Abstract
Past-oriented rumination and future-oriented worry are two aspects of perseverative negative thinking related to the neuroticism endophenotype and associated with depression and anxiety. Our present aim was to investigate the genomic background of these two aspects of perseverative negative thinking within separate groups of individuals with suboptimal versus optimal folate intake. We conducted a genome-wide association study in the UK Biobank database (n = 72,621) on the “rumination” and “worry” items of the Eysenck Personality Inventory Neuroticism scale in these separate groups. Optimal folate intake was related to lower worry, but unrelated to rumination. In contrast, genetic associations for worry did not implicate specific biological processes, while past-oriented rumination had a more specific genetic background, emphasizing its endophenotypic nature. Furthermore, biological pathways leading to rumination appeared to differ according to folate intake: purinergic signaling and circadian regulator gene ARNTL emerged in the whole sample, blastocyst development, DNA replication, and C-C chemokines in the suboptimal folate group, and prostaglandin response and K+ channel subunit gene KCNH3 in the optimal folate group. Our results point to possible benefits of folate in anxiety disorders, and to the importance of simultaneously taking into account genetic and environmental factors to determine personalized intervention in polygenic and multifactorial disorders.
1. Introduction
Neuroticism is a personality trait manifested as a tendency to experience negative, distressing emotions and cognitions [1,2]. The association of neuroticism with depression or anxiety symptoms has been demonstrated to be either fully or partially mediated by rumination, worry, or both [2,3,4,5]. Rumination and worry represent two types of repetitive thought or perseverative cognition. They are negatively valenced, attentive and frequent thoughts about oneself and one’s world which dysfunctionally prolong mental representations of a stressor [6,7,8]. While rumination is considered a past-oriented form of repetitive thought concerning themes of loss and worry is future-oriented negative thinking [7], they share a common latent factor in their variance which predicts future depression and anxiety levels in case of stress [9].
Neuroticism and rumination, in addition to their shared phenotypic variance that has important mental health correlates, have been revealed to have a partially overlapping genetic background, which explains a considerable portion of variance in internalizing—but not in externalizing—symptoms [10].
Among genetic associations for rumination [11,12,13], genes of folate metabolism [14] may warrant a special interest. They underpin several biological pathways, properties of which can easily be influenced by diet or supplementation. Low folate levels may be associated with a more severe and endogenous subtype of depression, with marked affective and motivational symptoms and a poorer response to antidepressants [15]. Low folate intake has indeed been associated with an increased risk of later depression [16]. Although results are contradictory [17], folate has been found to be an effective and safe adjuvant antidepressant treatment for major depressive disorder [18,19], especially at a dose of <5 mg/day (or 15 mg/day in case of methylfolate) and as an adjunct to selective serotonin reuptake inhibitor therapy [20]. As a gene-by-environment interaction, folate status may also moderate the effect of folate pathway genes on cognitive flexibility, such that these genetic effects could be detected only in the case of a low erythrocyte folate level [21].
Although a genome-wide association study (GWAS) was recently published on rumination and its two subtypes [11], the role of folate intake has never been investigated within this framework. Our present aim was a GWAS on the two perseverative negative thinking items of neuroticism scale, within separate groups of individuals with suboptimal versus optimal folate intake, in the UK Biobank database (http://biobank.ctsu.ox.ac.uk/, accessed on 27 November 2019). We expected distinct genetic associations in different folate intake groups. We also aimed to test the explanatory value of top risk variants on neuroticism score, rumination score and scores of the two subtypes of rumination in a separate database. We expected shared genetic factors between the two perseverative negative thinking items and the investigated comprehensive personality traits. Participants’ current depressive symptoms and lifetime depression status were controlled for in all analyses, to uncover the potential transdiagnostic relevance of the endophenotype of perseverative negative thinking beyond its importance in depression.
2. Materials and Methods
2.1. Participants
Invitation and recruitment in the UK Biobank study were based on NHS patient registers of people aged 40–69 years [22], with an ethical approval from the National Research Ethics Service Committee North West–Haydock [23].
In the NewMood study, participants aged 18–60 years were recruited through advertisements, general practices and a website, with ethical approvals from the North Manchester Local Research Ethics Committee, Manchester, United Kingdom, and the Scientific and Research Ethics Committee of the Medical Research Council, Budapest, Hungary [11].
