simulation of Gastrointestinal Digestion
2.6. Cell Adhesion Ability
2.7. DPPH Free Radical Scavenging Assay
2.8. Analysis of Antibacterial Activity
2.9. Analysis of Bile Salt Hydrolase Activity
2.10. Hemolytic Activity
2.11. Toxicity Experiment
2.12. Metabolomic Analysis
2.13. RNA-Seq Quantification and RT-PCR
2.14. ELISA
2.15. Cell Culture and Stimulation
2.16. Statistical Analysis
3. Results
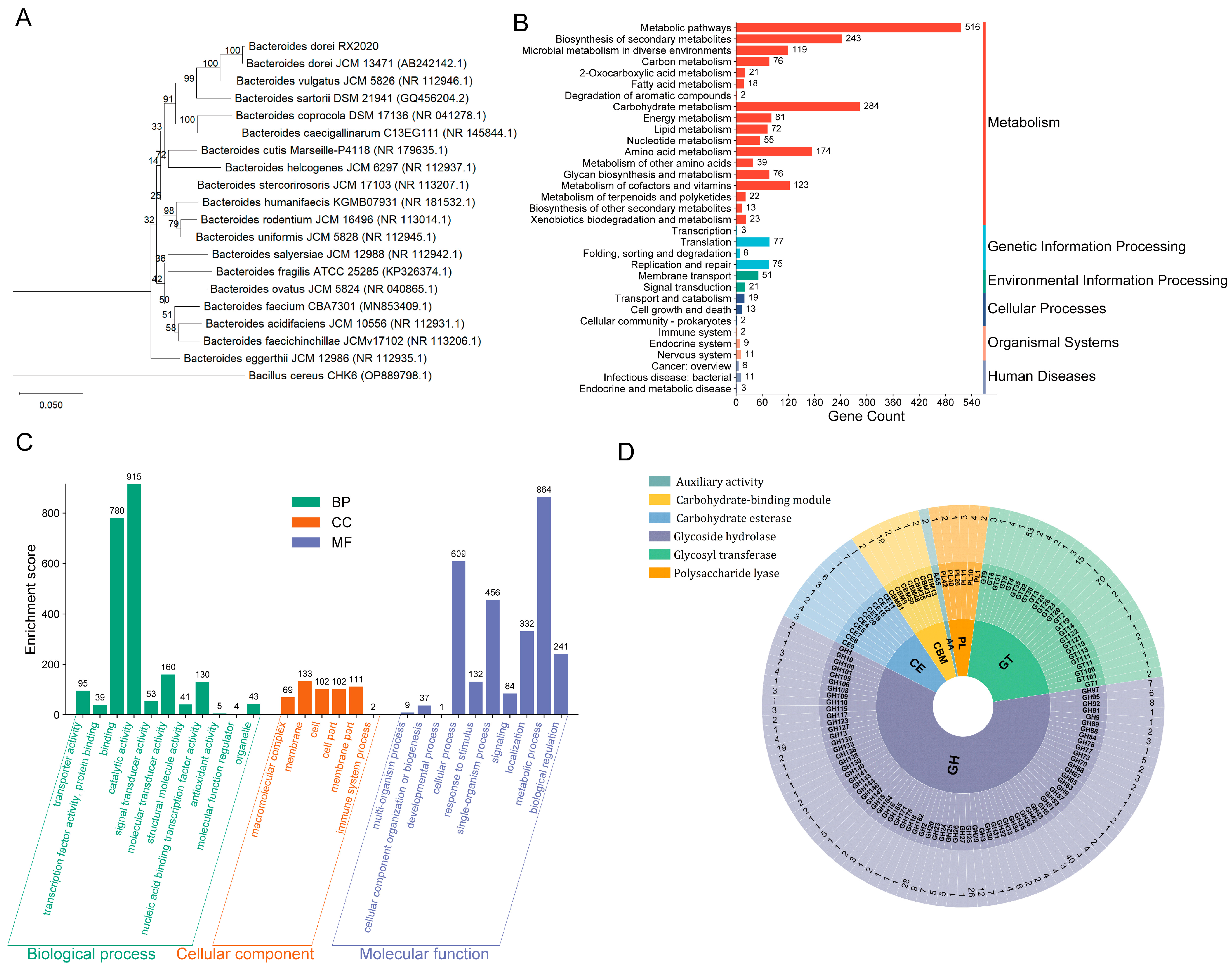
3.1. Genetic Characteristics of B. dorei Strain
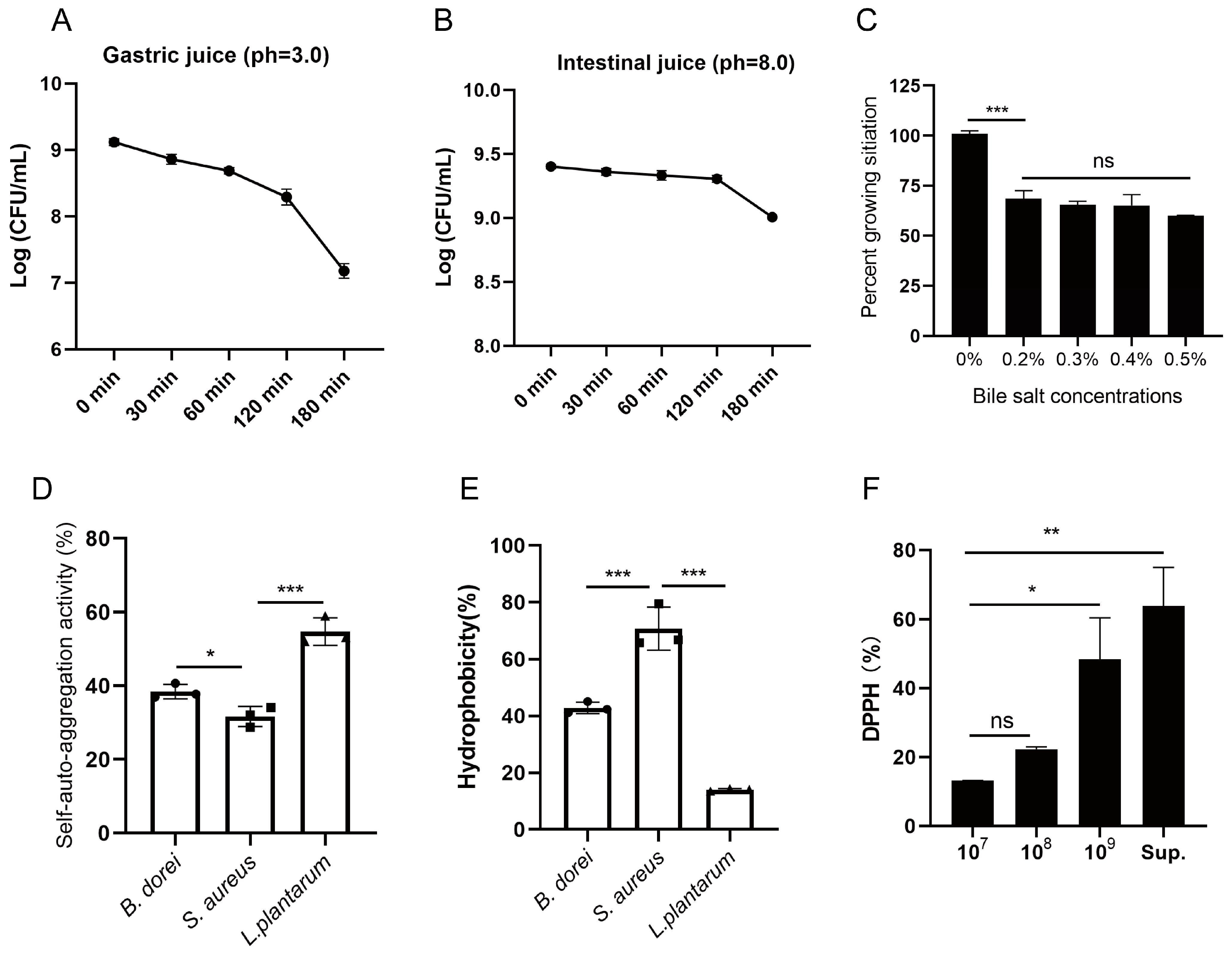
3.2. Probiotic Properties of B. dorei Strains
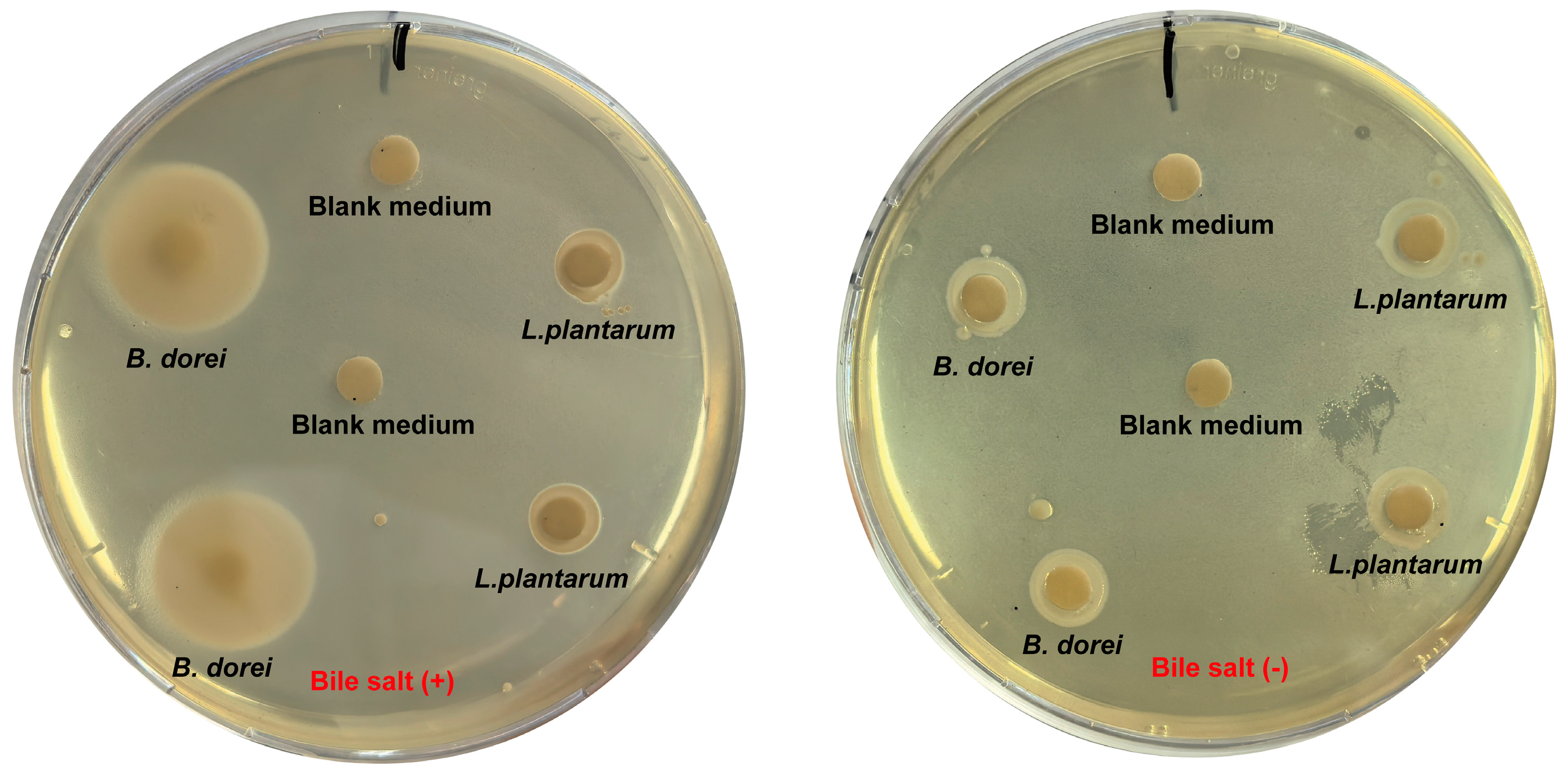
3.3. Antibacterial Properties
3.4. Bile Salt Hydrolase Activity
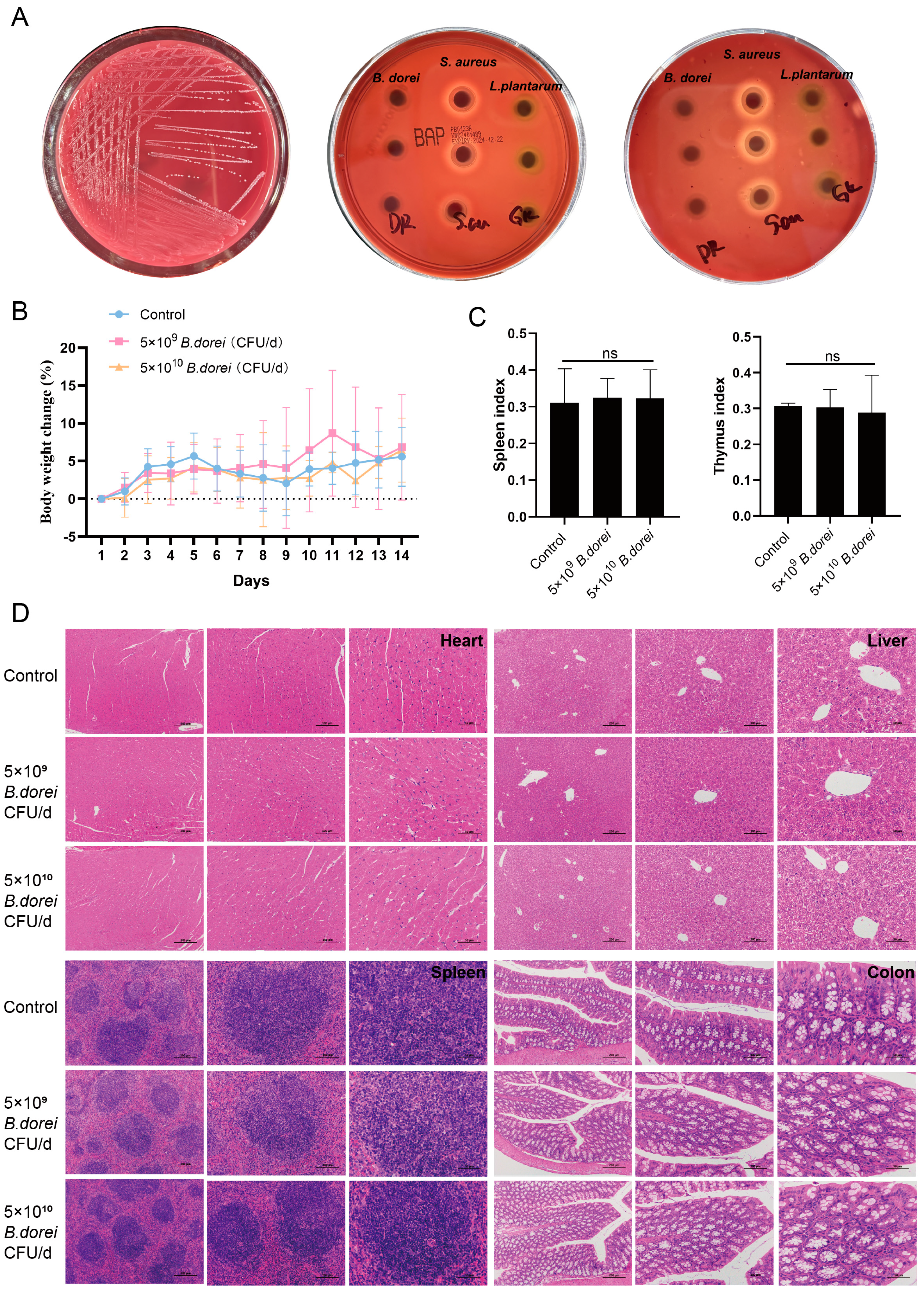
3.5. Safety Evaluation
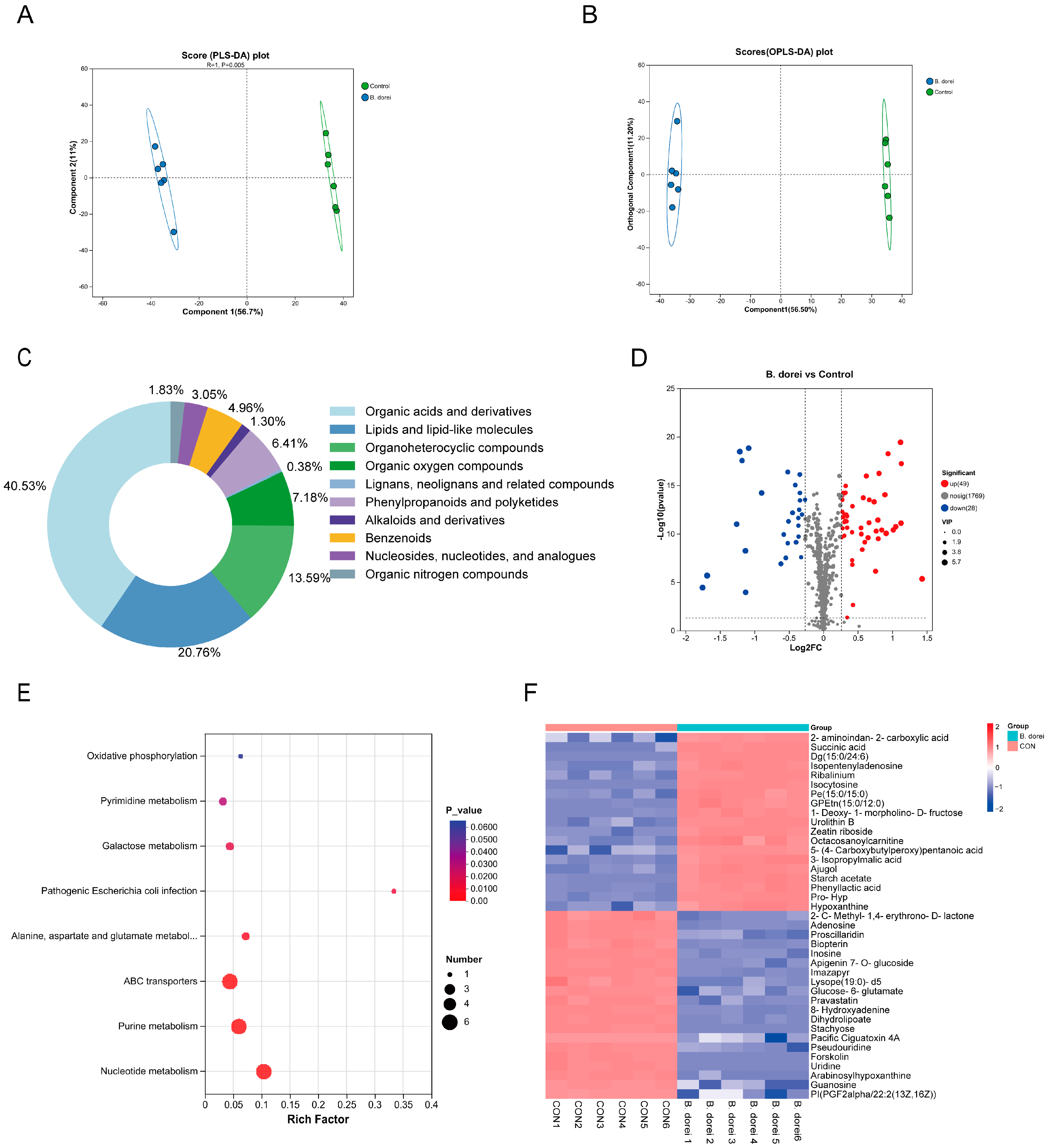
3.6. Metabolites of B. dorei
3.7. Immunomodulatory Activity of B. dorei
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Z.; Lv, J.; Pan, L.; Zhang, Y. Roles and Applications of Probiotic Lactobacillus Strains. Appl. Microbiol. Biotechnol. 2018, 102, 8135–8143. [Google Scholar] [CrossRef] [PubMed]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best Practices for Analysing Microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef]
- Gao, B.; Chi, L.; Zhu, Y.; Shi, X.; Tu, P.; Li, B.; Yin, J.; Gao, N.; Shen, W.; Schnabl, B. An Introduction to Next Generation Sequencing Bioinformatic Analysis in Gut Microbiome Studies. Biomolecules 2021, 11, 530. [Google Scholar] [CrossRef]
- O’Toole, P.W.; Marchesi, J.R.; Hill, C. Next-Generation Probiotics: The Spectrum from Probiotics to Live Biotherapeutics. Nat. Microbiol. 2017, 2, 17057. [Google Scholar] [CrossRef]
- Zhang, J.; Ni, Y.; Qian, L.; Fang, Q.; Zheng, T.; Zhang, M.; Gao, Q.; Zhang, Y.; Ni, J.; Hou, X.; et al. Decreased Abundance of Akkermansia Muciniphila Leads to the Impairment of Insulin Secretion and Glucose Homeostasis in Lean Type 2 Diabetes. Adv. Sci. 2021, 8, e2100536. [Google Scholar] [CrossRef]
- Liu, E.; Ji, X.; Zhou, K. Akkermansia Muciniphila for the Prevention of Type 2 Diabetes and Obesity: A Meta-Analysis of Animal Studies. Nutrients 2024, 16, 3440. [Google Scholar] [CrossRef] [PubMed]
