Abstract
The development and health of infants are intertwined with the protective and regulatory functions of different microorganisms in the gut known as the gut microbiota. Preterm infants born with an imbalanced gut microbiota are at substantial risk of several diseases including inflammatory intestinal diseases, necrotizing enterocolitis, late-onset sepsis, neurodevelopmental disorders, and allergies which can potentially persist throughout adulthood. In this review, we have evaluated the role of Bifidobacterium as commonly used probiotics in the development of gut microbiota and prevention of common diseases in preterm infants which is not fully understood yet. The application of Bifidobacterium as a therapeutical approach in the re-programming of the gut microbiota in preterm infants, the mechanisms of host-microbiome interaction, and the mechanism of action of this bacterium have also been investigated, aiming to provide new insights and opportunities in microbiome-targeted interventions in personalized medicine.
1. Introduction
The gastrointestinal tract, which houses trillions of microorganisms, is the most populated anatomical niche in the human body and plays a critical role in the development of the immune system, metabolism, cognitive development, and host physiology [1].
The gut microbiota structure is constantly changing during life in infancy and childhood and stabilizing through adulthood [2]. Different prenatal and postnatal factors can influence the structure and composition of the gut microbiota including delivery method, genetics, feeding method, maternal microbiota, antibiotics, and lifestyle.
Dysbiosis, or the disruption of the gut microbiota, has been associated with the development of a number of chronic illnesses in premature newborns, which may persist later in adulthood, including gastrointestinal disorders, neurodevelopmental and metabolic abnormalities, and allergies [3].
Although preterm infants’ health outcomes are equally relevant and important, the majority of research on gut microbiota has focused on full-term infants and adults. According to the World Health Organization (WHO), 15 million infants are delivered prematurely each year. Complications associated with prematurity are the major reason for 1 million deaths among children under 5 years of age each year, survivors may also face lifetime mental and physical challenges [4].
Preterm newborns are immunologically underdeveloped, making them vulnerable to bacterial infections. Neutropenia, deficiency of phagocytosis, chemotaxis, the cytolytic activity of NK cells, low expression of histocompatibility complex class II, and suppressed toll-like receptor (TLR) are the most common immunodeficiencies in preterm infants [5,6]. Preterm infants born before 37 weeks of gestational age (weight < 2500 g) may be exposed to different environmental factors including long-term stays in the neonatal intensive care units, use of broad-spectrum antibiotics, and monitored feeding regimens [7]. Although maternal milk contains several beneficial components such as antimicrobial peptides, immunoglobulins, essential nutrients such as proteins, Zinc, lactoferrin, natural probiotics, Fructooligosaccharides (FOS), short-chain galactooligosaccharides (GOS), and polydextrose, not all preterm infants can digest their mother’s milk and absorb its nutritional substances [8]. Therefore, preterm infants with underlying health conditions require additional nutritional support to maintain gastrointestinal health and absorption of essential nutrients [9].
The classic pattern of the gut microbiota in a full-term, vaginally born, and breastfed infant follows a general trend that includes initial colonization with facultative anaerobes including Enterobacteriaceae family (e.g., Escherichia coli, Klebsiella spp.), Enterococcus spp., Streptococcus spp., and Staphylococcus spp. After depletion of oxygen by facultative anaerobes in a matter of days after birth and diet shift to human milk, which is a rich source of oligosaccharides, obligate anaerobes and oligosaccharides metabolizers such as Bifidobacterium spp., Bacteroides spp., and Clostridium spp. dominate the gut [10]. Subsequently, solid food consumption by infants after the age of six months reduces Bifidobacterium abundance by 30% to 40%, and this decline persists throughout childhood and adolescence as a result of lifestyle, puberty, nutrition, and antibiotic administration [11]. In adulthood, Bifidobacterium abundance stabilizes between 0% to 18% and declines in elderlies which might be related to declined immune function in this group [12].
Recent investigations using culture-based and sequencing-based approaches have found a strong association between the function of Bifidobacterium in the development of inflammatory intestinal diseases, neurodevelopmental disorders, and allergies in premature infants [13]. In addition to the numerous correlations observed, a substantial body of evidence has shown the beneficial impact of Bifidobacterium in a range of preclinical and clinical models. However, it remains unclear how this interaction can lead to the regulation of immunological pathways and the improvement of the immature gastrointestinal tract.
To gain a mechanistic understanding of host-microbiome interaction and how bacterial metabolites can remotely regulate other organs and pathways, we discussed the impact of Bifidobacterium on host metabolism and physiology in pre-term infants, aiming to provide new insights and opportunities in microbiome-targeted interventions in personalized medicine in this population.
2. Common Gut-Microbiota-Associated Complications in Preterm Infants
2.1. Gastrointestinal Disorders
2.1.1. Necrotizing Enterocolitis (NEC)
Necrotizing enterocolitis (NEC) is the most common intestinal complication in preterm infants. NEC is a devastating condition defined as intestinal inflammation/perforation (ischemic necrosis of intestinal mucosa) that mainly occurs during the first two weeks of life in 10% of preterm infants. Preterm infants diagnosed with NEC may experience severe symptoms including lethargy, bloated stomach, vomiting, blood in stool, multiorgan failures such as slow heartbeat (bradycardia), difficulties in breathing (apnea), and even death [14] (Figure 1). According to a systematic and meta-analysis review of 574,692 premature infants, the global incidence of NEC was predicted in seven out of 100 preterm infants [15].
Figure 1.
Common gut-microbiota-associated complications in preterm infants. Figures in this manuscript were created specifically for this manuscript in BioRender.com, accessed on 15 August 2022.
Despite many research efforts on the management of NEC over the last decades, NEC risk in preterm infants still is high and survivors may experience long-term consequences. Current management of NEC includes a controlled diet through a nasogastric tube, administration of inotropes and intravenous fluids to maintain oxygen delivery to different organs, and prevention of enteric bacterial infection using broad-spectrum antibiotics. Severe cases may require abdominal surgery to resect the necrotic tissue and drainage of fluid from the peritoneal cavity [16].
Recently, gut microbiota dysbiosis has been identified as one of the main factors in the development of NEC in preterm infants. Several studies have shown the association of NEC incidence with a high abundance of Gram-negative facultative bacteria (e.g, Proteobacteria and Gammaproteobacteria (Enterobacteriaceae members (Klebsiella pneumoniae, E. coli, and Enterobacter cloacae), clostridia (C. neonatale, C. butyricum, and C. perfringens)), and a low abundance of obligate anaerobic bacteria such as Bifidobacterium (B. longum sp. Infantis), Bacteroides spp., and Clostridium spp. [13,17,18].
Uncertainty surrounds how dysbiosis in gut microbiota affects NEC pathogenesis, however, results of piglet, mice, and human studies suggest that stimulation of immature enterocytes by Gram-negative lipopolysaccharide through Toll-like receptor 4 (TLR4) can lead to over-activation of inflammatory responses in the intestines of premature infants and lead to bowel damage and NEC progression [19,20]. In a study conducted by Cynthia et al. [21], TLR4- deficient C3H/HeJ mice did not develop NEC, whereas wild-type C3H/HeOUJ genotypes had a significant chance of developing NEC. This may imply the impact of TLR4 over-expression in mucosal damage, death of enterocyte cells, and bacterial translocation into bodily fluids [22].
Other studies have also shown how TLR4 prevention factors such as nucleotide-binding oligomerization domain-containing 2 (NOD2) receptor (CARD15) could prevent NEC onset. TLR4-NOD2 inhibitory interaction in enterocytes protected intestinal mucosal from NEC development. In this study enterocytes without TLR4 or NOD2 were assessed in intestinal-specific wild-type mice or mice with intestinal-specific wild-type or dominant-negative TLR4 or NOD2, and in mice with NEC. The result showed that NOD2 could prevent TLR4 expression and enterocyte apoptosis in mice models [23]. Another study has also shown the impact of Recombination-activating gene 1 (RAG1) deficiency (an essential gene in T and B lymphocyte development) in the onset of NEC. In this study, (Rag1−/−) deficient mice were protected from NEC while transferring intestinal lymphocytes from NEC mice into naive mice triggered intestinal inflammation. Moreover, inhibition of IL-17 or STAT3 (an essential factor in the differentiation of TH17 helper) lowered the risk of enterocyte proliferation and NEC in this study [24]. Gram-negative bacteria such as Enterobacteriaceae members can also influence the activation of TLR4 in the enterocyte. Preterm infants with NEC have an overabundance of LPS-producing bacteria, which could lead to the over-stimulation of TLR4. LPS-enriched gut microbiota (particularly Enterobacteriaceae-dominated microbiota) has been associated with a higher risk of epithelial necrosis and NEC in preterm infants, while bacterial communities with lower CpG DNA (potent activator of TLR4 and TLR9) have been associated with a lower risk of NEC [25]. Other studies have also shown the association of NEC with Enterobacteriaceae dominance. In Greenwood et al.’s study on 74 preterm infants with and without antibiotic administration, preterm infants who received antibiotics showed a different microbial pattern compared to the control group. Early antibiotic exposure led to a higher abundance of Enterobacter in preterm infants which may be associated with the over-activation of TLR4 and a higher risk of NEC incidence [26].
2.1.2. Late-Onset Sepsis (LOS)
Sepsis is a medical emergency that requires early diagnosis and treatment in neonates. Sepsis defines as a blood infection by pathogenic microorganisms. According to a large neonatal population-based meta-analysis study from 12 middle-income and high-income countries on four continents, the number of neonatal sepsis for each 100,000 live births was estimated at 2202 cases with a mortality rate of 11–19% or 3.0 million cases annually [27]. Neonatal sepsis may occur during the first 72 h of life by mother-to-infant pathogen transmission before or during delivery (early-onset sepsis) or it can develop later in life through hospital-associated pathogen transmission or the translocation of pathogens from the gut to the bloodstream (late-onset sepsis) [28]. The current management approach of LOS is limited to antimicrobial therapy and adjunctive therapy by increasing neutrophil quantity (e.g., granulocyte-macrophage colony-stimulating factor (GM-CSF), granulocyte transfusions, and intravenous immune globulin (IVIG) [29]. Although no specific bacterial taxa have been detected as the causative agent of LOS, recent studies have shown the association of various bacterial species to the onset of LOS. The development of LOS has been linked to a low Bifidobacterium abundance and a high abundance of Gram-negative bacteria such as enteric bacteria (E. coli, Pseudomonas spp., and Klebsiella spp.), coagulative-negative Staphylococci (CoNS), and Gram-positive bacteria (Enterococcus spp. and Streptococcus spp.) [30,31].
2.2. Allergies
Inadequate early exposure to immune system modulator factors during the crucial newborn period may result in low immunological tolerance and an exaggerated immune response to endogenous and exogenous antigens and lead to the development of allergic diseases in preterm infants [32].
Atopic disease is a broad phrase for explaining various allergic diseases in children and atopy is the overactivation of the IgE-mediated immune response to allergens, which causes a variety of allergic disorders, including food allergy, asthma, atopic dermatitis, and rhinitis [33]. Pro-allergic pathways, which are activated as the result of imbalanced Th1, Th2, and Treg phenotypes, increased secretion of IL-4, IL-5, IL-13, and low secretion of IFN-γ by Th2, can lead to the development of different allergic disorders. Activation of pro-allergic pathways can be controlled by the gut microbiome, which maintains the Th1-Th2 balance and regulates Th17 and Treg cells [34].
In healthy conditions, mature Th1 and Treg can regulate the Th2 phenotype and prevent the activation of proinflammatory cytokines [35]. Even though dysbiosis can lead to the development of allergic diseases in preterm infants, the supplementation of different Bifidobacterium strains in the regulation of anti-allergic pathways has shown promising results in the prevention of allergies.
Studies applying sequencing-based approaches have shown the association of allergic diseases with lower gut microbial diversity and lower abundance of Bifidobacterium strains between non-allergic and allergic infants [36,37,38,39,40]. For instance, Guo et al.’s study has shown that infants with cow’s milk protein allergy had lower Bifidobacterium diversity, which may explain the key role of Bifidobacterium in the digestion of essential components in milk and the gut-immune system crosstalk in infants. Moreover, a case-controlled investigation on 21 toddlers revealed different gut bacterial compositions between children with and without atopic dermatitis (AD). This study demonstrated the considerable long-term effects of immature gut microbiota on the development of allergies even after infancy by demonstrating significantly decreased Bifidobacterium abundance in children aged 3 to 5 with eczema [41]. The association of allergic diseases such as atopy and asthma with a low abundance of Bifidobacterium, Faecalibacterium, Akkermansia, and Faecalibacterium was also reported in a follow-up study on 308 children aged 1–11 months [42].
Abrahamsson et al. investigated the microbial diversity of 47 infants during the first year of life and school-age at 7 years old. This study showed that lower bacterial diversity was associated with an increased risk of subsequent allergic disease, while bacterial phyla/genera abundance did not differ significantly in children with and without allergic diseases [43]. The author has also previously shown the association of IgE-associated eczema with low gut microbial diversity in the same study population [44]. In a larger sample size, a meta-analysis study on 147,252 children showed that preterm infants with younger gestational age were at a high risk of preschool wheezing and school-age asthma. The risk of allergic diseases such as food allergies was also investigated on 13,980 preterm infants [45]. However, this study did not report any significant statistical difference in the risk of food allergy with prematurity.
2.3. Neurodevelopmental Diseases
Gut-brain axis is shaped during prenatal and postnatal life, therefore, imbalanced gut microbiota can have a significant effect on the nervous system and brain development [46]. Imbalanced gut microbiota can impact different domains of cognitive trajectories such as learning and memory, complex attention, social cognition, and executive function [47]. Among different microbial metabolites, short-chain fatty acids seem to be the main mediators in the gut-brain crosstalk [48]. However, reciprocal interaction and pathways involved in this crosstalk have not been fully understood yet.