All participants in both databases provided written informed consent. All procedures were carried out in accordance with the Declaration of Helsinki.
Our study was based on UK Biobank Application Number 1602, which focused on the Oxford WEbQ dietary questionnaire data, completed by a subset of UK Biobank participants. In our present analyses, we included white British UK Biobank participants who had passed genomic quality control (QC) and had non-missing values on all of the variables of interest (both perseverative negative thinking items, folate intake, sex, age and both depression measures, as detailed below). White British ancestry was defined by a self-report and genetic ancestry in data-field 22006, and we decided to analyze only this population because it is the one that constitutes the vast majority of the UK Biobank database [24]. All of these restrictions yielded a total of 72,621 participants in our analyses. Of these, 10,638 subjects belonged to the suboptimal folate intake group and 61,983 belonged to the optimal folate intake group.
Within our NewMood database, we restricted our present analyses to a set of 1746 subjects that were very similar to those in our previous rumination GWAS paper [11], specifically, white Europeans from Manchester, United Kingdom and Budapest, Hungary, who did not overlap UK Biobank participants, passed genomic QC and provided information on sex, age, both depression phenotypes (detailed below), as well as on rumination and neuroticism.
2.2. Phenotypes
In UK Biobank, the two phenotypes on perseverative negative thinking were two items from the neuroticism scale of the Eysenck Personality Inventory, assessed at baseline [22,23,25]: “Do you worry too long after an embarrassing experience?” (data field 2000; rumination item, because of its orientation towards the past) and “Are you a worrier?” (data field 1980; worry item, because its content is restricted to this trait). Both items are used as dichotomous variables: their values can be either yes or no.
In UK Biobank, current depression level was determined by the sum of four item scores [26] detailed in Supplementary File S1, each one measured at a four-point Likert scale at the baseline time point. Lifetime depression status was assessed also at baseline, by a verbal interview (data field 20002).
Folate intake in UK Biobank was estimated via food and beverage consumption the day before, excluding any supplements (data field 100014). It was based on Oxford WEbQ, a 24 h dietary recall questionnaire, which was validated against an interviewer-administered 24 h dietary recall [27]. To control for seasonal variability of dietary intake, more time points of assessment were used, with a maximum of five. 41.82% of participants provided dietary data at only one time point, 20.28% had two, 18.29% had three, 14.45% had four, and 5.15% had five instances of assessment. 59.78% of all participants provided dietary data at the baseline time point, others only at online follow-up instance(s). Values over multiple instances were averaged for each subject, and nutrient calculation has been detailed elsewhere [28]. Cut-off point of a healthy level of folate intake was set to 200 µg per day. This value was based on the recommendation of the British government, as detailed in Supplementary File S1.
In our NewMood database, folate intake was not measured. However, we had measures on comprehensive scales of personality traits. Neuroticism was measured by the 44-item Big Five Inventory (BFI-44) [29]. Rumination and its two subtypes were assessed by the 10-item Ruminative Response Scale (RRS) [30]. Five items belong to the brooding subscale, which denotes a “moody pondering”, passive comparisons of the person’s current situation with unachieved standards [30]. The other five items of RRS belong to the reflection subscale, denoting a purposeful turning inward with the aim of problem solving and alleviating depression [30].
In NewMood, current depression level was addressed by the sum of depression item scores and additional item scores of Brief Symptom Inventory (BSI) [31]. Lifetime depression status was measured by a self-reported question within the background questionnaire and was validated with diagnostic interview within a subsample [32].
In order to handle item-level missingness properly for neuroticism, rumination, brooding, reflection, and BSI depression in NewMood, and current depression score in UK Biobank, the sums of item scores were divided by the number of responded items.
2.3. Genotyping, Imputation and Genomic Quality Control
In UK Biobank, we selected participants with a genetically defined white British ancestry subset (data field 22006), without putative sex chromosome aneuploidy (data field 22019), and, as part of a further QC process, those involved in the maximal set of unrelated individuals (data field 22020) [24].
In both UK Biobank release v3 and our NewMood database, genetic variants were restricted to biallelic single-nucleotide polymorphisms (SNPs), analyzing directly genotyped and imputed variants as well. QC steps are detailed in Supplementary File S1.