- Manor, O.; Dai, C.L.; Kornilov, S.A.; Smith, B.; Price, N.D.; Lovejoy, J.C.; Gibbons, S.M.; Magis, A.T. Health and Disease Markers Correlate with Gut Microbiome Composition across Thousands of People. Nat. Commun. 2020, 11, 5206. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhao, J.; Zhang, H.; Lee, Y.-K.; Zhai, Q.; Chen, W. Roles of Intestinal Bacteroides in Human Health and Diseases. Crit. Rev. Food Sci. Nutr. 2021, 61, 3518–3536. [Google Scholar] [CrossRef]
- Yan, Y.; Lei, Y.; Qu, Y.; Fan, Z.; Zhang, T.; Xu, Y.; Du, Q.; Brugger, D.; Chen, Y.; Zhang, K.; et al. Bacteroides Uniformis-Induced Perturbations in Colonic Microbiota and Bile Acid Levels Inhibit TH17 Differentiation and Ameliorate Colitis Developments. NPJ Biofilms Microbiomes 2023, 9, 56. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e8. [Google Scholar] [CrossRef]
- Steed, A.L.; Christophi, G.P.; Kaiko, G.E.; Sun, L.; Goodwin, V.M.; Jain, U.; Esaulova, E.; Artyomov, M.N.; Morales, D.J.; Holtzman, M.J.; et al. The Microbial Metabolite Desaminotyrosine Protects from Influenza through Type I Interferon. Science 2017, 357, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Huang, Y.; Liu, G.; Li, X.; Xiao, Y.; Liu, C.; Zhang, Y.; Li, J.; Xu, J.; Lu, S.; et al. A Novel Immunobiotics Bacteroides Dorei Ameliorates Influenza Virus Infection in Mice. Front. Immunol. 2022, 12, 828887. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, R.; Zhu, H.; Ruan, J.; Qian, W.; Fang, X.; Shi, Z.; Li, Y.; Li, S.; Shan, G.; Kristiansen, K.; et al. De Novo Assembly of Human Genomes with Massively Parallel Short Read Sequencing. Genome Res. 2010, 20, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.M.; Birol, I. ABySS: A Parallel Assembler for Short Read Sequence Data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-H.; Liao, Y.-C. CISA: Contig Integrator for Sequence Assembly of Bacterial Genomes. PLoS ONE 2013, 8, e60843. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A Self-Training Method for Prediction of Gene Starts in Microbial Genomes. Implications for Finding Sequence Motifs in Regulatory Regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From Genomics to Chemical Genomics: New Developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes Database (CAZy): An Expert Resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Famiglietti, M.L.; Estreicher, A.; Gos, A.; Bolleman, J.; Géhant, S.; Breuza, L.; Bridge, A.; Poux, S.; Redaschi, N.; Bougueleret, L.; et al. Genetic Variations and Diseases in UniProtKB/Swiss-Prot: The Ins and Outs of Expert Manual Curation. Hum. Mutat. 