2.3.1. Attention Deficit Hyperactivity Disorder
Attention deficit hyperactivity disorder (ADHD) is largely a heritable mental disorder, however, recent findings have shown the association of environmental factors such as nutrition and gut microbiota on the onset of ADHD. Studies have shown that pro-inflammatory inducer molecules of gut microbiota such as TNF, IL-6, and IL-1β could stimulate the brain’s innate immune system and lead to neuroinflammation and neurodevelopmental abnormalities [49]. Mouse transformation models with preterm infants’ gut microbiota induced systematic pro-inflammatory mediators such as TNF, IL-1β, IFNγ, and NOS1 in the brain, which emphasized the impact of gut microbial structure and its metabolites on neuroinflammation and brain development [50]. Furthermore, studies on adults diagnosed with attention deficit hyperactivity disorder (ADHD) have shown a different gut microbial composition in the ADHD population compared to healthy individuals. For example, Aarts et al. showed that ADHD cases had an increased abundance of Actinobacteria genus (particularly Bifidobacterium; controls: 12.66% to ADHD: 20.47%; p = 0.002). Nevertheless, this study did not investigate the functional effect of Bifidobacterium metabolites on the onset of neurodevelopmental disorders, which should be taken into account in the management of neurological disorders using gut microbial signature [51].
2.3.2. Schizophrenia Spectrum Disorder
Prospective research on neurodevelopmental outcomes in preterm infants has shown that premature infants are at a higher risk of psychotic disorders such as schizophrenia. In addition, they have a 2.9 times higher risk of developing serious depression and 7 times higher risk of bipolar illness, and a 3.5 times greater chance of developing eating disorders in their childhood and adulthood [52]. According to the Nosarti et al., study, infants born prematurely are at higher risk of hospitalization due to different psychiatric disorders [53]. Although limited studies are available on the investigation of gut microbiota with schizophrenia in preterm infants, it has been shown that patients with the first episode of psychosis showed a higher abundance of Lactobacillus, Bifidobacterium, and Ascomycota [54,55].
2.3.3. Autism Spectrum Disorder
Poor social communication skills and restricted patterns of repetitive behavior known as autism spectrum disorders (ASD) are other adverse neurodevelopmental outcomes that may develop in preterm infants. Preterm infants have a 3.3 times higher chance of autism diagnosis than full-term infants [56]. Clinical studies have shown an imbalanced gut microbial composition and metabolites in preterm infants with ASD. However, there are discrepancies in the findings which may be related to the antibiotics administration as well as different study designs and methodologies.
A systematic review conducted on 15 cross-sectional studies showed incompatible findings on gut microbial composition between ASD and non-ASD populations. Based on this study three major phyla; Firmicutes, Bacteroidetes, and Proteobacteria showed the highest variations between ASD and non-ASD populations. This study has shown a lower abundance of Bifidobacterium in the ASD group [56]. Recent metabolomics studies have also shown higher concentrations of short-chain fatty acids and lower concentrations of phenylacetylglutamine, hippurate, and 4-cresol sulfate in the ASD group compared to non-ASD controls [57].
3. General Characteristics of Bifidobacterium
Members of the Bifidobacterium genus are the most prevalent bacterial community forming 40 to 90% of the total gut microbiota at different developmental ages. Bifidobacteria are gram-positive, non-spore-forming anaerobic bacteria with pleomorphic rod morphology [58]. Bifidobacterium was first isolated from fecal samples in healthy breastfed infants by Henri Tissier at the Pasteur Institute in France in 1899 [59]. Bifidobacterium belonging to the Actinobacteria phylum has 94 recognized (sub) species classified in seven clusters including Bifidobacterium longum, Bifidobacterium bifidum, Bifidobacterium adolescentis, Bifidobacterium boum, Bifidobacterium pullorum, Bifidobacterium asteroids, and Bifidobacterium pseudolongum [60,61]. Bifidobacterium longum (subsp. Infantis), Bifidobacterium breve, and Bifidobacterium bifidum are common colonizers in the early stages of life, while Bifidobacterium adolescentis are associated with adulthood [62,63].
Successful adaptation of Bifidobacterium to the human gastrointestinal tract from infancy to adulthood may be explained by the presence of many genes attributed to stomach acid tolerance, metabolism of carbohydrates, and transport systems in the Bifidobacterium genome [64]. The average genome size of Bifidobacterium is 2.44 Mb with an average of 58.91% G + C content containing a large number of genes involved in the complex metabolism of human milk oligosaccharides (HMOs) [61]. Fermentation of HMOs by Bifidobacteria using glycosyl hydrolases produces short-chain fatty acids (SCFAs), which have many health-promoting properties including maintenance of intestinal barrier integrity and anti-inflammatory functions [65]. Moreover, the metabolism of aromatic amino acids (phenylalanine, tryptophan, and tyrosine) by Bifidobacterium produces aromatic lactic acids (4-hydroxyphenyl acetic acid (4-OH-PLA), indolelactic acid (ILA), and phenyllactic acid (PLA)), which have anti-inflammatory and antibacterial activities [66].
Even though recent studies have shown promising results in the administration of Bifidobacterium as a probiotic in the development of the gut microbiota in preterm infants, it is still unclear how Bifidobacterium abundance and its metabolites are inversely associated with the development of several life-threatening diseases in prematurely born infants.
4. Immunomodulatory Effects of Bifidobacterium
The gut-associated lymphoid tissue (GALT) is the largest mass of lymphoid tissue in the human body and contains a variety of immune cells, including B and T lymphocytes, as well as antigen-presenting cells such as dendritic cells (DC) and macrophages.
Intestinal epithelial cells provide a protective layer between intestinal mucosa and luminal microorganisms (Figure 2). For instance, Goblet and Paneth cells secrete mucus layer and antimicrobial peptides, respectively, to enhance protective effects against luminal microorganisms in the gastrointestinal tract. Secretory immunoglobulin A (sIgA) secreted by B cells have also protective roles against luminal microbiota [67].
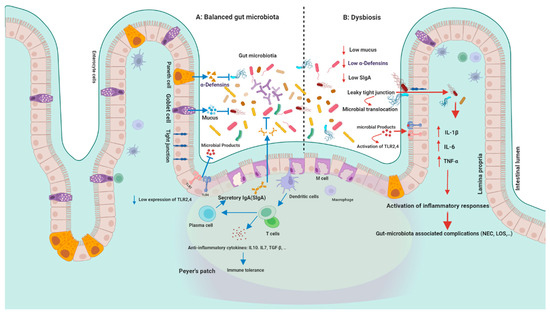
Figure 2.
Gut microbiota and immunity: (A) in healthy conditions: intestinal epithelial cells provide a protective layer between intestinal mucosa and luminal microorganisms. Goblet and Paneth cells secrete mucus layer and antimicrobial peptides. Secretory immunoglobulin A (sIgA) secreted by B cells have protective roles against luminal microbiota. While controlling permeability and microbial translocation, epithelial tight junctions (TJs) between intestinal cells maintain the integrity of the intestinal barrier. Gut microbiota-immunity crosstalk can activate different immunological pathways either in a regulated or exaggerated way and lead to the development of several diseases. Host-microbe interaction is activated through different recognition receptors, which are highly expressed in intestinal epithelial cells (IECs) such as TLRs. PRRs activation can lead to the production of different antimicrobial peptides such as α-defensins. Host-microbe interaction can influence T cell differentiation into Th1, Th2, Th17, and Treg cells, which are regulated by pro-inflammatory and anti-inflammatory cytokines such as TGF β and IL-10. (B) Dysbiosis; recognition of microbial compounds (such as Gram-negative lipopolysaccharide) by TLRs leads to the activation of MYD88 and the production of several inflammatory cytokines. TLR4 stimulation by Gram-negative bacteria causes enterocyte death and mucosal injury. TLR2 (TLR1 and TLR6) can also recognize Gram-positive bacteria. The interaction of TLRs and microbial signals leads to the activation of a cascade of immune responses. Increased intestinal permeability, TJ disruption, and subsequent uncontrolled translocation of microbial pathogens (leaky gut) can lead to several gastrointestinal diseases. Figures in this manuscript were created specifically for this manuscript in BioRender.com, accessed on 15 August 2022.
Gut microbiota-immunity crosstalk can activate different immunological pathways either in a regulated or exaggerated way and lead to the development of several diseases including gastrointestinal and dermatological disorders, allergies, and host behavioral changes [68]. This interaction is activated by beneficial microbiota and pathogens through different recognition receptors which are highly expressed in intestinal epithelial cells (IECs) including pattern-recognition receptors (PRRs), Nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs), Toll-like receptors (TLRs), C-type lectin receptors (CLRs), RIG-I-like receptor (RLR), Absent in melanoma 2 (AIM2)-like receptors (ALRs), and the oligoadenylate synthase receptor (OAS) [69]. Activation of PRRs can lead to the production of different antimicrobial peptides (AMPs) such as α- defensins (HD5, HD6) and regenerating islet-derived protein III (REGIII α, β, and γ) by immune cells and intestinal Paneth cells and restrict the access of pathogens to the mucosal epithelium [70]. Host-microbe interaction can also influence T cell differentiation into Th1, Th2, Th17, and regulatory (Treg) cells, which are regulated by pro-inflammatory and anti-inflammatory cytokines such as transforming growth factor-β (TGF β) and interleukin-10 (IL-10) [71].
The antibacterial and antiviral effects of Bifidobacterium against various pathogenic microorganisms have been the subject of numerous studies. For instance, using human colorectal adenocarcinoma cell lines (HT-29), B. longum has been proven to have an inhibitory effect against Gram-negative bacteria including Salmonella typhi STN12, Salmonella enteritidis SEN6, Escherichia coli EC4219, and Escherichia coli EC3960. Although this investigation has primarily focused on the prevention effect of B. longum on adhesion activities of Gram-negative pathogens, under in vivo conditions, a variety of contributing factors, such as intestinal epithelial cells (IECs) and IECs’ tight junction can determine how Bifidobacterium acts antagonistically [72]. Other studies have also shown the inhibitory effect of Bifidobacterium strains such as B. longum, B. adolescentis, and B. pseudocatenulatum against multidrug-resistant pathogens (e.g., E. coli), Vancomycin-resistant bacteria (Enterococcus and Staphylococcus aureus) using in vitro human cell line models and animal models [73,74,75].
Bifidobacteria has also been proven in numerous studies to have antiviral effects in mice models and colonic cells [76,77,78,79,80]. For instance, in Caco-2 and HT-29 cells, B. thermophilum RBL67 showed anti-rotaviral activities. According to this investigation, B. thermophilum RBL67 had greater adhesion indices on Caco-2 and HT-29 cells than B. thermacidophilum isolated from newborn fecal samples (RBL69 and RBL70). However, to confirm the inhibitory effects of Bifidobacterium strains on bacterial and viral infections further studies in human-like models are needed [76].
Bifidobacterium strains can also contribute to the regulation of pro-inflammatory and anti-inflammatory cytokines; in a case-control study, the intervention population who consumed dairy products containing B. lactis and other beneficial strains showed higher serum levels of pro-inflammatory cytokines (interferon-γ (IFN-γ), interleukin 12 ((IL12), and immunoglobulin (Ig)) and higher activity in natural killer cells, which may suggest the effectiveness of the Bifidobacterium in the improvement of immune responses and NK cell functions [81]. Bifidobacterium strains can also induce macrophage mediators and modulate host immune responses. It was also shown that B. pseudocatenulatum SPM1204 isolated from fecal samples cultured with dendritic cells and macrophages increased histocompatibility complex (MHC) class I and induced the production of nitric oxide (NO), tumor necrosis factor (TNF), and IL1 [82]. Table 1 shows the Bifidobacterium role as a probiotic in preterm infants in human studies.

Table 1.
Clinical trials on the effects of Bifidobacterium strains in preterm infants.
5. Bifidobacterium as Probiotic
According to the definition introduced by the Food and Agriculture Organization of the United Nations and the World Health Organization (FAO/WHO), probiotic is live microorganisms that when administered in adequate amounts confer a health benefit on the host [118].
Nutrition has been identified as the main approach in the regulation of host physiology and programming of the gut microbiota in the early stages of life. The significant impact of environmental factors including diet on gut microbiota brought the new concept of “Early-life Nutritional Programming” theory during the first three years of life which has long-lasting consequences throughout the lifespan [119]. Reprogramming of gut microbiota by maintaining the balance of beneficial bacterial species through the administration of probiotic strains can prevent several microbiota-associated infections in preterm infants and protect survivors from severe morbidity.
Owing to the protective and immunomodulatory effects of Bifidobacterium in early life, the European Food Safety Authority (EFSA) has approved the Qualified Presumption of Safety (QPS) status of different species of Bifidobacterium including B. longum, B. breve, B. bifidum, B. adolescentis, and B. animalis [120].
Human milk is the first source of Bifidobacterium species and predominates in breastfed infants during the first three years of life. The metabolism of human milk oligosaccharides by Bifidobacterium species can alter gut microbial composition and promote immune system development. B. longum subsp. Infantis, B. longum subsp. Longum, B. bifidum, and B. breve are the most commonly identified species in newborn infants [121]. The primary protective function of Bifidobacterium species is their overabundance in human milk, which can result in higher Bifidobacterium colonization in the gastrointestinal tract, particularly in the colon, as the most suitable niche for this bacterial community [122]. Additionally, in vivo investigations have demonstrated that Bifidobacterium species grown on HMO have excellent adhesion abilities to intestinal epithelial cells, which is essential to compete with opportunistic pathogens [123]. Additionally, prebiotics such as HMO and lactoferrin in human milk which are human non-digestible beneficial components can be metabolized by the gut microbiota and promote the growth of beneficial microorganisms and prevent the overgrowth of pathogenic microorganisms in the gastrointestinal tract [124].