2.4. Analyses
Descriptive statistical analyses, as well as calculation of standardized residuals for polygenic risk score (PRS) testing were performed with SPSS 25. LD Score regression [33] was used to calculate the SNP heritability of rumination and worry; namely, the phenotypic variance explained by the whole set of SNPs in each of the three groups of UK Biobank participants (whole study sample, suboptimal folate intake group, optimal folate intake group). In each group, the top ten principal components of the genome were calculated with the approximative method [24,34] implemented in Plink2 (www.cog-genomics.org/plink/2.0/, accessed on 26 March 2020) [35], after respective linkage disequilibrium (LD) pruning in that group. Plink v1.9 (www.cog-genomics.org/plink/1.9/ (accessed on 26 June 2020)) [35] was used to explore identity-by-descent parameter to detect potential overlap between UK Biobank and NewMood participants.
For SNP-level analyses, logistic regression models were run in Plink v1.9, for each of the rumination or worry items as outcome, in all three groups according to folate intake. We applied the strictest method to correct for multiple testing: the Bonferroni correction. The number of tests entailed a p ≤ 1.38 × 10−9 significance threshold (detailed in Supplementary File S1).
To go further with these SNP-level results, FUMA v1.3.6 [36] was used. Within FUMA, MAGMA v1.07 [37] was used for gene-based and gene set-based analyses, and for gene property analysis that assessed tissue specificity. Gene-based testing assigned SNPs to protein-coding genes based on position, with gene boundaries extended by 10,000 base pairs, and entailed a p ≤ 4.36 × 10−7 as a significance threshold by the strictest Bonferroni method. To further analyze the resulting gene p-values, gene set testing used 15,496 gene sets from MsigDB v7.0: 5500 C2 curated gene sets and 9996 C5 GO terms, yielding a p ≤ 5.38 × 10−7 significance threshold, again by the strictest Bonferroni method. To explore associations between gene p-values and tissue specificity of genes, expression databases used for testing included the 30 general tissue types of GTEx v8 [38] and the 11 general developmental stages of BrainSpan’s developmental brain samples [39], entailing a 2.03 × 10−4 p-value threshold with the strictest Bonferroni method.
In additional FUMA analyses, to overcome the limitations of using only position-based assignment and protein-coding genes within MAGMA, top SNPs of SNP-level tests were mapped to genes based not only on position, but also on functional annotations: expression quantitative trait loci (eQTL) and 3D chromatin interaction. These mapped genes were then further analyzed with hypergeometric tests if overrepresented in any pre-defined gene set of the above detailed MsigDB gene set collections. As a built-in function of FUMA “GENE2FUNC” analysis, a Benjamini–Hochberg false discovery rate (FDR); p < 0.05 significance criterion was applied within each subcategory of gene sets.
Further details of FUMA analyses are provided in Supplementary File S1.
PRSice-2 [40] was used for PRS analyses. Explanatory value of PRS composed of each of UK Biobank’s six SNP-level logistic regression analyses was tested for standardized residuals of NewMood’s neuroticism, rumination, brooding and reflection scores. To control for false positive results due to multiple testing, the built-in permutation function of PRSice-2 was used to run 10,000 permutations for each of these 24 models, and models with a resulting empirical p ≤ 0.05 were considered significant.
PRS calculation, using the same procedure, was also applied to test the explanatory value of top SNPs within the suboptimal folate intake group for the same perseverative negative thinking item within optimal folate intake group, and vice versa.
A more detailed description on PRS calculation is provided in Supplementary File S1.
Figures on results were exported from FUMA or PRSice-2, and labels were added to them in Microsoft Word and PowerPoint.
3. Results
3.1. Descriptive Statistics and Single-Nucleotide Polymorphism (SNP) Heritability
In the UK Biobank, folate intake was inversely related to worry, but unrelated to rumination. The NewMood sample was younger, more depressed and more predominated by females than the UK Biobank sample. Descriptive statistics and relationships between phenotypes are further detailed in Supplementary Tables S1–S4 of Supplementary File S1.
Table 1 depicts proportions of phenotypic variance explained by the whole set of SNPs.

Table 1.
Proportion of variance of “rumination” and “worry” items explained by all SNPs (SNP heritability) in the whole study sample, in the suboptimal folate intake group, and in the optimal folate intake group.
3.2. SNP-Based Results
Supplementary Figure S1 of Supplementary File S1 shows that no SNP survived genome-level Bonferroni correction for multiple testing. QQ plots and lambda values for each analysis are provided in Supplementary Figure S2 of Supplementary File S1.