2014, 35, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Fei, Y.; Li, L.; Zheng, Y.; Liu, D.; Zhou, Q.; Fu, L. Characterization of Lactobacillus Amylolyticus L6 as Potential Probiotics Based on Genome Sequence and Corresponding Phenotypes. LWT 2018, 90, 460–468. [Google Scholar] [CrossRef]
- Shen, H.; Wang, T.; Dong, W.; Sun, G.; Liu, J.; Peng, N.; Zhao, S. Metagenome-Assembled Genome Reveals Species and Functional Composition of Jianghan Chicken Gut Microbiota and Isolation of Pediococcus Acidilactic with Probiotic Properties. Microbiome 2024, 12, 25. [Google Scholar] [CrossRef]
- He, Z.; Wang, T.; Zhang, S.; Shi, K.; Wang, F.; Li, Y.; Lin, C.; Chen, J. Evaluation of Cholesterol Transformation Abilities and Probiotic Properties of Bacteroides Dorei YGMCC0564. Front. Microbiol. 2023, 14, 1279996. [Google Scholar] [CrossRef]
- Cheruvari, A.; Kammara, R. Genomic Characterization and Probiotic Properties of Lactiplantibacillus Pentosus Isolated from Fermented Rice. Probiotics Antimicrob. Proteins 2024, 1–23. [Google Scholar] [CrossRef]
- Drumond, M.M.; Tapia-Costa, A.P.; Neumann, E.; Nunes, Á.C.; Barbosa, J.W.; Kassuha, D.E.; Mancha-Agresti, P. Cell-Free Supernatant of Probiotic Bacteria Exerted Antibiofilm and Antibacterial Activities against Pseudomonas Aeruginosa: A Novel Biotic Therapy. Front. Pharmacol. 2023, 14, 1152588. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Haleem, H.; Kheadr, E.; Dabour, N.; El-Saadany, K.; El-Ziney, M. Potential Anti-Hypercholesterolemic Activity and Acidogenic Ability of Probiotic Lactic Acid Bacteria Isolated From Camel Milk. J. Appl. Sci. Res. 2018, 14, 12–18. [Google Scholar]
- He, S.; Lu, S.; Huang, Y.; Song, L.; Yang, T.; Xiao, Y.; Ren, Z. Next-Generation Probiotic Bacteroides. Dorei: Improving the Efficacy of COVID-19 Vaccinations. J. Funct. Foods 2024, 116, 106190. [Google Scholar] [CrossRef]
- Cui, Y.; Chen, C.; Tang, Z.; Yuan, W.; Yue, K.; Cui, P.; Qiu, X.; Zhang, H.; Li, T.; Zhu, X.; et al. TREM2 Deficiency Aggravates Renal Injury by Promoting Macrophage Apoptosis and Polarization via the JAK-STAT Pathway in Mice. Cell Death Dis. 2024, 15, 401. [Google Scholar] [CrossRef] [PubMed]
- Gregory, G.J.; Morreale, D.P.; Boyd, E.F. CosR Is a Global Regulator of the Osmotic Stress Response with Widespread Distribution among Bacteria. Appl. Environ. Microbiol. 2020, 86, e00120-20. [Google Scholar] [CrossRef]