5.1. Bifidobacterium and Prevention of NEC and LOS
Recent findings suggested probiotics as the most effective human intervention in the management of LOS and NEC. According to a systematic analysis of 44 observational, randomized controlled, and RCTs studies, probiotics could reduce the sepsis rate by 12% in RCTs and 19% in observational studies in preterm infants. This study has also shown a slight reduction in NEC incidence in observational studies. The results suggested the beneficial effect of probiotics in the prevention of late-onset sepsis, NEC, and mortality rate in preterm infants [125]. Another meta-analysis review of 16 studies including 2842 preterm infants revealed a significant impact of probiotic supplementation on NEC incidence (typical RR 0.35, 95% CI 0.24 to 0.52) and mortality rate (typical RR 0.40, 95% CI 0.27 to 0.60), while no significant reduction was reported on sepsis incidence (typical RR 0.90, 95% CI 0.76 to 1.07) [126]. The effectiveness of specific probiotic strains in NEC prevention was also evaluated in a meta-analysis review of 26 studies. In this study findings from 6605 infants (placebo: 3281 and probiotic: 3324) showed that the relative risk of NEC was significantly lower in infants receiving probiotics compared to the placebo group (0.47 (95% CI 0.36–0.60) p < 0.00001). Studies using Lactobacillus GG [127,128], Lactobacillus reuteri [129,130], Lactobacillus sporogenes, and Saccharomyces boulardii [131,132,133] showed no significant reduction in NEC incidence (0.62 (95% CI 0.37–1.05), p = 0.07) [134]. In contrast, studies using B. lactis [86,87,92,101], B. breve [83,97], B. bifidum [135] showed a significant reduction in relative risk of NEC in the probiotic group (0.24 (95% CI 0.10–0.54), p = 0.0006).
Investigation of Bifidobacteria’s role in the prevention of gastrointestinal disorders in animal models has also shown encouraging results. The prevention role of Bifidobacteria on intestinal microbes’ invasion from mucosa to internal organs showed that bacterial translocation in Peyer’s patches in mice models decreased by higher Bifidobacteria colonization in caecum and colon and prevented blood, liver, and lungs infections, while colonization of other pathogenic microorganisms such as Bacteroides fragilis and clostridia were associated with increased risk of bacteremia and lung infection in these models [136]. Also, the transcriptional activity of enterocytes and regulation of innate immune-mediated inflammation in mice models has shown that administration of B. infantis downregulated the expression of IL8, IL6, TNFα, IL23, iNOS, and antimicrobial peptides and altered the expression of intestinal mucus-related proteins and led to the low incidence of NEC in animal models [137]. B. infantis administration has also been associated with enhanced expression of tight junction proteins (4 Claudin and occludin), and a low incidence of NEC in the neonatal mouse NEC model [138].
Some studies have also shown the higher effectiveness of multiple species compared to single-species probiotics. For instance, the comparison of daily administration of a single strain of B. breve M-16V (5 × 108; one-species group) and a combination of three species B. breve M-16V, B. longum subsp. infantis M-63 and B. longum subsp. longum BB536 (5 × 108 of each strain; three-species group) for one week has shown that Bifidobacterial fecal count was significantly higher in preterm infants who received three-species probiotics compared to the one-species group. Moreover, the abundance of pathogenic bacterial species such as Clostridium and Enterobacteriaceae was significantly lower in preterm infants who received three-species probiotics [95]. Combination of probiotic strains including B. longum subsp. infantis BB-02, B. animalis subsp. lactis BB-12, and S. thermophilus TH-4 in 459 preterm infants (probiotic: 229 and placebo: 230) could also increase the abundance of probiotic species in the gut microbiota of preterm infants which may imply the importance of early administration of multi-strain probiotics on the abundance of beneficial bacterial species in preterm infants [139]. Likewise, a comparison of 119 preterm infants who received human milk with probiotics (combined supplementation of B. breve and Lactobacillus casei) with 112 preterm infants receiving human milk without probiotics showed that supplementation of B. breve and L. casei reduced the NEC occurrence [93].
Metabolomic studies have also shown the association of probiotic supplementation with variation in concentration of beneficial health indicators such as short-chain fatty acids (SCFAs) (acetate and lactate) in preterm infants. Short-chain fatty acids are one of the primary microbial byproducts of the breakdown of human milk oligosaccharides and indigestible fiber [140]. Primary colonization of gut microbiota with lactate-producing bacteria (e.g., Bifidobacterium, Lactobacillus, and Bacteroides) in infants has beneficial effects on the maturation of epithelial cells and mucosal dendritic cells. As a result, the level of fecal SCFAs can indicate microbial structure and state of health in infants. According to an observational longitudinal study on 234 preterm infants (probiotic:101 and placebo:133), supplementation of Bifidobacterium and Lactobacillus was associated with higher fecal acetate and lactate and a lower fecal pH in the probiotic group compared to the placebo group. A higher concentration of acetate and lactate may show the exceptional ability of the Bifidobacterium strain in metabolizing human milk oligosaccharides into SCFAs [110]. Another study investigating the role of B. lactis Bb12 supplementation on health indicators of preterm infants showed that in preterm infants receiving probiotic fecal pH and calprotectin (an indicator of gastrointestinal disorders) were significantly lower compared to the placebo group, while fecal concentrations of acetate, lactate, and IgA were significantly higher in the probiotic group compared to the placebo group [86].
While some research found encouraging results in the prevention of common complications in preterm infants using probiotic strains, other studies have shown no association in this regard. For instance, a single-center retrospective study of 293 preterm infants (37 NEC cases) who were routinely supplemented with a multispecies probiotic for 4 years prior to and 5 years after probiotic administration (n = 14, n = 23, respectively) showed no significant difference in NEC rate [115]. However, these findings may show an already low rate of NEC rate in this center, and a multi-center retrospective analysis is needed to determine the beneficial effects of probiotics in NEC reduction and mortality. Similarly, a randomized controlled study on 1315 preterm infants (probiotics:650 and placebos: 660) showed that preterm infants who received daily B. breve BBG-001 over 6 weeks showed no significant reduction in NEC rate and late-onset sepsis compared to the placebo [99]. Routine administration of B. breve M-16V (1 mL = 1.5 billion CFU) in preterm infants and full-term infants also did not reduce the NEC and LOS rate between preterm (n = 162) and full-term infants (n = 1218) in a similar study [114]. Underestimation of the beneficial effects of probiotic strains may also be caused by cross-contamination of the placebo and probiotic participants or unsuccessful colonization of probiotic strains in the probiotic group due to antibiotic use or gastrointestinal immaturity. However, ignoring the effectiveness of probiotic strains might be a simple conclusion; larger randomized controlled trials are needed to evaluate the impact of probiotics, prebiotics, or a combination of both known as symbiotics on the prevention of common complications in preterm infants.
5.2. Bifidobacterium and Prevention of Neurodevelopmental Diseases
Due to the anti-inflammatory effects of probiotics including the prevention of brain tissue infection such as white matter infection and modulation of brain development through regulation of immune cytokines, hormones, and neurotransmitters, probiotics may have neuroprotective effects in preterm infants. Recent findings suggest that early exposure to probiotics in preterm infants may be protective against neurodevelopmental disorders such as attention deficit hyperactivity disorder (ADHD) and autism spectrum disorder (ASD) [141]. For instance, Partty et al., have investigated the association of early probiotic intervention with neuropsychiatric disorders. In this study, 75 eligible infants (probiotic:40 and placebo:35) have been recruited. This study has shown that infants who received probiotics (Lactobacillus rhamnosus GG (ATCC 53103)) during the first 6 months of life which were followed up for 13 years had a lower rate of ADHD disorder compared to the placebo group 6/35 (17.1%). In contrast, none of the infants receiving probiotics was diagnosed with ADHD (p = 0.008). Bifidobacterium abundance was also lower in children diagnosed with ADHD during their infancy than in children without any neurodevelopmental disorder [142]. Another long-term follow-up study on 67 preterm infants (probiotic:36 and placebo:31) has also found that supplementation of B. breve M-16V (commonly isolated from human milk) did not have any significant effect on different developmental skills (e.g., language, learning, and memory, executive ability ad attention, social skills, sensorimotor functioning, and visuospatial processing) at 3 to 5 years age in preterm infants [111]. Also, combined probiotic treatment using B. infantis, B. lactis, and Streptococcus thermophilus on 1099 very preterm infants (probiotic:548, and placebo:551) over 2 to 5 years, showed no adverse neurodevelopment and behavior changes later in childhood. In this study, the development of infants was assessed across cognitive, language, and motor development domains following the Bayley-III tool [106]. Although some of these studies have been limited to a low number of participants and a low follow-up rate, the findings may be useful in designing long-term follow-up studies on the safety and long-term effects of probiotic administration in preterm infants.
6. Bifidobacterium: Mechanism of Action
Probiotic strains can modulate the host immune system through several mechanisms (Figure 3). Major mechanisms of action include modulation of adaptive and innate immunity, enhancement of intestinal epithelial barrier, prevention of pathogen adhesion, and production of antimicrobial compounds, which have been discussed in detail as follows.
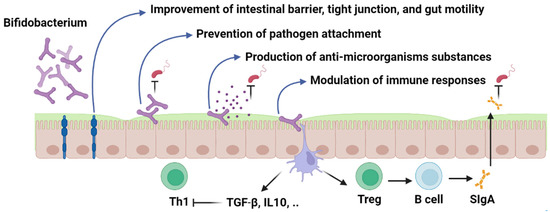
Figure 3.
Bifidobacterium; mechanism of action. Figures in this manuscript were created specifically for this manuscript in BioRender.com, accessed on 15 August 2022.
6.1. Modulation of the Immune System
The innate immune system also known as the nonspecific immune system is the first line of defense in the human body including the protective effects of skin and mucosal membrane and immune system cells. While the adaptive immune system is a specific immunity to identifying pathogens by specialized immune cells including B and T lymphocyte cells [143]. Probiotics can modulate innate and adaptive immunity and lead to the enhancement of intestinal epithelium through immune mediators such as Toll-like receptors (TLRs), cytosolic signaling receptors such as nucleotide-binding oligomerization domain leucine-rich repeat-containing and pyrin domain-containing (NLRP), and anti-inflammatory cytokines.
6.2. Intracellular Immune Receptors (TLRs, NLRs) and Anti-Inflammatory Mediators
Intracellular immune receptors have a remarkable role in recognizing pathogen-associated molecular patterns (PAMPs) and microbial signals. Toll-like receptors (TLRs) are highly expressed in immune cells (dendritic cells, macrophages, and Natural killer cells (NK)) and non-immune cells (endothelial and epithelial cells). Recognition of microbial compounds by TLRs leads to the activation of downstream immune responses and the production of several inflammatory cytokines and other immune mediators which lead to innate and adaptive immune responses [144]. Enterocytes or intestinal absorptive cells line the inner surface of the intestine and express TLR4 as abundant proteins on their outer surface which are in close contact with microbial compounds in the gut lumen. TLR4 can recognize Lipopolysaccharide (LPS) in Gram-negative bacteria and activate MYD88 protein (myeloid differentiation primary response 88). Activation of MYD88 leads to kinase activation and degradation of NFκB/IKB dimer (Nuclear factor kappa-light-chain-enhancer of activated B cells)/(an inhibitory protein bound to NFκB). After the degradation of the NF-κB/IKB dimer, NFκB complex is translocated to the nucleus where the gene transcription of many pro-inflammatory cytokines, tumor necrosis factor-alpha (TNFα), and interleukin occur [145]. As previously mentioned, TLR4 stimulation by Gram-negative bacteria causes enterocyte death and mucosal injury, both of which have been related to the etiology of NEC in several studies.
It has been shown that Bifidobacterium probiotics and their metabolites can alter the transcriptional activity of enterocytes and modulate the intestinal innate immune response. For instance, probiotic-conditioned media (PCM) with a single probiotic strain or combined probiotic strains including B. infantis and L. acidophilus could lead to a significant decrease in the expression of IL-1β, IL-8, IL-6, TLR2 mRNA, and TLR4 mRNA and high expression of inflammatory inhibitors (Tollip and SIGIRR). Exposure of PCM with primary enterocyte cultures of NEC tissue has also led to down-regulation of IL-6, IL-8, and TLR2 and up-regulation of Tollip and SIGIRR [146].
Similar to this, transcription profiling of immature human fetal intestinal epithelial cells exposed to B. infantis and L. acidophilus revealed modification of several genes involved in immune responses and cell survival pathways. Probiotic conditioned media (PCM)-exposed cells displayed decreased NF-B pathway gene expression as well as IL-6 and IL-8 levels. As a result of PCM exposure, genes involved in remodeling the extracellular matrix were also downregulated [147]. Given the strong influence of probiotic strains on the regulation of NF-κB pathways, it can be a potential therapeutic strategy to manipulate receptors and cytokines which leads to the activation of this pathway in the functionally immature intestinal tract in preterm infants. TLR2 detected on the surface of several immune cells has also the same function as TLR4. Since immature enterocytes in preterm infants have been associated with high expression of TLR2, probiotic administrations have shown a significant impact on the regulation of TLR2-ligand interaction. The heterodimeric complex of TLR2 with TLR1 and TLR6 can recognize Gram-positive bacteria compounds such as lipoteichoic acids, peptidoglycan, and lipopeptides. The interaction of TLRs and microbial signals leads to the activation of a cascade of immune responses [148].