3.3. MAGMA’s Gene-Based Results
Only two genes survived correction for the six tests: ARNTL, for rumination in the whole sample (Figure 1a), and KCNH3, also for rumination, in the optimal folate intake group (Figure 1b). Manhattan and QQ plots of gene-based p-values for the two items in the three groups, are shown in Supplementary Figures S3 and S4 of Supplementary File S1. Gene-based results are detailed fully in Sheet 1 of Supplementary Files S2–S7.
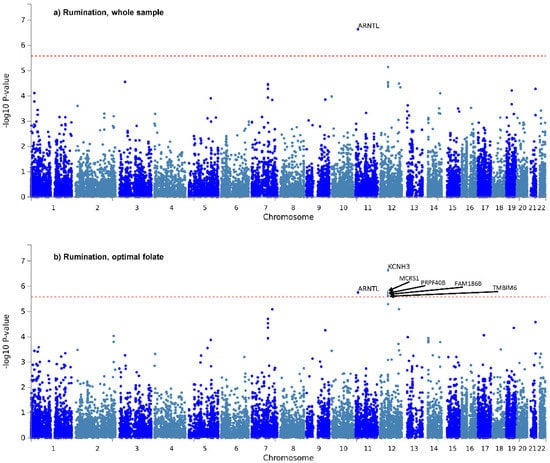
Figure 1.
Manhattan plots of gene-based tests for “rumination” item in the whole study sample (a) and in the optimal folate intake group (b). −log10 of p-value is displayed in function of genomic position. Red line denotes significance threshold, corrected within only one analysis. In the whole sample (a) ARNTL, and in the optimal folate intake group (b) KCNH3, MCRS1, ARNTL, PRPF40B, FAM186B and TMBIM6 survived this less strict correction.
3.4. MAGMA’s Gene Set-Based Results
No gene set survived multiple testing correction. Sheet 2 of Supplementary Files S2–S7 detail full gene set-based results for the six analyses. Among folate-related pathways, only the two gene sets related to transmembrane folate transport emerged in the top few ones, and only for rumination in the whole sample. Specifically, these ranked at positions 24 and 34 (Sheet 2 of Supplementary File S2).
Homocysteine metabolism is closely linked to folate metabolism and it emerged in top ten pathways—but only of rumination and only in the suboptimal folate group, as Table 2 points out, along with ranks of these top ten pathways in the other five groups.

Table 2.
Most significant ten gene sets by MAGMA analysis, for “rumination” in the suboptimal folate intake group, and their ranks in the other five groups. GO_bp: biological processes, and GO_mf: molecular function subcategories of MsigDB C5 gene set collections.
3.5. MAGMA’s Tissue-Specific Upregulation of Genes Relevant in Rumination or Worry
Gene property analysis for tissue-specific upregulation uncovered no significant results. However, at a nominally significant level, Figure 2 depicts results indicating that genes that showed strong association with rumination in the suboptimal folate group were highly expressed during late-mid and late prenatal brain development. Moreover, also at a nominally significant level, genes with strong association with worry in the whole study sample were highly expressed in the pancreas (Supplementary Figure S6b of Supplementary File S1). Results are fully detailed in Supplementary Figures S5 and S6.
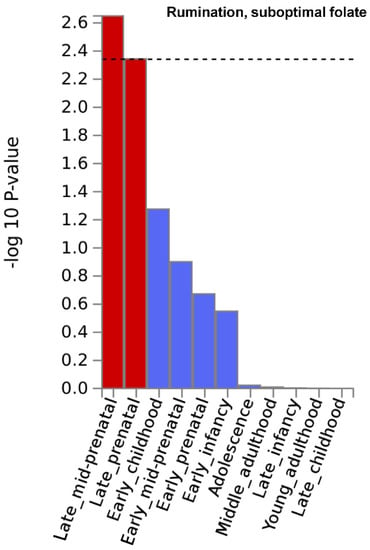
Figure 2.
Brain developmental stage-specific expression of genes that are strongly associated with “rumination” in the suboptimal folate intake group. −log10 p-value is displayed for each of the 11 general developmental stages of BrainSpan’s brain development samples. Dashed line denotes significance threshold corrected for the 11 tissues only within this analysis. Results suggest that genes that showed strong association with rumination in the suboptimal folate group are highly expressed during late-mid and late prenatal brain development.