- Li, X.-D.; Lin, Y.-C.; Yang, R.-S.; Kang, X.; Xin, W.-G.; Wang, F.; Zhang, Q.-L.; Zhang, W.-P.; Lin, L.-B. Genomic and In-Vitro Characteristics of a Novel Strain Lacticaseibacillus Chiayiensis AACE3 Isolated from Fermented Blueberry. Front. Microbiol. 2023, 14, 1168378. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, S.; Li, H.; Zhu, J.; Liu, Z.; Hu, X.; Yi, J. Assessments of Probiotic Potentials of Lactiplantibacillus Plantarum Strains Isolated From Chinese Traditional Fermented Food: Phenotypic and Genomic Analysis. Front. Microbiol. 2022, 13, 895132. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Liu, J.; Wen, X.; Zhang, G.; Cai, J.; Qiao, Z.; An, Z.; Zheng, J.; Li, L. Unique Probiotic Properties and Bioactive Metabolites of Saccharomyces Boulardii. Probiotics Antimicrob. Proteins 2023, 15, 967–982. [Google Scholar] [CrossRef]
- Mather, A.E.; Gilmour, M.W.; Reid, S.W.J.; French, N.P. Foodborne Bacterial Pathogens: Genome-Based Approaches for Enduring and Emerging Threats in a Complex and Changing World. Nat. Rev. Microbiol. 2024, 22, 543–555. [Google Scholar] [CrossRef]
- Hu, C.-B.; Malaphan, W.; Zendo, T.; Nakayama, J.; Sonomoto, K. Enterocin X, a Novel Two-Peptide Bacteriocin from Enterococcus Faecium KU-B5, Has an Antibacterial Spectrum Entirely Different from Those of Its Component Peptides. Appl. Environ. Microbiol. 2010, 76, 4542–4545. [Google Scholar] [CrossRef] [PubMed]
- van Belkum, M.J.; Worobo, R.W.; Stiles, M.E. Double-Glycine-Type Leader Peptides Direct Secretion of Bacteriocins by ABC Transporters: Colicin V Secretion in Lactococcus Lactis. Mol. Microbiol. 1997, 23, 1293–1301. [Google Scholar] [CrossRef]
- Hudson, G.A.; Mitchell, D.A. RiPP Antibiotics: Biosynthesis and Engineering Potential. Curr. Opin. Microbiol. 2018, 45, 61–69. [Google Scholar] [CrossRef]
- Collins, S.L.; Stine, J.G.; Bisanz, J.E.; Okafor, C.D.; Patterson, A.D. Bile Acids and the Gut Microbiota: Metabolic Interactions and Impacts on Disease. Nat. Rev. Microbiol. 2023, 21, 236–247. [Google Scholar] [CrossRef]