Modulation of TLR2 and TLR4 expression and development of the immune system through probiotic strain activities have been investigated in several animal-based studies. For instance, an investigation of Bifidobacterium administration in intestinal epithelial cells in rat models showed that TLR2 expression was significantly lower in intestinal epithelial cells treated with different strains of Bifidobacterium (B. longum, B. infantis, and B. youth). While cells infected by E. coli endotoxin showed higher expression of TLR2 and TLR4. Also, intestinal barrier function measured by transepithelial/transendothelial electrical resistance (TEER) was significantly higher in Bifidobacterium-treated cells compared to cells infected by E. coli endotoxin [149].
Human cytokine synthesis inhibitory factor (CSIF) or interleukin 10 (IL-10) mainly produced by monocytes and other immune cells such as Th2, Treg, mast cells, and B cells, is another anti-inflammatory cytokine that can be regulated by probiotics. IL-10 can suppress the production of several pro-inflammatory cytokines including TNFα, IFN-γ, GM-CSF, IL-2, and IL-3 [150]. Animal-based studies have also confirmed the regulatory effect of Bifidobacterium strains on IL-10 and subsequently the prevention of inflammatory bowel diseases. For instance, L. casei and B. breve-treated mouse models could selectively enhance the amount of IL-10-producing CD4+ T cells in the large intestine by twofold without altering intestinal microbiota [151]. B. adolescentis supplementation in preterm rat models could also decrease the development of NEC through the modulation of inhibitory adaptor proteins such as TOLLIP, and inhibitory receptor toll interleukin-1R 8 (SIGIRR) and expression of TLR4 [152].
Inhibition of NLRP3 inflammasome (NOD-, LRR- and pyrin domain-containing protein 3) has also shown promising results in the prevention of gastrointestinal disorders. NLRP3 inflammasome is a cytosolic multiprotein oligomer in the innate immune system belonging to the nucleotide-binding oligomerization domain-like receptors (NOD-like receptors: NLRs).
NLRP3 acts as a pattern recognition receptor (PRR) and can detect microbial signals and lead to the production of proinflammatory cytokines (IL-1β) and caspase 1 [153]. Overactivation of NLRP3 has been associated with the development of different inflammatory diseases which can be regulated by the inhibitory effects of probiotic strains [154]. Investigation of NLRP3 inflammasome in NEC mouse models treated with NLRP3 inhibitor MCC950 showed that the NEC mouse model showed higher expression of NLRP3 in the intestine and brain and mature IL-1β compared to mice receiving NLRP3 inhibitor (MCC950). As a result, inflammatory cytokines, NEC survival rate, and histological damage in the brain and gut were all dramatically decreased by MCC950 treatment, demonstrating the significance of blocking the NLRP3 pathway in the prevention of inflammatory bowel disorders [150].
6.3. Regulation of Intestinal Epithelium Function
The gastrointestinal barrier provides a vast surface for interacting with microbial signals and environmental stimuli. This contact has a substantial impact on the host’s physiology and may trigger a regulated and normal immunological response or infection development, depending on the initial stimulus. The outermost layer of the intestinal epithelium is made up of enterocytes, Paneth cells, goblet cells, intraepithelial lymphocytes, and enteroendocrine cells. While controlling permeability and microbial translocation, epithelial tight junctions (TJs) between intestinal cells maintain the integrity of the intestinal barrier [155]. Increased intestinal permeability, TJ disruption, and subsequent uncontrolled translocation of microbial pathogens (leaky gut) may occur in preterm newborns with an underdeveloped gut barrier and lead to gastrointestinal diseases.
Human and animal trials have shown the prophylactic effects of Bifidobacterium strains on the intestinal barrier [156]. Investigation of Bifidobacterium’s role on the TJ and intestinal barrier in animal models and human intestinal cell models (Caco-2) has shown that Bifidobacterium administration can down-regulate the expression of proinflammatory cytokines and improve transepithelial electrical resistance and permeability of Caco-2. Bifidobacterium in 108 CFU could also increase the expression of ZO-1, occludin, and claudins (TJ proteins) (p < 0.01) compared to Caco-2 monolayers treated with LPS. Moreover, compared to the LPS-induced enterocyte barrier injury of Caco-2 monolayers (E. coli 055), and LPS-fed mice models, Bifidobacterium significantly suppressed the expression of TNF-α and IL-6 and decreased the NEC rate from 88 to 47% (p < 0.05) in controls [157]. Another study showed a different approach in regulation of intestinal barrier by Bifidobacterium strains. This study has demonstrated that B. bifidum (108 CFU) might improve the intestinal epithelial tight junction barrier in Caco-2 monolayers by targeting the TLR2 pathway in an NF-B-Independent manner (attachment with enterocyte TLR-2 receptors and stimulation of p38 kinase pathway) [158].
6.4. Competitive Exclusion and Adhesion Properties
Elimination of pathogens with identical needs for resources by probiotic strains known as competitive exclusion is a common strategy applied by probiotic microorganisms in the gastrointestinal tract [159]. Adhesion of probiotics to the intestinal epithelium can prevent the attachment and colonization of bacterial pathogens, especially enteropathogens, and resultant infections. Probiotics adhesion can also enhance host-probiotic interaction which leads to longer transient colonization time and provide sufficient time to express their immunomodulatory effects while attached to the epithelial receptors [160].
Serine protease inhibitor (serpin) produced by B. longum subsp. Longum NCC2705, B. longum subsp. Infantis, B. dentium, and B. breve and pentapeptide (CHWPR) in B. animalis are common extracellular proteins that facilitate host-probiotic interaction. Neutrophil and pancreatic elastases which are produced during inflammation by immune cells can be prevented by Bifidobacterium serin and suppress inflammatory responses and immune cell recruitment [161]. CHWPR can also pass through the cytoplasmic membrane and reach the nucleus and upregulate c-myc and il-6 genes, which are involved in many cellular metabolisms including gastrointestinal tract physiology [162].
Several in vitro and in vivo studies have investigated different extracellular proteins in probiotics using intestinal cell lines to evaluate the antagonistic interactions between pathogens and probiotics. In a study assessing the adhesion ability of 12 commonly used probiotic strains and antagonistic interactions with enteropathogens (Enterobacter, Clostridium, Staphylococcus, and Bacteroides), all tested probiotic strains could prevent bacterial pathogen colonization in the intestinal epithelium models [163]. Tight adhesion (Tad) pili (Type IVb pili) in B. breve UCC2003 has also been found to be a critical element for gut colonization (202) and has a proliferation impact on intestinal epithelial cells in mice models [164]. A comparative study on the physiological characteristics and acid-resistant phenotype of B. longum and B. catenulatum has shown that acid-resistant Bifidobacterium strains showed a greater adhesion to the human intestinal mucus and a higher displacement ability (competitive exclusion) on E. coli, Salmonella enterica serovar Typhimurium, Listeria monocytogenes, Enterobacter sakazakii, and Clostridium difficile from adhering to human intestinal mucus compared to the acid-sensitive strains. These results highlight the significance of carefully evaluating the safety, effectiveness, and phenotypic traits of probiotic strains before clinical trial research [165].
The human plasminogen-binding activity of different species of Bifidobacterium (B. bifidum, B. longum, and Bifidobacterium lactis) has also shown that Bifidobacterium has a unique adhesion ability through degradation of the extracellular matrix which allows Bifidobacterium-host interaction [166].
In another study, the phenotypic characteristics of B. breve and B. longum isolated from preterm and full-term infants were examined. This study revealed a significant variation across different isolates in terms of Caco-2 cells adhesion, surface hydrophobicity, and autoaggregation properties which may show strain-specific phenotypic traits that should be considered when choosing the probiotic candidate for modifying the gut microbiota in preterm newborns [98]. It might also explain why, despite probiotic treatment, some investigations have not shown successful competitive exclusion or fecal detection of Bifidobacterium. These findings may point to the need for a case-by-case comparison of probiotic strains and infectious agents in order to identify the optimal probiotic candidate with the potential to adhere to and colonize the gastrointestinal tract while also improving disease outcomes.
6.5. Synthesis of Antimicrobial Compounds
Another successful tactic against Gram-positive and Gram-negative bacteria is the production of antibacterial compounds by probiotic strains. There have been several low molecular weight compounds (LMWs) found in Bifidobacterium strains that show inhibitory properties against pathogens. For instance, short-chain fatty acids (such as acetate, butyrate, and propionate) are the end-products of the metabolism of human undigestible carbohydrates produced by gut microbiota and probiotic strains and have been used as health indicators in the diagnosis of gastrointestinal diseases. In multiple human and animal investigations, the administration of Bifidobacterium was linked to greater levels of short-chain fatty acids and a reduction in intestinal damage [85,90,167]. Numerous studies have also linked LMW lipophilic compounds to the inhibitory actions of Bifidobacterium [168,169]. In Caco-2 cells and mouse models, for example, the antibacterial activity of 14 Bifidobacterium strains isolated from newborn fecal samples against S Typhimurium SL1344 revealed antagonistic action of Bifidobacterium strains either through cell entry prevention or intracellular inhibition [168].
7. Safety of Bifidobacterium Probiotic
Despite the common use of probiotics in preterm infants and the low rate of adverse effects, controversies remain around the safety, short-term and long-term effects of probiotic administration. The safe use of probiotics in preterm infants has been documented in numerous studies, but there is no guarantee of their absolute safety, which calls for ongoing observation and case-by-case evaluation.
For instance, in a preterm infant with surgery for omphalocele four hours after birth and treated with Bifidobacterium breve BBG-01 probiotic on day 2, the blood culture was positive for Bifidobacterium breve BBG-01 (resistant to meropenem, and susceptible to Ampicillin/Sulbactam and penicillin in vitro), which was genetically identical to orally administered probiotic strain [170]. This may raise the importance of case-by-case evaluation and potential risk factors of probiotic strains in preterm infants with particular medical conditions as other preterm infants had been treated with a similar probiotic strain without displaying any systematic consequence in this study. Another case study reported LOS diagnosis in a preterm infant with laparotomy and probiotic treatment (Lactobacillus rhamnosus). It is important to note that in these two case studies, both infants were diagnosed with underlying intestinal diseases prior to probiotic treatment [171].
There are still a lot of unanswered questions surrounding the target population, the choice of efficient probiotic strains, the length of therapy, and the dosage. For instance, a systematic review and meta-analysis of 51 randomized controlled trials with 11231 preterm infants revealed that not all research used the same probiotic strains and same dosage in preterm newborns, making it challenging to evaluate the safety, effectiveness, and optimal dosage of different probiotic strains. In this systematic and meta-analysis review, three combined probiotic therapy out of 25 studies showed a mortality reduction rate, seven therapies decreased NEC, two reduced LOS, and three treatments reduced enteral feeding time [172]. However, this study was unable to draw any definitive conclusions regarding the most effective probiotic strains for various clinical outcomes which might be due to a limited number of studies and lack of a standardized method of probiotic treatment. Another meta-analysis review with 24 studies showed a significant association between probiotic administration and NEC reduction rate and mortality, with no remarkable impact on LOS and without any reported systematic infection after using a single probiotic strain (lactobacillus) or in combination with Bifidobacterium strains [173]. These findings show that a lack of adherence to a standard protocol in probiotic therapy could result in the inappropriate or even unsafe administration of probiotics to premature infants.
From the manufacturing point of view, probiotic strains undergo five main phases: strain selection, culture, fermentation, centrifuge, and blending. As probiotic manufacturing has a long history in the food industry, probiotic strains are mainly certified for dietary use as a food supplement, not for medical purposes which is of the utmost importance, particularly in vulnerable individuals with medical conditions. Also, the Qualified Presumption of Safety provided by the European Food Standards Agency (EFSA) and the Good Manufacturing Practice (GMP) provided by the Food and Drug Administration (FDA) in the USA do not require the medical efficiency and quality of probiotic strains [174]. Additionally, some beneficial properties of probiotics are just strain-specific and shouldn’t be generalized to a formulation. Potential phenotypic and genotypic variations in probiotic strains under in vivo and in vivo conditions should also be noted when determining the beneficial effects of probiotic strains [174]. For example, a comparison of Bifidobacterium strains in 16 probiotic products showed that in over 90% of the cases, Bifidobacterium strains in the product did not show the same descriptions and properties claimed on the package label [175,176]. These findings highlight the urgent need for a regulated and consistent protocol from probiotic strain production to delivery, especially for medical uses.
Though there are not many case studies reporting the adverse effect of Bifidobacterium probiotics, the potential risks should not be discounted because some adverse effects may remain unreported due to difficulty in isolation of probiotic strains which are usually anaerobes and hard to grow. Given the difficulties in isolating probiotic strains from clinical specimens, research and diagnostic laboratories should be equipped with proper methods and tools to accurately evaluate probiotic strains. Benefits and risk considerations should be assessed in critically ill populations, even though a consistent protocol can enhance benefits and decrease adverse effects. Also, studies utilizing different probiotic strains should take further measures to identify, assess, and report any relevant risk factors.
8. Conclusions
Bifidobacterium is one of the initial and dominant colonizers of the gastrointestinal tract with protective and immunomodulatory roles. Many preclinical and clinical studies have shown the effectiveness of Bifidobacterium probiotics as a therapeutic approach in the prevention and treatment of preterm infant complications including inflammatory intestinal diseases, neurodevelopmental diseases, and allergies. However, many studies discussed here had limitations, including a possible bias in the study design, small sample size, cross-contamination, low follow-up rate, single-center comparison, and lack of a standardized method in terms of probiotic dose and treatment duration. Therefore, well-designed studies with larger sample sizes are required to fully evaluate the reciprocal interaction between the host and Bifidobacterium probiotic. Also, further investigations in human and animal trials are needed to fully evaluate the effectiveness of Bifidobacterium (single or combined probiotics) as a microbiome-targeted intervention for the re-programming of the gut microbiota and treatment of gut-microbiota-associated diseases in preterm infants and other vulnerable populations.