3.6. Genes Mapped Both by Position and Functional Annotations, and Their Enrichment in MsigDB C2 and C5 Gene Sets
For each of the six analyses, Sheets 3 and 4 of Supplementary Files S2–S7 (and Supplementary File S8) detail genes mapped to top SNPs, based on expression quantitative trait loci (eQTL) and 3D chromatin interaction databases, respectively. These full results are sorted according to tissue or cell type and gene names within each tissue or cell type.
Enrichment tests of mapped genes revealed a possible significance of purinergic signaling in rumination in the whole study sample (Supplementary Figure S7 of Supplementary File S1), although the involved genes (P2RY1, P2RY12, P2RY13, P2RY14, GPR87 and GPR171) all reside in the q25.1–2 regions of chromosome 3 (https://genome.ucsc.edu/ (accessed on 14 May 2020)). Similarly, for rumination in the suboptimal folate group, diverse immune system gene sets, ERK cascade, regulation of DNA replication, and blastocyst development might be implicated (Supplementary Figure S8 of Supplementary File S1), but most of these genes (CCL2, CCL7, CCL11, CCL8, CCL13, CCL1, MMP28, ZNF830, LIG3, SLFN11, NLE1, HNF1B, RFFL) are located in the same region of chromosome 17q12. For rumination in the optimal folate group, some general pathways emerged, such as responses to fatty acids and prostaglandin (Supplementary Figure S9 of Supplementary File S1) involving PTGDR, PTGER4, GNG2, APOB and CREB1, among others.
For worry, significantly enriched gene sets are detailed in Supplementary Figures S10–S12 of Supplementary File S1. The majority of these very few hits were related to cancer diseases.
3.7. Explanatory Value of UK Biobank’s Risk SNPs in NewMood’s Phenotypes
Polygenic risk scores (PRS) that had been calculated based on UK Biobank’s risk SNPs for rumination and worry items in the three (whole sample, suboptimal and optimal folate intake) groups, were tested for neuroticism, rumination, brooding and reflection scores of the NewMood sample. Sheets 5–8 of Supplementary Files S2–S7 show that from these 24 analyses, only one result survived permutation: the most significant (with a p-value inclusion threshold of 5.01 × 10−5) variants (n = 23) that denoted a risk for rumination item in the optimal folate group significantly explained 0.56% of variance in the rumination scale (Supplementary Figure S13).
3.8. Potential Overlap in Risk SNPs of “Rumination” or “Worry” between Suboptimal and Optimal Folate Intake Groups
PRS based on the suboptimal folate intake group explained 0.054% of rumination and 0.051% of worry within the optimal folate intake group. In contrast, PRS based on the optimal folate intake group explained 0.3% of rumination and 0.35% of worry within the suboptimal folate intake group (Sheet 9 of Supplementary Files S4–S7). All these results are significant at a permuted p = 1 × 10−4 level.
4. Discussion
Perseverative negative thinking is a complex cognitive process leading to several somatic and mental disorders. Our study demonstrated that the past-focused rumination component of perseverative negative thinking has a more specific genetic background compared to general worry, which seems more heterogeneous. Namely, our data, in line with previous findings, suggested that the ARNTL gene and purinergic genetic pathway contributed to rumination in the whole sample. However, when we focused on the subgroup of suboptimal folate intake, well-known folate-dependent pathways emerged in the background of rumination, such as regulation of DNA replication, blastocyst development, homocysteine metabolism, and diverse immune responses including chemotaxis, ERK cascade, interferon-, interleukin 1- and tumor necrosis factor-response. Furthermore, genes that were strongly associated to rumination in the suboptimal folate intake subgroup are involved in brain development during late-mid and late prenatal period, emphasizing the role of these genes not only in brain development but in adult cognitive processes. Polygenic genetic compositions of rumination within different folate intake groups explained much less variance from each other than the total set of SNPs from rumination within each group. These observations suggested that lack of adequate folate intake can divert biological processes in the background of rumination.
4.1. Time Perspective and Event-Specificity of Perseverative Negative Thinking
In our study, significant and plausible results emerged only for rumination, i.e., no such results were found for worry. Nagel et al. [23] conducted analyses on the same neuroticism items within the same UK Biobank database as our present work, and with a considerable overlap in analysis methods. Results for rumination seemed more consistent across the two studies of different sample sizes and different covariates than results for worry (discussed in detail in Supplementary File S1).