- Song, Z.; Cai, Y.; Lao, X.; Wang, X.; Lin, X.; Cui, Y.; Kalavagunta, P.K.; Liao, J.; Jin, L.; Shang, J.; et al. Taxonomic Profiling and Populational Patterns of Bacterial Bile Salt Hydrolase (BSH) Genes Based on Worldwide Human Gut Microbiome. Microbiome 2019, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.M.; Clardy, J.; Xavier, R.J. Gut Microbiome Lipid Metabolism and Its Impact on Host Physiology. Cell Host Microbe 2023, 31, 173–186. [Google Scholar] [CrossRef]
- Li, M.; Han, X.; Sun, L.; Liu, X.; Zhang, W.; Hao, J. Indole-3-Acetic Acid Alleviates DSS-Induced Colitis by Promoting the Production of R-Equol from Bifidobacterium Pseudolongum. Gut Microbes 2024, 16, 2329147. [Google Scholar] [CrossRef] [PubMed]
- Min, B.H.; Devi, S.; Kwon, G.H.; Gupta, H.; Jeong, J.-J.; Sharma, S.P.; Won, S.-M.; Oh, K.-K.; Yoon, S.J.; Park, H.J.; et al. Gut Microbiota-Derived Indole Compounds Attenuate Metabolic Dysfunction-Associated Steatotic Liver Disease by Improving Fat Metabolism and Inflammation. Gut Microbes 2024, 16, 2307568. [Google Scholar] [CrossRef]
- Winkler, E.S.; Shrihari, S.; Hykes, B.L.; Handley, S.A.; Andhey, P.S.; Huang, Y.-J.S.; Swain, A.; Droit, L.; Chebrolu, K.K.; Mack, M.; et al. The Intestinal Microbiome Restricts Alphavirus Infection and Dissemination through a Bile Acid-Type I IFN Signaling Axis. Cell 2020, 182, 901–918.e18. [Google Scholar] [CrossRef]
- Geva-Zatorsky, N.; Sefik, E.; Kua, L.; Pasman, L.; Tan, T.G.; Ortiz-Lopez, A.; Yanortsang, T.B.; Yang, L.; Jupp, R.; Mathis, D.; et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 2017, 168, 928–943.e11. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, G.R.; Chang, P.V. Deciphering the Chemical Lexicon of Host-Gut Microbiota Interactions. Trends Pharmacol. Sci. 2019, 40, 430–445. [Google Scholar] [CrossRef]
- Perry, E.K.; Meirelles, L.A.; Newman, D.K. From the Soil to the Clinic: The Impact of Microbial Secondary Metabolites on Antibiotic Tolerance and Resistance. Nat. Rev. Microbiol. 2022, 20, 129–142. [Google Scholar] [CrossRef]
- Wardman, J.F.; Bains, R.K.; Rahfeld, P.; Withers, S.G. Carbohydrate-Active Enzymes (CAZymes) in the Gut Microbiome. Nat. Rev. Microbiol. 2022, 20, 542–556. [Google Scholar] [CrossRef]
- Onyango, S.O.; Juma, J.; De Paepe, K.; Van de Wiele, T. Oral and Gut Microbial Carbohydrate-Active Enzymes Landscape in Health and Disease. Front. Microbiol. 2021, 12, 653448. [Google Scholar] [CrossRef]