Author Contributions
F.S.H. study design, data extraction, and writing the manuscript, H.H. reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| TLR | toll-like receptor |
| FOS | Fructooligosaccharides |
| GOS | galactooligosaccharides |
| NEC | Necrotizing enterocolitis |
| NOD2 | nucleotide-binding oligomerization domain-containing 2 |
| LOS | Late-onset sepsis |
| AD | atopic dermatitis |
| ADHD | Attention deficit hyperactivity disorder |
| ASD | autism spectrum disorders |
| HMOs | human milk oligosaccharides |
| GALT | gut-associated lymphoid tissue |
| DC | dendritic cells |
| sIgA | Secretory immunoglobulin A |
| IECs | intestinal epithelial cells |
| PRRs | pattern-recognition receptors |
| CLRs | C-type lectin receptors |
| RLR | RIG-I-like receptor |
| AIM2 | Absent in melanoma 2 like receptors |
| OAS | oligoadenylate synthase receptor |
| AMPs | antimicrobial peptides |
| TGF β | transforming growth factor-β |
| IL | interleukin |
| IFN-γ | interferon-γ |
| Ig | immunoglobulin |
| TNF | tumor necrosis factor |
| QPS | Qualified Presumption of Safety |
| EFSA | European Food Safety Authority |
| FDA | Food and Drug Administration |
| PAMPs | pathogen-associated molecular patterns |
| TJs | tight junctions |
| LMWs | low molecular weight compounds |
References
- Kumbhare, S.V.; Patangia, D.V.; Patil, R.H.; Shouche, Y.S.; Patil, N.P. Factors influencing the gut microbiome in children: From infancy to childhood. J. Biosci. 2019, 44, 49. [Google Scholar] [CrossRef] [PubMed]
- Kapourchali, F.R.; Cresci, G.A. Early-Life Gut Microbiome—The Importance of Maternal and Infant Factors in Its Establishment. Nutr. Clin. Pract. 2020, 35, 386–405. [Google Scholar] [CrossRef] [PubMed]
- Walker, W. Dysbiosis. In The Microbiota in Gastrointestinal Pathophysiology; Elsevier: Amsterdam, The Netherlands, 2017; pp. 227–232. [Google Scholar]
- WHO. Available online: https://www.who.int/news-room/fact-sheets/detail/preterm-birth (accessed on 15 August 2022).
- Schmutz, N.; Henry, E.; Jopling, J.; Christensen, R. Expected ranges for blood neutrophil concentrations of neonates: The Manroe and Mouzinho charts revisited. J. Perinatol. 2008, 28, 275–281. [Google Scholar] [CrossRef]
- Li, J.; Li, H.; Mao, H.; Yu, M.; Yang, F.; Feng, T.; Fan, Y.; Lu, Q.; Shen, C.; Yin, Z. Impaired NK cell antiviral cytokine response against influenza virus in small-for-gestational-age neonates. Cell. Mol. Immunol. 2013, 10, 437–443. [Google Scholar] [CrossRef]
- Pammi, M.; Cope, J.; Tarr, P.I.; Warner, B.B.; Morrow, A.L.; Mai, V.; Gregory, K.E.; Kroll, J.S.; McMurtry, V.; Ferris, M.J. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: A systematic review and meta-analysis. Microbiome 2017, 5, 31. [Google Scholar] [CrossRef]
- Pietzak, M. Bacterial colonization of the neonatal gut. J. Craniofacial Surg. 2004, 38, 389–391. [Google Scholar] [CrossRef]
- Carbone, F.; Montecucco, F.; Sahebkar, A. Current and emerging treatments for neonatal sepsis. Expert Opin. Pharmacother. 2020, 21, 549–556. [Google Scholar] [CrossRef]
- Harmsen, H.J.; Wildeboer–Veloo, A.C.; Raangs, G.C.; Wagendorp, A.A.; Klijn, N.; Bindels, J.G.; Welling, G.W. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 2000, 30, 61–67. [Google Scholar] [CrossRef]
- Tannock, G.W. Analysis of bifidobacterial populations in bowel ecology studies. Bifidobacteria. Genom. Mol. Asp. 2010, 1–15. [Google Scholar]
- Avershina, E.; Storrø, O.; Øien, T.; Johnsen, R.; Wilson, R.; Egeland, T.; Rudi, K. Bifidobacterial succession and correlation networks in a large unselected cohort of mothers and their children. Appl. Environ. Microbiol. 2013, 79, 497–507. [Google Scholar] [CrossRef]
- Masi, A.C.; Embleton, N.D.; Lamb, C.A.; Young, G.; Granger, C.L.; Najera, J.; Smith, D.P.; Hoffman, K.L.; Petrosino, J.F.; Bode, L. Human milk oligosaccharide DSLNT and gut microbiome in preterm infants predicts necrotising enterocolitis. Gut 2021, 70, 2273–2282. [Google Scholar] [CrossRef] [PubMed]
- Kliegman, R.; Fanaroff, A. Necrotizing enterocolitis. New Engl. J. Med. 1984, 310, 1093–1103. [Google Scholar] [CrossRef] [PubMed]
- Alsaied, A.; Islam, N.; Thalib, L. Global incidence of necrotizing enterocolitis: A systematic review and meta-analysis. BMC Pediatr. 2020, 20, 344. [Google Scholar] [CrossRef] [PubMed]
- Hackam, D.J. Necrotizing Enterocolitis: Pathogenesis, Diagnosis and Treatment; CRC Press: Boca Raton, FL, USA, 2021. [Google Scholar]
- Brown, C.T.; Xiong, W.; Olm, M.R.; Thomas, B.C.; Baker, R.; Firek, B.; Morowitz, M.J.; Hettich, R.L.; Banfield, J.F. Hospitalized premature infants are colonized by related bacterial strains with distinct proteomic profiles. MBio 2018, 9, e00441-18. [Google Scholar] [CrossRef]
- Kaelin, E.A.; Rodriguez, C.; Hall-Moore, C.; Hoffmann, J.A.; Linneman, L.A.; Ndao, I.M.; Warner, B.B.; Tarr, P.I.; Holtz, L.R.; Lim, E.S. Longitudinal gut virome analysis identifies specific viral signatures that precede necrotizing enterocolitis onset in preterm infants. Nat. Microbiol. 2022, 7, 653–662. [Google Scholar] [CrossRef]
- Fundora, J.B.; Guha, P.; Shores, D.R.; Pammi, M.; Maheshwari, A. Intestinal dysbiosis and necrotizing enterocolitis: Assessment for causality using Bradford Hill criteria. Pediatr. Res. 2020, 87, 235–248. [Google Scholar] [CrossRef]
- Jilling, T.; Simon, D.; Lu, J.; Meng, F.J.; Li, D.; Schy, R.; Thomson, R.B.; Soliman, A.; Arditi, M.; Caplan, M.S. The roles of bacteria and TLR4 in rat and murine models of necrotizing enterocolitis. J. Immunol. 2006, 177, 3273–3282. [Google Scholar] [CrossRef]
- Leaphart, C.L.; Cavallo, J.; Gribar, S.C.; Cetin, S.; Li, J.; Branca, M.F.; Dubowski, T.D.; Sodhi, C.P.; Hackam, D.J. A critical role for TLR4 in the pathogenesis of necrotizing enterocolitis by modulating intestinal injury and repair. J. Immunol. 2007, 179, 4808–4820. [Google Scholar] [CrossRef]
- Hackam, D.J.; Sodhi, C.P. Bench to bedside—New insights into the pathogenesis of necrotizing enterocolitis. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 468–479. [Google Scholar] [CrossRef]
- Richardson, W.M.; Sodhi, C.P.; Russo, A.; Siggers, R.H.; Afrazi, A.; Gribar, S.C.; Neal, M.D.; Dai, S.; Prindle, T., Jr.; Branca, M. Nucleotide-binding oligomerization domain-2 inhibits toll-like receptor-4 signaling in the intestinal epithelium. Gastroenterology 2010, 139, 904–917.e906. [Google Scholar] [CrossRef]
- Egan, C.E.; Sodhi, C.P.; Good, M.; Lin, J.; Jia, H.; Yamaguchi, Y.; Lu, P.; Ma, C.; Branca, M.F.; Weyandt, S. Toll-like receptor 4–mediated lymphocyte influx induces neonatal necrotizing enterocolitis. J. Clin. Investig. 2016, 126, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.G.; Sim, K.; Rose, G.; Wooldridge, D.J.; Li, M.-S.; Misra, R.V.; Gharbia, S.; Kroll, J.S. Premature neonatal gut microbial community patterns supporting an epithelial TLR-mediated pathway for necrotizing enterocolitis. BMC Microbiol. 2021, 21, 225. [Google Scholar] [CrossRef]
- Greenwood, C.; Morrow, A.L.; Lagomarcino, A.J.; Altaye, M.; Taft, D.H.; Yu, Z.; Newburg, D.S.; Ward, D.V.; Schibler, K.R. Early empiric antibiotic use in preterm infants is associated with lower bacterial diversity and higher relative abundance of Enterobacter. J. Pediatr. 2014, 165, 23–29. [Google Scholar] [CrossRef]
- Fleischmann-Struzek, C.; Goldfarb, D.M.; Schlattmann, P.; Schlapbach, L.J.; Reinhart, K.; Kissoon, N. The global burden of paediatric and neonatal sepsis: A systematic review. Lancet Respir. Med. 2018, 6, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Chi, C.; Buys, N.; Li, C.; Sun, J.; Yin, C. Effects of prebiotics on sepsis, necrotizing enterocolitis, mortality, feeding intolerance, time to full enteral feeding, length of hospital stay, and stool frequency in preterm infants: A meta-analysis. Eur. J. Clin. Nutr. 2019, 73, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Castagnola, E.; Dufour, C. Role of G-CSF GM-CSF in the management of infections in preterm newborns: An update. Early Hum. Dev. 2014, 90, S15–S17. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.G.; Sim, K.; Randell, P.; Cox, M.J.; McClure, Z.E.; Li, M.-S.; Donaldson, H.; Langford, P.R.; Cookson, W.O.; Moffatt, M.F. Late-onset bloodstream infection and perturbed maturation of the gastrointestinal microbiota in premature infants. PloS One 2015, 10, e0132923. [Google Scholar] [CrossRef] [PubMed]
- Mai, V.; Torrazza, R.M.; Ukhanova, M.; Wang, X.; Sun, Y.; Li, N.; Shuster, J.; Sharma, R.; Hudak, M.L.; Neu, J. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS ONE 2013, 8, e52876. [Google Scholar] [CrossRef]
- Okada, H.; Kuhn, C.; Feillet, H.; Bach, J.-F. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: An update. Clin. Exp. Immunol. 2010, 160, 1–9. [Google Scholar] [CrossRef]
- Wold, A.E. The hygiene hypotheslis revised: Is the rising frequency of allergy due to changes in the intestinal flora? Allergy 1998, 53, 20–25. [Google Scholar] [CrossRef]
- O’Mahony, C.; Scully, P.; O’Mahony, D.; Murphy, S.; O’Brien, F.; Lyons, A.; Sherlock, G.; MacSharry, J.; Kiely, B.; Shanahan, F. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-κB activation. PLoS Pathog. 2008, 4, e1000112. [Google Scholar] [CrossRef] [PubMed]
- Cukrowska, B. Microbial and nutritional programming—The importance of the microbiome and early exposure to potential food allergens in the development of allergies. Nutrients 2018, 10, 1541. [Google Scholar] [CrossRef] [PubMed]
- Garland, S.M.; Tobin, J.M.; Pirotta, M.; Tabrizi, S.N.; Opie, G.; Donath, S.; Tang, M.L.; Morley, C.J.; Hickey, L.; Ung, L. The ProPrems trial: Investigating the effects of probiotics on late onset sepsis in very preterm infants. BMC Infect. Dis. 2011, 11, 210. [Google Scholar] [CrossRef]
- Low, J.; Soh, S.-E.; Lee, Y.; Kwek, K.; Holbrook, J.; Van der Beek, E.; Shek, L.; Goh, A.; Teoh, O.; Godfrey, K. Ratio of Klebsiella/Bifidobacterium in early life correlates with later development of paediatric allergy. Benef. Microbes 2017, 8, 681. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jin, S.; Wang, J.; Zhang, L.; Mu, Y.; Huang, K.; Zhao, B.; Zhang, K.; Cui, Y.; Li, S. Variations in early gut microbiome are associated with childhood eczema. FEMS Microbiol. Lett. 2019, 366, fnz020. [Google Scholar] [CrossRef] [PubMed]
- Simonyté Sjödin, K.; Hammarström, M.L.; Rydén, P.; Sjödin, A.; Hernell, O.; Engstrand, L.; West, C.E. Temporal and long-term gut microbiota variation in allergic disease: A prospective study from infancy to school age. Allergy 2019, 74, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Melli, L.C.F.L.; do Carmo-Rodrigues, M.S.; Araújo-Filho, H.B.; Mello, C.S.; Tahan, S.; Pignatari, A.C.C.; Solé, D.; de Morais, M.B. Gut microbiota of children with atopic dermatitis: Controlled study in the metropolitan region of São Paulo, Brazil. Allergol. Et Immunopathol. 2020, 48, 107–115. [Google Scholar] [CrossRef]
- Mah, K.; Björkstén, B.; Lee, B.; Van Bever, H.; Shek, L.; Tan, T.; Lee, Y.; Chua, K. Distinct pattern of commensal gut microbiota in toddlers with eczema. Int. Arch. Allergy Immunol. 2006, 140, 157–163. [Google Scholar] [CrossRef]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; Levan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef]
- Blümer, N.; Herz, U.; Renz, H. Das pränatale/frühkindliche Immunsystem und Allergie–Ergebnisse humaner und tierexperimenteller Studien/Prenatal and early postnatal immune system and allergy-outcomes of human and animal studies. LaboratoriumsMedizin 2004, 28, 273–278. [Google Scholar] [CrossRef]