- Pavalakumar, D.; Undugoda, L.J.S.; Gunathunga, C.J.; Manage, P.M.; Nugara, R.N.; Kannangara, S.; Lankasena, B.N.S.; Patabendige, C.N.K. Evaluating the Probiotic Profile, Antioxidant Properties, and Safety of Indigenous Lactobacillus Spp. Inhabiting Fermented Green Tender Coconut Water. Probiotics Antimicrob. Proteins 2024, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kusada, H.; Morinaga, K.; Tamaki, H. Identification of Bile Salt Hydrolase and Bile Salt Resistance in a Probiotic Bacterium Lactobacillus Gasseri JCM1131T. Microorganisms 2021, 9, 1011. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Tohda, H.; Okuda, K.; Takazoe, I. Adherence and Hydrophobicity of Invasive and Noninvasive Strains of Porphyromonas Gingivalis. Oral Microbiol. Immunol. 1993, 8, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Mining Lactobacillus and Bifidobacterium for Organisms with Long-Term Gut Colonization Potential. Clin. Nutr. 2020, 39, 1315–1323. [Google Scholar] [CrossRef]
- Ma, X.; Tian, M.; Yu, X.; Liu, M.; Li, B.; Ren, D.; Wang, W. Characterization and Preliminary Safety Evaluation of Akkermansia Muciniphila PROBIO. Foods 2024, 13, 442. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, N.; Su, X.; Gao, Y.; Yang, R. Gut-Microbiota-Derived Metabolites Maintain Gut and Systemic Immune Homeostasis. Cells 2023, 12, 793. [Google Scholar] [CrossRef]
- Li, J.; Chen, Y.; Li, R.; Zhang, X.; Chen, T.; Mei, F.; Liu, R.; Chen, M.; Ge, Y.; Hu, H.; et al. Gut Microbial Metabolite Hyodeoxycholic Acid Targets the TLR4/MD2 Complex to Attenuate Inflammation and Protect against Sepsis. Mol. Ther. 2023, 31, 1017–1032. [Google Scholar] [CrossRef]
- Hu, J.; Wang, C.; Huang, X.; Yi, S.; Pan, S.; Zhang, Y.; Yuan, G.; Cao, Q.; Ye, X.; Li, H. Gut Microbiota-Mediated Secondary Bile Acids Regulate Dendritic Cells to Attenuate Autoimmune Uveitis through TGR5 Signaling. Cell Rep. 2021, 36, 109726. [Google Scholar] [CrossRef]
- Yu, X.; Ou, J.; Wang, L.; Li, Z.; Ren, Y.; Xie, L.; Chen, Z.; Liang, J.; Shen, G.; Zou, Z.; et al. Gut Microbiota Modulate CD8+ T Cell Immunity in Gastric Cancer through Butyrate/GPR109A/HOPX. Gut Microbes 2024, 16, 2307542. [Google Scholar] [CrossRef]

| Attribute | Values |
|---|---|
| Genome size (bp) | 7,588,798 |
| GC content (%) | 40.81 |
| Total genes | 6783 |
| Gene total length (bp) | 6,789,711 |
| Gene average length (bp) | 1001 |
| Gene length/Genome (%) | 89.47 |
| tRNA genes | 124 |
| rRNA genes | 14 |
| sRNA genes | 8 |
| Gene Name | Annotation | Annotation No. |
|---|---|---|
| Temperature stress | ||
| groEL | Chaperonin GroEL | A6KXA0; Q03VC3 |
| groES | Co-chaperonin GroES | A6KXA1; Q03VC2 |
| hslO | Heat shock protein 33 homolog | Q88Z30 |
| hsp18 | 18 kDa heat shock protein | Q03928 |
| dnaJ | Chaperone protein | O34136; Q5LED4 |
| dnaK | Chaperone protein | A6L2 × 7; Q03WI2 |
| clpB | Chaperone protein | Q8EU05; Q89YY3 |
| grpE | Co-chaperone GrpE (heat shock protein) | Q03WI1; Q8A8C4 |
| cspLA | Cold shock-like protein | P0A356 |
| cspG | Cold shock-like protein | Q9S170 |
| PH stress | ||
| atpA | ATP synthase subunit alpha | A6L4M4 |
| atpB | ATP synthase subunit a | B3QZE8 |
| atpC | ATP synthase epsilon chain | Q04S19 |
| atpD | ATP synthase subunit beta | A6L4L7 |
| atpE | ATP synthase subunit c | A6L4M1 |
| atpF | ATP synthase subunit b | A6L4M2 |
| atpG | ATP synthase gamma chain | A6L4M5 |
| atpH | ATP synthase subunit delta | A6L4M3 |
| clpA | ATP-dependent Clp protease ATP-binding subunit | P0ABI1 |
| clpE | ATP-dependent Clp protease ATP-binding subunit | Q9S5Z2 |
| clpX | ATP-dependent Clp protease ATP-binding subunit | Q03W09; Q5L8L7 |
| clpP | ATP-dependent Clp protease proteolytic subunit | B1MXG9; Q8A129 |
| clpS | ATP-dependent Clp protease adapter protein | A3DER9 |
| nhaA | Na(+)/H(+) antiporter | A6L743 |
| nhaP2 | K(+)/H(+) antiporter | A4XPR4 |
| nhaS4 | Na(+)/H(+) antiporter | P72973 |
| Osmotic stress | ||
| opuAA | Glycine betaine transport ATP-binding protein | Q9KIF7 |
| gbuA | Glycine betaine/carnitine transport ATP-binding protein | Q9RR46 |
| gbuB | Glycine betaine/carnitine transport permease protein | Q9RR45 |
| gbuC | Glycine betaine/carnitine transport binding protein | Q9RR44 |
| proW | Glycine betaine/proline betaine transport system permease protein | P14176 |
| Oxidative stress | ||
| gpo | Glutathione peroxidase | Q9CFV1 |
| gpx2 | Glutathione peroxidase-like peroxiredoxin 2 | P38143 |
| nox | NADH oxidase | P37061 |
| tpx | Thiol peroxidase | Q71Z84; Q8KED5 |
| npr | NADH peroxidase | P37062 |
| trxA | Thioredoxin | O51088; P0A4L4 |
| trxB | Thioredoxin reductase | O32823; P50971 |
| ntrc | Thioredoxin reductase | Q70G58 |
| trx2 | Thioredoxin H2 | Q38879 |
| Gene Name | Annotation | Annotation No. |
|---|---|---|
| malP | Maltose phosphorylase | E6ENP7 |