- Abrahamsson, T.; Jakobsson, H.; Andersson, A.F.; Björkstén, B.; Engstrand, L.; Jenmalm, M. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy 2014, 44, 842–850. [Google Scholar] [CrossRef] [PubMed]
- Kogevinas, M.; Sunyer Deu, J.; Vrijheid, M.; Duijts, L. Preterm birth, infant weight gain, and childhood asthma risk: A meta-analysis of 147,000 European children. J. Allergy Clin. Immunol. 2014, 133, 1317–1329. [Google Scholar]
- Woodward, L.J.; Edgin, J.O.; Thompson, D.; Inder, T.E. Object working memory deficits predicted by early brain injury and development in the preterm infant. Brain 2005, 128, 2578–2587. [Google Scholar] [CrossRef] [PubMed]
- Bhutta, A.T.; Cleves, M.A.; Casey, P.H.; Cradock, M.M.; Anand, K.J. Cognitive and behavioral outcomes of school-aged children who were born preterm: A meta-analysis. Jama 2002, 288, 728–737. [Google Scholar] [CrossRef] [PubMed]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef]
- Ginsberg, Y.; Khatib, N.; Weiner, Z.; Beloosesky, R. Maternal inflammation, fetal brain implications and suggested neuroprotection: A summary of 10 years of research in animal models. Rambam Maimonides Med. J. 2017, 8, e0028. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Yu, Y.; Guo, Y.; Wang, Y.; Chang, E.B.; Claud, E.C. Transcriptional modulation of intestinal innate defense/inflammation genes by preterm infant microbiota in a humanized gnotobiotic mouse model. PloS One 2015, 10, e0124504. [Google Scholar] [CrossRef]
- Aarts, E.; Ederveen, T.H.; Naaijen, J.; Zwiers, M.P.; Boekhorst, J.; Timmerman, H.M.; Smeekens, S.P.; Netea, M.G.; Buitelaar, J.K.; Franke, B. Gut microbiome in ADHD and its relation to neural reward anticipation. PloS One 2017, 12, e0183509. [Google Scholar] [CrossRef]
- Vanes, L.D.; Murray, R.M.; Nosarti, C. Adult outcome of preterm birth: Implications for neurodevelopmental theories of psychosis. Schizophr. Res. 2021, 247, 41–54. [Google Scholar] [CrossRef]
- Nosarti, C.; Reichenberg, A.; Murray, R.M.; Cnattingius, S.; Lambe, M.P.; Yin, L.; MacCabe, J.; Rifkin, L.; Hultman, C.M. Preterm birth and psychiatric disorders in young adult life. Arch. Gen. Psychiatry 2012, 69, 610–617. [Google Scholar] [CrossRef]
- Schwarz, E.; Maukonen, J.; Hyytiäinen, T.; Kieseppä, T.; Orešič, M.; Sabunciyan, S.; Mantere, O.; Saarela, M.; Yolken, R.; Suvisaari, J. Analysis of microbiota in first episode psychosis identifies preliminary associations with symptom severity and treatment response. Schizophr. Res. 2018, 192, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Castro-Nallar, E.; Bendall, M.L.; Pérez-Losada, M.; Sabuncyan, S.; Severance, E.G.; Dickerson, F.B.; Schroeder, J.R.; Yolken, R.H.; Crandall, K.A. Composition, taxonomy and functional diversity of the oropharynx microbiome in individuals with schizophrenia and controls. PeerJ 2015, 3, e1140. [Google Scholar] [CrossRef]
- Laverty, C.; Surtees, A.; O’Sullivan, R.; Sutherland, D.; Jones, C.; Richards, C. The prevalence and profile of autism in individuals born preterm: A systematic review and meta-analysis. J. Neurodev. Disord. 2021, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Yap, I.K.; Angley, M.; Veselkov, K.A.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Urinary metabolic phenotyping differentiates children with autism from their unaffected siblings and age-matched controls. J. Proteome Res. 2010, 9, 2996–3004. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Mangifesta, M.; Mancabelli, L.; Lugli, G.A.; James, K.; Duranti, S.; Turroni, F.; Ferrario, C.; Ossiprandi, M.C.; van Sinderen, D. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J. 2017, 11, 2834–2847. [Google Scholar] [CrossRef]
- Tissier, H. Le bacterium coli et la reaction chromophile d’escherich. Crit. Rev. Soc. Biol. 1899, 51, 943–945. [Google Scholar]
- Alessandri, G.; van Sinderen, D.; Ventura, M. The genus Bifidobacterium: From genomics to functionality of an important component of the mammalian gut microbiota. Comput. Struct. Biotechnol. J. 2021, 19, 1472–1487. [Google Scholar] [CrossRef] [PubMed]
- Lugli, G.A.; Milani, C.; Turroni, F.; Duranti, S.; Mancabelli, L.; Mangifesta, M.; Ferrario, C.; Modesto, M.; Mattarelli, P.; van Sinderen, D. Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genom. 2017, 18, 568. [Google Scholar] [CrossRef]
- Turroni, F.; Peano, C.; Pass, D.A.; Foroni, E.; Severgnini, M.; Claesson, M.J.; Kerr, C.; Hourihane, J.; Murray, D.; Fuligni, F. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 2012, 7, e36957. [Google Scholar] [CrossRef]
- Turroni, F.; Foroni, E.; Pizzetti, P.; Giubellini, V.; Ribbera, A.; Merusi, P.; Cagnasso, P.; Bizzarri, B.; de’Angelis, G.L.; Shanahan, F. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 2009, 75, 1534–1545. [Google Scholar] [CrossRef]
- Sela, D.A.; Price, N.P.; Mills, D.A. Metabolism of bifidobacteria. Bifidobact. Genom. Mol. Asp. 2010, 45–70. [Google Scholar]
- Stewart, C.J. Breastfeeding promotes bifidobacterial immunomodulatory metabolites. Nat. Microbiol. 2021, 6, 1335–1336. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Ding, Y.; Saedi, N.; Choi, M.; Sridharan, G.V.; Sherr, D.H.; Yarmush, M.L.; Alaniz, R.C.; Jayaraman, A.; Lee, K. Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep. 2018, 23, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Bron, P.A.; Van Baarlen, P.; Kleerebezem, M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat. Rev. Microbiol. 2012, 10, 66–78. [Google Scholar] [CrossRef]
- Roberts, D.J. Molecular mechanisms of development of the gastrointestinal tract. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2000, 219, 109–120. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Levy, M.; Itav, S.; Elinav, E. Integration of innate immune signaling. Trends Immunol. 2016, 37, 84–101. [Google Scholar] [CrossRef]
- Macpherson, A.J.; Slack, E.; Geuking, M.B.; McCoy, K.D. The mucosal firewalls against commensal intestinal microbes. Semin. Immunopathol. 2009, 31, 145–149. [Google Scholar] [CrossRef]
- Owaga, E.; Hsieh, R.-H.; Mugendi, B.; Masuku, S.; Shih, C.-K.; Chang, J.-S. Th17 cells as potential probiotic therapeutic targets in inflammatory bowel diseases. Int. J. Mol. Sci. 2015, 16, 20841–20858. [Google Scholar] [CrossRef]
- Inturri, R.; Stivala, A.; Furneri, P.; Blandino, G. Growth and adhesion to HT-29 cells inhibition of Gram-negatives by Bifidobacterium longum BB536 e Lactobacillus rhamnosus HN001 alone and in combination. Eur. Rev. Med. Pharmacol. Sci 2016, 20, 4943–4949. [Google Scholar]
- Abdelhamid, A.G.; Esaam, A.; Hazaa, M.M. Cell free preparations of probiotics exerted antibacterial and antibiofilm activities against multidrug resistant E. coli. Saudi Pharm. J. 2018, 26, 603–607. [Google Scholar] [CrossRef]
- Yun, J.-H.; Kim, Y.-A.; Song, M.-S.; Kang, B.-Y.; Ha, N.-J. Lactic acid bacteria isolated from healthy Korean having antimicrobial activity against VISA and VRE. Yakhak Hoeji 2006, 50, 78–83. [Google Scholar]
- Lkhagvadorj, E.; Nagata, S.; Wada, M.; Bian, L.; Wang, C.; Chiba, Y.; Yamashiro, Y.; Shimizu, T.; Asahara, T.; Nomoto, K. Anti-infectious activity of synbiotics in a novel mouse model of methicillin-resistant Staphylococcus aureus infection. Microbiol. Immunol. 2010, 54, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, M.; Vimont, A.; Darveau, A.; Fliss, I.; Jean, J. Study of the ability of bifidobacteria of human origin to prevent and treat rotavirus infection using colonic cell and mouse models. PLoS ONE 2016, 11, e0164512. [Google Scholar] [CrossRef]
- Ishizuka, T.; Kanmani, P.; Kobayashi, H.; Miyazaki, A.; Soma, J.; Suda, Y.; Aso, H.; Nochi, T.; Iwabuchi, N.; Xiao, J.-z. Immunobiotic bifidobacteria strains modulate rotavirus immune response in porcine intestinal epitheliocytes via pattern recognition receptor signaling. PLoS ONE 2016, 11, e0152416. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Chattha, K.S.; Kandasamy, S.; Liu, Z.; Esseili, M.; Shao, L.; Rajashekara, G.; Saif, L.J. Lactobacilli and bifidobacteria promote immune homeostasis by modulating innate immune responses to human rotavirus in neonatal gnotobiotic pigs. PLoS ONE 2013, 8, e76962. [Google Scholar] [CrossRef] [PubMed]
- Moreno Muñoz, J.A.; Chenoll, E.; Casinos, B.; Bataller, E.; Ramón, D.; Genovés, S.; Montava, R.; Ribes, J.M.; Buesa, J.; Fàbrega, J. Novel probiotic Bifidobacterium longum subsp. infantis CECT 7210 strain active against rotavirus infections. Appl. Environ. Microbiol. 2011, 77, 8775–8783. [Google Scholar] [CrossRef]
- Holscher, H.D.; Czerkies, L.A.; Cekola, P.; Litov, R.; Benbow, M.; Santema, S.; Alexander, D.D.; Perez, V.; Sun, S.; Saavedra, J.M. Bifidobacterium lactis Bb12 enhances intestinal antibody response in formula-fed infants: A randomized, double-blind, controlled trial. J. Parenter. Enter. Nutr. 2012, 36, 106S–117S. [Google Scholar] [CrossRef]
- Lee, A.; Lee, Y.J.; Yoo, H.J.; Kim, M.; Chang, Y.; Lee, D.S.; Lee, J.H. Consumption of dairy yogurt containing Lactobacillus paracasei ssp. paracasei, Bifidobacterium animalis ssp. lactis and heat-treated Lactobacillus plantarum improves immune function including natural killer cell activity. Nutrients 2017, 9, 558. [Google Scholar] [CrossRef]
- HAN, S.; CHO, K.; LEE, C.-K.; SONG, Y.; Park, S.H.; HA, N.-J.; KIM, K. Enhancement of antigen presentation capability of dendritic cells and activation of macrophages by the components of Bifidobacterium pseudocatenulatum SPM 1204. Biomol. Ther. 2005, 13, 174–180. [Google Scholar]
- Kitajima, H.; Sumida, Y.; Tanaka, R.; Yuki, N.; Takayama, H.; Fujimura, M. Early administration of Bifidobacterium breve to preterm infants: Randomised controlled trial. Arch. Dis. Child. Fetal Neonatal Ed. 1997, 76, F101–F107. [Google Scholar] [CrossRef]
- Li, Y.; Shimizu, T.; Hosaka, A.; Kaneko, N.; Ohtsuka, Y.; Yamashiro, Y. Effects of Bifidobacterium breve supplementation on intestinal flora of low birth weight infants. Pediatr. Int. 2004, 46, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Shoji, H.; Sato, H.; Nagata, S.; Ohtsuka, Y.; Shimizu, T.; Yamashiro, Y. Effects of oral administration of Bifidobacterium breve on fecal lactic acid and short-chain fatty acids in low birth weight infants. J. Pediatr. Gastroenterol. Nutr. 2007, 44, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Mohan, R.; Koebnick, C.; Schildt, J.; Schmidt, S.; Mueller, M.; Possner, M.; Radke, M.; Blaut, M. Effects of Bifidobacterium lactis Bb12 supplementation on intestinal microbiota of preterm infants: A double-blind, placebo-controlled, randomized study. J. Clin. Microbiol. 2006, 44, 4025–4031. [Google Scholar] [CrossRef]
- Stratiki, Z.; Costalos, C.; Sevastiadou, S.; Kastanidou, O.; Skouroliakou, M.; Giakoumatou, A.; Petrohilou, V. The effect of a bifidobacter supplemented bovine milk on intestinal permeability of preterm infants. Early Hum. Dev. 2007, 83, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Ohtsuka, Y.; Lee, T.; Kudo, T.; Shoji, H.; Sato, H.; Nagata, S.; Shimizu, T.; Yamashiro, Y. Bifidobacterium breve enhances transforming growth factor β1 signaling by regulating smad7 expression in preterm infants. J. Pediatr. Gastroenterol. Nutr. 2006, 43, 83–88. [Google Scholar] [CrossRef]
- Butel, M.-J.; Suau, A.; Campeotto, F.; Magne, F.; Aires, J.; Ferraris, L.; Kalach, N.; Leroux, B.; Dupont, C. Conditions of bifidobacterial colonization in preterm infants: A prospective analysis. J. Pediatr. Gastroenterol. Nutr. 2007, 44, 577–582. [Google Scholar] [CrossRef]
- Mohan, R.; Koebnick, C.; Schildt, J.; Mueller, M.; Radke, M.; Blaut, M. Effects of Bifidobacterium lactis Bb12 supplementation on body weight, fecal pH, acetate, lactate, calprotectin, and IgA in preterm infants. Pediatr. Res. 2008, 64, 418–422. [Google Scholar] [CrossRef]
- Lin, H.-C.; Hsu, C.-H.; Chen, H.-L.; Chung, M.-Y.; Hsu, J.-F.; Lien, R.-i.; Tsao, L.-Y.; Chen, C.-H.; Su, B.-H. Oral probiotics prevent necrotizing enterocolitis in very low birth weight preterm infants: A multicenter, randomized, controlled trial. Pediatrics 2008, 122, 693–700. [Google Scholar] [CrossRef]
- Mihatsch, W.A.; Vossbeck, S.; Eikmanns, B.; Högel, J.; Pohlandt, F. Effect of Bifidobacterium lactis on the incidence of nosocomial infections in very-low-birth-weight infants: A randomized controlled trial. Neonatology 2010, 98, 156–163. [Google Scholar] [CrossRef]
- Braga, T.D.; da Silva, G.A.P.; de Lira, P.I.C.; de Carvalho Lima, M. Efficacy of Bifidobacterium breve and Lactobacillus casei oral supplementation on necrotizing enterocolitis in very-low-birth-weight preterm infants: A double-blind, randomized, controlled trial. Am. J. Clin. Nutr. 2011, 93, 81–86. [Google Scholar] [CrossRef]
- Yamasaki, C.; Totsu, S.; Uchiyama, A.; Nakanishi, H.; Masumoto, K.; Washio, Y.; Shuri, K.; Ishida, S.; Imai, K.; Kusuda, S. Effect of Bifidobacterium administration on very-low-birthweight infants. Pediatr. Int. 2012, 54, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Ishizeki, S.; Sugita, M.; Takata, M.; Yaeshima, T. Effect of administration of bifidobacteria on intestinal microbiota in low-birth-weight infants and transition of administered bifidobacteria: A comparison between one-species and three-species administration. Anaerobe 2013, 23, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, S.E.; Tobin, J.M.; Opie, G.F.; Donath, S.; Tabrizi, S.N.; Pirotta, M.; Morley, C.J.; Garland, S.M. Probiotic effects on late-onset sepsis in very preterm infants: A randomized controlled trial. Pediatrics 2013, 132, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Patole, S.; Keil, A.D.; Chang, A.; Nathan, E.; Doherty, D.; Simmer, K.; Esvaran, M.; Conway, P. Effect of Bifidobacterium breve M-16V supplementation on fecal bifidobacteria in preterm neonates-a randomised double blind placebo controlled trial. PLoS ONE 2014, 9, e89511. [Google Scholar] [CrossRef]
- Andriantsoanirina, V.; Teolis, A.-C.; Xin, L.X.; Butel, M.J.; Aires, J. Bifidobacterium longum and Bifidobacterium breve isolates from preterm and full term neonates: Comparison of cell surface properties. Anaerobe 2014, 28, 212–215. [Google Scholar] [CrossRef]
- Costeloe, K.; Hardy, P.; Juszczak, E.; Wilks, M.; Millar, M.R. Bifidobacterium breve BBG-001 in very preterm infants: A randomised controlled phase 3 trial. Lancet 2016, 387, 649–660. [Google Scholar] [CrossRef]
- Grześkowiak, Ł.; Teixeira, T.F.S.; Bigonha, S.M.; Lobo, G.; Salminen, S.; Ferreira, C.L.d.L.F. Gut Bifidobacterium microbiota in one-month-old Brazilian newborns. Anaerobe 2015, 35, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Dilli, D.; Aydin, B.; Fettah, N.D.; Özyazıcı, E.; Beken, S.; Zenciroğlu, A.; Okumuş, N.; Özyurt, B.M.; İpek, M.Ş.; Akdağ, A. The propre-save study: Effects of probiotics and prebiotics alone or combined on necrotizing enterocolitis in very low birth weight infants. J. Pediatr. 2015, 166, 545–551.e541. [Google Scholar] [CrossRef]
- Patole, S.K.; Rao, S.C.; Keil, A.D.; Nathan, E.A.; Doherty, D.A.; Simmer, K.N. Benefits of Bifidobacterium breve M-16V supplementation in preterm neonates-a retrospective cohort study. PloS One 2016, 11, e0150775. [Google Scholar] [CrossRef]
- Patole, S.K.; Keil, A.D.; Nathan, E.; Doherty, D.; Esvaran, M.; Simmer, K.N.; Conway, P. Effect of Bifidobacterium breve M-16V supplementation on faecal bifidobacteria in growth restricted very preterm infants–analysis from a randomised trial. J. Matern. Fetal Neonatal Med. 2016, 29, 3751–3755. [Google Scholar] [CrossRef]
- Zhang, S.-F.; Tang, Z.-S.; Tong, L.; Tao, X.-X.; Suo, Q.-F.; Xu, X.-M. Effects of clostridium butyricum and bifidobacterium on BTLA expression on CD4+ T cells and lymphocyte differentiation in late preterm infants. Microb. Pathog. 2016, 100, 112–118. [Google Scholar] [CrossRef]
- Härtel, C.; Pagel, J.; Spiegler, J.; Buma, J.; Henneke, P.; Zemlin, M.; Viemann, D.; Gille, C.; Gehring, S.; Frommhold, D. Lactobacillus acidophilus/Bifidobacterium infantis probiotics are associated with increased growth of VLBWI among those exposed to antibiotics. Sci. Rep. 2017, 7, 5633. [Google Scholar] [CrossRef]
- Jacobs, S.E.; Hickey, L.; Donath, S.; Opie, G.F.; Anderson, P.J.; Garland, S.M.; Cheong, J.L.; Groups, P. Probiotics, prematurity and neurodevelopment: Follow-up of a randomised trial. BMJ Paediatr. Open 2017, 1, e000176. [Google Scholar] [CrossRef] [PubMed]
- Plummer, E.L.; Bulach, D.M.; Murray, G.L.; Jacobs, S.E.; Tabrizi, S.N.; Garland, S.M. Gut microbiota of preterm infants supplemented with probiotics: Sub-study of the ProPrems trial. BMC Microbiol. 2018, 18, 184. [Google Scholar] [CrossRef] [PubMed]
- Robertson, C.; Savva, G.M.; Clapuci, R.; Jones, J.; Maimouni, H.; Brown, E.; Minocha, A.; Hall, L.J.; Clarke, P. Incidence of necrotising enterocolitis before and after introducing routine prophylactic Lactobacillus and Bifidobacterium probiotics. Arch. Dis. Child. Fetal Neonatal Ed. 2020, 105, 380–386. [Google Scholar] [CrossRef]
- Tanaka, K.; Nakamura, Y.; Terahara, M.; Yanagi, T.; Nakahara, S.; Furukawa, O.; Tsutsui, H.; Inoue, R.; Tsukahara, T.; Koshida, S. Poor Bifidobacterial colonization is associated with late provision of colostrum and improved with probiotic supplementation in low birth weight infants. Nutrients 2019, 11, 839. [Google Scholar] [CrossRef]
- Alcon-Giner, C.; Dalby, M.J.; Caim, S.; Ketskemety, J.; Shaw, A.; Sim, K.; Lawson, M.A.; Kiu, R.; Leclaire, C.; Chalklen, L. Microbiota supplementation with Bifidobacterium and Lactobacillus modifies the preterm infant gut microbiota and metabolome: An observational study. Cell Rep. Med. 2020, 1, 100077. [Google Scholar] [CrossRef]
- Agrawal, S.; Pestell, C.; Granich, J.; Rao, S.; Nathan, E.; Wray, J.; Whitehouse, A.; Patole, S. Difficulties in developmental follow-up of preterm neonates in a randomised-controlled trial of Bifidobacterium breve M16-V—Experience from Western Australia. Early Hum. Dev. 2020, 151, 105165. [Google Scholar] [CrossRef]
- Arboleya, S.; Saturio, S.; Suárez, M.; Fernández, N.; Mancabelli, L.; de Los Reyes-Gavilán, C.G.; Ventura, M.; Solís, G.; Gueimonde, M. Donated human milk as a determinant factor for the gut bifidobacterial ecology in premature babies. Microorganisms 2020, 8, 760. [Google Scholar] [CrossRef] [PubMed]
- Fortmann, I.; Marißen, J.; Siller, B.; Spiegler, J.; Humberg, A.; Hanke, K.; Faust, K.; Pagel, J.; Eyvazzadeh, L.; Brenner, K. Lactobacillus acidophilus/bifidobacterium infantis probiotics are beneficial to extremely low gestational age infants fed human milk. Nutrients 2020, 12, 850. [Google Scholar] [CrossRef]
- Athalye-Jape, G.; Minaee, N.; Nathan, E.; Simmer, K.; Patole, S. Outcomes in preterm small versus appropriate for gestation infants after Bifidobacterium breve M-16 V supplementation. J. Matern. Fetal Neonatal Med. 2020, 33, 2209–2215. [Google Scholar] [CrossRef] [PubMed]
- Juber, B.A.; Boly, T.J.; Pitcher, G.J.; McElroy, S.J. Routine Administration of a Multispecies Probiotic Containing Bifidobacterium and Lactobacillus to Very Low Birth Weight Infants Had No Significant Impact on the Incidence of Necrotizing Enterocolitis. Front. Pediatr. 2021, 1237. [Google Scholar] [CrossRef]
- Tobias, J.; Olyaei, A.; Laraway, B.; Jordan, B.K.; Dickinson, S.; Arroyo, L.G.; Fialkowski, E.; Owora, A.; Scottoline, B. Feeding Activated Bifidobacterium infantis EVC001 to Very Low Birth Weight Infants is Associated with Significant Reduction in Rates of Necrotizing Enterocolitis. MedRxiv 2021. [Google Scholar]
- Wydau-Dematteis, S.; Delannoy, J.; Téolis, A.-C.; Giuseppi, A.; Campeotto, F.; Lapillonne, A.; Butel, M.-J.; Aires, J. Isolation and Characterization of Commensal Bifidobacteria Strains in Gut Microbiota of Neonates Born Preterm: A Prospective Longitudinal Study. Microorganisms 2022, 10, 654. [Google Scholar] [CrossRef] [PubMed]
- Hotel, A.C.P.; Cordoba, A. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Prevention 2001, 5, 1–10. [Google Scholar]
- Barker, D.J. The origins of the developmental origins theory. J. Intern. Med. 2007, 261, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Authority, E.F.S. Opinion of the Scientific Panel on biological hazards (BIOHAZ) on microbiological criteria and targets based on risk analysis. EFSA J. 2007, 5, 462. [Google Scholar]
- Henrick, B.M.; Rodriguez, L.; Lakshmikanth, T.; Pou, C.; Henckel, E.; Arzoomand, A.; Olin, A.; Wang, J.; Mikes, J.; Tan, Z. Bifidobacteria-mediated immune system imprinting early in life. Cell 2021, 184, 3884–3898.e3811. [Google Scholar] [CrossRef]
- Kavanaugh, D.; O’Callaghan, J.; Butto, L.; Slattery, H.; Lane, J.; Clyne, M.; Kane, M.; Joshi, L.; Hickey, R. Exposure of subsp. To milk oligosaccharides increases adhesion to epithelial cells and induces a substantial transcriptional response. PLoS ONE 2013, 8, e67224. [Google Scholar] [CrossRef]
- Chichlowski, M.; De Lartigue, G.; German, J.B.; Raybould, H.E.; Mills, D.A. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 321. [Google Scholar] [CrossRef]
- Underwood, M.A.; Gaerlan, S.; De Leoz, M.L.A.; Dimapasoc, L.; Kalanetra, K.M.; Lemay, D.G.; German, J.B.; Mills, D.A.; Lebrilla, C.B. Human milk oligosaccharides in premature infants: Absorption, excretion, and influence on the intestinal microbiota. Pediatr. Res. 2015, 78, 670–677. [Google Scholar] [CrossRef]
- Dermyshi, E.; Wang, Y.; Yan, C.; Hong, W.; Qiu, G.; Gong, X.; Zhang, T. The “golden age” of probiotics: A systematic review and meta-analysis of randomized and observational studies in preterm infants. Neonatology 2017, 112, 9–23. [Google Scholar] [CrossRef] [PubMed]
- AlFaleh, K.; Anabrees, J. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Evid. Based Child Health Cochrane Rev. J. 2014, 9, 584–671. [Google Scholar] [CrossRef] [PubMed]
- Dani, C.; Biadaioli, R.; Bertini, G.; Martelli, E.; Rubaltelli, F.F. Probiotics feeding in prevention of urinary tract infection, bacterial sepsis and necrotizing enterocolitis in preterm infants. Neonatology 2002, 82, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Manzoni, P.; Mostert, M.; Leonessa, M.; Priolo, C.; Farina, D.; Monetti, C.; Latino, M.; Gomirato, G. Oral supplementation with Lactobacillus casei subspecies rhamnosus prevents enteric colonization by Candida species in preterm neonates: A randomized study. Clin. Infect. Dis. 2006, 42, 1735–1742. [Google Scholar] [CrossRef] [PubMed]
- Oncel, M.Y.; Sari, F.N.; Arayici, S.; Guzoglu, N.; Erdeve, O.; Uras, N.; Oguz, S.S.; Dilmen, U. Lactobacillus reuteri for the prevention of necrotising enterocolitis in very low birthweight infants: A randomised controlled trial. Arch. Dis. Child. Fetal Neonatal Ed. 2014, 99, F110–F115. [Google Scholar] [CrossRef]
- Rojas, M.A.; Lozano, J.M.; Rojas, M.X.; Rodriguez, V.A.; Rondon, M.A.; Bastidas, J.A.; Perez, L.A.; Rojas, C.; Ovalle, O.; Garcia-Harker, J.E. Prophylactic probiotics to prevent death and nosocomial infection in preterm infants. Pediatrics 2012, 130, e1113–e1120. [Google Scholar] [CrossRef]