| lspA | Lipoprotein signal peptidase | B2RI39 |
| tuf | Elongation factor Tu | A6KYK9; Q03YI2 |
| gpr | L-glyceraldehyde 3-phosphate reductase | Q8 × 529 |
| gapc | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic | Q01558 |
| tpiA | Triosephosphate isomerase | Q03SL6; A6KXL2 |
| groEL | Chaperonin GroEL | A6KXA0; Q03VC3 |
| groES | Co-chaperonin GroES | A6KXA1; Q03VC2 |
| Strains | Blank Medium | B. dorei Supernatant | |
|---|---|---|---|
| Log CFU/mL | Log CFU/mL | Survival Rate | |
| E. coli (EHEC) | 8.05 ± 0.05 | 7.69 ± 0.12 a | 43.76% |
| S. aureus | 7.75 ± 0.19 | 7.16 ± 0.35 b | 30.20% |
| S. typhimurium | 8.15 ± 0.02 | 7.96 ± 0.04 a | 65.41% |
| E. faecalis | 7.61 ± 0.16 | 7.63 ± 0.09 | 103.25% |
| Type | Genus | Description | Query | E Value | Per. Ident | Acc. Len |
|---|---|---|---|---|---|---|
| BSH-T0 | Enterococcus | 674_gene_2740 | 89% | 6 × 10−33 | 28.75% | 359 |
| BSH-T1 | Eubacterium | 674_gene_2740 | 94% | 1 × 10−34 | 30.51% | 359 |
| BSH-T2 | Streptococcus | 674_gene_2740 | 97% | 1 × 10−36 | 30.03% | 359 |
| BSH-T3 | Lactobacillus | 674_gene_2740 | 98% | 2 × 10−33 | 26.04% | 359 |
| BSH-T4 | Bifidobacterium | 674_gene_2740 | 92% | 1 × 10−38 | 30.72% | 359 |
| BSH-T5 | Bacteroides | 674_gene_2740 | 94% | 4 × 10−153 | 59.18% | 359 |
| BSH-T6 | Bacteroides | 651_gene_1492 | 97% | 0 | 86.26% | 352 |
| BSH-T7 | Blautia | No significant similarity found | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, S.; Song, L.; Xiao, Y.; Huang, Y.; Ren, Z. Genomic, Probiotic, and Functional Properties of Bacteroides dorei RX2020 Isolated from Gut Microbiota. Nutrients 2025, 17, 1066. https://doi.org/10.3390/nu17061066
He S, Song L, Xiao Y, Huang Y, Ren Z. Genomic, Probiotic, and Functional Properties of Bacteroides dorei RX2020 Isolated from Gut Microbiota. Nutrients. 2025; 17(6):1066. https://doi.org/10.3390/nu17061066
Chicago/Turabian StyleHe, Siqin, Liqiong Song, Yuchun Xiao, Yuanming Huang, and Zhihong Ren. 2025. "Genomic, Probiotic, and Functional Properties of Bacteroides dorei RX2020 Isolated from Gut Microbiota" Nutrients 17, no. 6: 1066. https://doi.org/10.3390/nu17061066
APA StyleHe, S., Song, L., Xiao, Y., Huang, Y., & Ren, Z. (2025). Genomic, Probiotic, and Functional Properties of Bacteroides dorei RX2020 Isolated from Gut Microbiota. Nutrients, 17(6), 1066. https://doi.org/10.3390/nu17061066