- Serce, O.; Benzer, D.; Gursoy, T.; Karatekin, G.; Ovali, F. Efficacy of Saccharomyces boulardii on necrotizing enterocolitis or sepsis in very low birth weight infants: A randomised controlled trial. Early Hum. Dev. 2013, 89, 1033–1036. [Google Scholar] [CrossRef]
- Demirel, G.; Erdeve, O.; Celik, I.H.; Dilmen, U. Saccharomyces boulardii for prevention of necrotizing enterocolitis in preterm infants: A randomized, controlled study. Acta Paediatr. 2013, 102, e560–e565. [Google Scholar] [CrossRef]
- Costalos, C.; Skouteri, V.; Gounaris, A.; Sevastiadou, S.; Triandafilidou, A.; Ekonomidou, C.; Kontaxaki, F.; Petrochilou, V. Enteral feeding of premature infants with Saccharomyces boulardii. Early Hum. Dev. 2003, 74, 89–96. [Google Scholar] [CrossRef]
- Sari, F.; Dizdar, E.; Oguz, S.; Erdeve, O.; Uras, N.; Dilmen, U. Oral probiotics: Lactobacillus sporogenes for prevention of necrotizing enterocolitis in very low-birth weight infants: A randomized, controlled trial. Eur. J. Clin. Nutr. 2011, 65, 434–439. [Google Scholar] [CrossRef]
- Totsu, S.; Yamasaki, C.; Terahara, M.; Uchiyama, A.; Kusuda, S.; Japan, P.S.G.i. bifidobacterium and enteral feeding in preterm infants: Cluster-randomized trial. Pediatr. Int. 2014, 56, 714–719. [Google Scholar] [CrossRef]
- Romond, M.-B.; Colavizza, M.; Mullié, C.; Kalach, N.; Kremp, O.; Mielcarek, C.; Izard, D. Does the intestinal bifidobacterial colonisation affect bacterial translocation? Anaerobe 2008, 14, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Underwood, M.A.; Arriola, J.; Gerber, C.W.; Kaveti, A.; Kalanetra, K.M.; Kananurak, A.; Bevins, C.L.; Mills, D.A.; Dvorak, B. Bifidobacterium longum subsp. infantis in experimental necrotizing enterocolitis: Alterations in inflammation, innate immune response, and the microbiota. Pediatr. Res. 2014, 76, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, K.R.; Liu, S.X.; Tian, R.; Kushnir, A.; Turner, J.R.; Li, H.-L.; Chou, P.M.; Weber, C.R.; De Plaen, I.G. Bifidobacteria stabilize claudins at tight junctions and prevent intestinal barrier dysfunction in mouse necrotizing enterocolitis. Am. J. Pathol. 2013, 182, 1595–1606. [Google Scholar] [CrossRef]
- Plummer, E.L.; Danielewski, J.A.; Garland, S.M.; Su, J.; Jacobs, S.E.; Murray, G.L. The effect of probiotic supplementation on the gut microbiota of preterm infants. J. Med. Microbiol. 2021, 70, 001403. [Google Scholar] [CrossRef] [PubMed]
- Frazer, L.C.; Yakah, W.; Martin, C.R. Decreased Acetic Acid in the Stool of Preterm Infants Is Associated with an Increased Risk of Bronchopulmonary Dysplasia. Nutrients 2022, 14, 2412. [Google Scholar] [CrossRef] [PubMed]
- Cormack, B.E.; Harding, J.E.; Miller, S.P.; Bloomfield, F.H. The influence of early nutrition on brain growth and neurodevelopment in extremely preterm babies: A narrative review. Nutrients 2019, 11, 2029. [Google Scholar] [CrossRef]
- Pärtty, A.; Kalliomäki, M.; Wacklin, P.; Salminen, S.; Isolauri, E. A possible link between early probiotic intervention and the risk of neuropsychiatric disorders later in childhood: A randomized trial. Pediatr. Res. 2015, 77, 823–828. [Google Scholar] [CrossRef]
- McComb, S.; Thiriot, A.; Akache, B.; Krishnan, L.; Stark, F. Introduction to the immune system. In Immunoproteomics; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–24. [Google Scholar]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 2014, 461. [Google Scholar] [CrossRef]
- Dąbek, J.; Kułach, A.; Gąsior, Z. Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB): A new potential therapeutic target in atherosclerosis? Pharmacol. Rep. 2010, 62, 778–783. [Google Scholar] [CrossRef]
- Ganguli, K.; Meng, D.; Rautava, S.; Lu, L.; Walker, W.A.; Nanthakumar, N. Probiotics prevent necrotizing enterocolitis by modulating enterocyte genes that regulate innate immune-mediated inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G132–G141. [Google Scholar] [CrossRef]
- Guo, S.; Guo, Y.; Ergun, A.; Lu, L.; Walker, W.A.; Ganguli, K. Secreted metabolites of Bifidobacterium infantis and Lactobacillus acidophilus protect immature human enterocytes from IL-1β-induced inflammation: A transcription profiling analysis. PLoS ONE 2015, 10, e0124549. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Nascimento, L.; Massari, P.; Wetzler, L.M. The role of TLR2 in infection and immunity. Front. Immunol. 2012, 3, 79. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Gao, X.-C.; Liu, J.; Ren, H.-Y. Effect of EPEC endotoxin and bifidobacteria on intestinal barrier function through modulation of toll-like receptor 2 and toll-like receptor 4 expression in intestinal epithelial cell-18. World J. Gastroenterol. 2017, 23, 4744. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Wang, L.; Gong, Z.; Wang, Y.; Gao, Y.; Cai, W.; Wu, J. Blockage of NLRP3 inflammasome activation ameliorates acute inflammatory injury and long-term cognitive impairment induced by necrotizing enterocolitis in mice. J. Neuroinflammation 2021, 18, 66. [Google Scholar] [CrossRef]
- Jeon, S.G.; Kayama, H.; Ueda, Y.; Takahashi, T.; Asahara, T.; Tsuji, H.; Tsuji, N.M.; Kiyono, H.; Ma, J.S.; Kusu, T. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012, 8, e1002714. [Google Scholar] [CrossRef]
- Wu, W.; Wang, Y.; Zou, J.; Long, F.; Yan, H.; Zeng, L.; Chen, Y. Bifidobacterium adolescentis protects against necrotizing enterocolitis and upregulates TOLLIP and SIGIRR in premature neonatal rats. BMC Pediatr. 2017, 17, 1. [Google Scholar] [CrossRef]
- JIANG, H.; YiQing, Y.; JIANG, W.; RongBin, Z. NLRP3 inflammasome: Activation, regulation, and role in diseases. Sci. Sin. Vitae 2017, 47, 125–131. [Google Scholar]
- Gomez-Lopez, N.; Romero, R.; Garcia-Flores, V.; Leng, Y.; Miller, D.; Hassan, S.S.; Hsu, C.-D.; Panaitescu, B. Inhibition of the NLRP3 inflammasome can prevent sterile intra-amniotic inflammation, preterm labor/birth, and adverse neonatal outcomes. Biol. Reprod. 2019, 100, 1306–1318. [Google Scholar] [CrossRef]
- Groschwitz, K.R.; Hogan, S.P. Intestinal barrier function: Molecular regulation and disease pathogenesis. J. Allergy Clin. Immunol. 2009, 124, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.Y.; Osaka, T.; Moriyama, E.; Date, Y.; Kikuchi, J.; Tsuneda, S. Strengthening of the intestinal epithelial tight junction by Bifidobacterium bifidum. Physiol. Rep. 2015, 3, e12327. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Linglong, P.; Weixia, D.; Hong, W. Protective effects of bifidobacterium on intestinal barrier function in LPS-induced enterocyte barrier injury of Caco-2 monolayers and in a rat NEC model. PLoS ONE 2016, 11, e0161635. [Google Scholar] [CrossRef] [PubMed]
- Al-Sadi, R.; Dharmaprakash, V.; Nighot, P.; Guo, S.; Nighot, M.; Do, T.; Ma, T.Y. Bifidobacterium bifidum Enhances the Intestinal Epithelial Tight Junction Barrier and Protects against Intestinal Inflammation by Targeting the Toll-like Receptor-2 Pathway in an NF-κB-Independent Manner. Int. J. Mol. Sci. 2021, 22, 8070. [Google Scholar] [CrossRef] [PubMed]
- Gareau, M.G.; Sherman, P.M.; Walker, W.A. Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 503–514. [Google Scholar] [CrossRef]
- Liu, Q.; Yu, Z.; Tian, F.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Surface components and metabolites of probiotics for regulation of intestinal epithelial barrier. Microb. Cell Factories 2020, 19, 23. [Google Scholar] [CrossRef]
- Ivanov, D.; Emonet, C.; Foata, F.; Affolter, M.; Delley, M.; Fisseha, M.; Blum-Sperisen, S.; Kochhar, S.; Arigoni, F. A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J. Biol. Chem. 2006, 281, 17246–17252. [Google Scholar] [CrossRef]
- Mitsuma, T.; Odajima, H.; Momiyama, Z.; Watanabe, K.; Masuguchi, M.; Sekine, T.; Shidara, S.; Hirano, S. Enhancement of gene expression by a peptide p (CHWPR) produced by Bifidobacterium lactis BB-12. Microbiol. Immunol. 2008, 52, 144–155. [Google Scholar] [CrossRef]
- Collado, M.; Meriluoto, J.; Salminen, S. Role of commercial probiotic strains against human pathogen adhesion to intestinal mucus. Lett. Appl. Microbiol. 2007, 45, 454–460. [Google Scholar] [CrossRef]
- O’Connell Motherway, M.; Houston, A.; O’Callaghan, G.; Reunanen, J.; O’Brien, F.; O’Driscoll, T.; Casey, P.G.; de Vos, W.M.; van Sinderen, D.; Shanahan, F. A Bifidobacterial pilus-associated protein promotes colonic epithelial proliferation. Mol. Microbiol. 2019, 111, 287–301. [Google Scholar] [CrossRef]
- Collado, M.C.; Gueimonde, M.; Sanz, Y.; Salminen, S. Adhesion properties and competitive pathogen exclusion ability of bifidobacteria with acquired acid resistance. J. Food Prot. 2006, 69, 1675–1679. [Google Scholar] [CrossRef] [PubMed]
- Candela, M.; Bergmann, S.; Vici, M.; Vitali, B.; Turroni, S.; Eikmanns, B.J.; Hammerschmidt, S.; Brigidi, P. Binding of human plasminogen to Bifidobacterium. J. Bacteriol. 2007, 189, 5929–5936. [Google Scholar] [CrossRef] [PubMed]
- Trindade, L.; Martins, V.; Rodrigues, N.; Souza, E.; Martins, F.; Costa, G.; Almeida-Leite, C.; Faria, A.; Cardoso, V.; Maioli, T. Oral administration of Simbioflora®(synbiotic) attenuates intestinal damage in a mouse model of 5-fluorouracil-induced mucositis. Benef. Microbes 2018, 9, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Liévin, V.; Peiffer, I.; Hudault, S.; Rochat, F.; Brassart, D.; Neeser, J.; Servin, A. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut 2000, 47, 646–652. [Google Scholar] [CrossRef]
- Gibson, G.R.; Wang, X. Regulatory effects of bifidobacteria on the growth of other colonic bacteria. J. Appl. Bacteriol. 1994, 77, 412–420. [Google Scholar] [CrossRef]
- Ohishi, A.; Takahashi, S.; Ito, Y.; Ohishi, Y.; Tsukamoto, K.; Nanba, Y.; Ito, N.; Kakiuchi, S.; Saitoh, A.; Morotomi, M. Bifidobacterium septicemia associated with postoperative probiotic therapy in a neonate with omphalocele. J. Pediatr. 2010, 156, 679–681. [Google Scholar] [CrossRef]
- Brecht, M.; Garg, A.; Longstaff, K.; Cooper, C.; Andersen, C. Lactobacillus sepsis following a laparotomy in a preterm infant: A note of caution. Neonatology 2016, 109, 186–189. [Google Scholar] [CrossRef]
- van den Akker, C.H.; Van Goudoever, J.B.; Szajewska, H.; Embleton, N.D.; Hojsak, I.; Reid, D.; Shamir, R. Probiotics for preterm infants: A strain-specific systematic review and network meta-analysis. J. Pediatr. Gastroenterol. Nutr. 2018, 67, 103–122. [Google Scholar] [CrossRef]
- Aceti, A.; Gori, D.; Barone, G.; Callegari, M.L.; Di Mauro, A.; Fantini, M.P.; Indrio, F.; Maggio, L.; Meneghin, F.; Morelli, L. Probiotics for prevention of necrotizing enterocolitis in preterm infants: Systematic review and meta-analysis. Ital. J. Pediatr. 2015, 41, 89. [Google Scholar] [CrossRef]
- De Simone, C. The unregulated probiotic market. Clin. Gastroenterol. Hepatol. 2019, 17, 809–817. [Google Scholar] [CrossRef]
- Lewis, Z.T.; Shani, G.; Masarweh, C.F.; Popovic, M.; Frese, S.A.; Sela, D.A.; Underwood, M.A.; Mills, D.A. Validating bifidobacterial species and subspecies identity in commercial probiotic products. Pediatr. Res. 2016, 79, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.A.; Schoeni, J.L.; Vegge, C.; Pane, M.; Stahl, B.; Bradley, M.; Goldman, V.S.; Burguière, P.; Atwater, J.B.; Sanders, M.E. Improving end-user trust in the quality of commercial probiotic products. Front. Microbiol. 2019, 10, 739. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).