Content of the two items (“Do you worry too long after an embarrassing experience?” and “Are you a worrier?”) differ from each other only in that the former has a focus on specifically defined past events, while the latter has no specific time or event focus. The “Are you a worrier?” item could itself possibly be further decomposed into distinct elements with divergent underlying cognitive processes, thus somewhat impeding the delineation of a compact, well-defined genetic background. The rumination item, in contrast, represents a well-defined target and mode of perseverative negative thinking, and may involve specific deficits of cognitive control and memory—namely, difficulties in discarding no longer relevant negative information from working memory [13,41]. Because of these specific cognitive underpinnings, in addition to more robust results across studies, it may be a better endophenotype [42] than the more general and wide concept of worry.
A further difficulty in the detection of replicable genetic underpinnings of worry may be that worry or its distinct components might be more dependent on interactions between different genetic variants with each other or with external factors.
Nevertheless, folate intake itself was inversely associated only with the more general item of worry, but not with the more specific form of perseverative negative thinking, rumination. Previous studies have yielded contradictory results on the associations between folate levels and anxiety disorders. While an early study found decreased serum folate level in adult patients with obsessive-compulsive disorder (OCD) compared to controls [43], no later studies could replicate this finding in pediatric OCD [44], in generalized anxiety disorder (GAD) [45], or in higher anxiety score within a cohort study [46]. On the other hand, the same studies found higher homocysteine levels in patient groups of adult OCD [43], pediatric OCD [44], and GAD [45]—but again, no association with anxiety level within a cohort study [46]. Similar to results on folate levels in patient groups, results have also been contradictory on the beneficial effects of adjuvant folates in treatment. While a retrospective study suggested the benefits of l-methylfolate calcium in multimorbid pediatric patients [47], a double-blind controlled study found no differences between placebo group and folic acid group in OCD patients treated with fluoxetine [48]. Further studies are needed to clarify the associations between folate and the emergence (as well as treatment) of different kinds of anxiety disorders, and our results within a large population study may fuel these investigations.
4.2. Genetic Determinants of Past-Focused Perseverative Negative Thinking
Nagel et al.’s [23] rumination results were in line with our whole study sample’s results. In our study, despite the lack of association between folate intake and rumination, nominal significance of gene sets related to transmembrane folate transport for rumination in the whole sample suggested the importance of folate metabolism in this form of perseverative negative thinking. Moreover, we could reveal different biological mechanisms in the background of rumination in different groups stratified according to folate intake. Herein, we proposed a summary model to depict the complexity of the pathomechanisms behind rumination, detailing its currently identified aspects and integrating our results with the results of former studies (Figure 3). Nevertheless, future studies are also needed to uncover causal directions in these gene-by-diet interactions in the background of rumination.
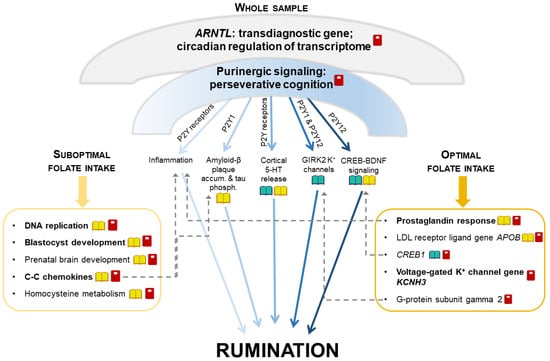
Figure 3.
A proposed model of the impact of multifaceted mechanisms on rumination via additive effects of distinct narrower pathways. Umbrella-like biological mechanisms, being significant for rumination in our whole sample, can be decomposed into separate components that are affected by folate intake in a distinct extent. Umbrella-like mechanisms include ARNTL (that regulates more than 10% of the transcriptome in a circadian rhythm) and purinergic signaling. Separate components influenced by purinergic signaling are displayed below the umbrella, between the two boxes. The left box represents components significant in our suboptimal folate intake group, and the right box represents components significant in our optimal folate intake group. Some of the components represented in the boxes can be biologically related to the components influenced by purinergic signaling displayed between the two boxes (dashed arrows). Dark-light shading of arrows represents the extent to which each component influenced by purinergic signaling might be sensitive to folate intake.  : biological pathways previously having been related to rumination;
: biological pathways previously having been related to rumination;  : biological pathways modulated by folate level;
: biological pathways modulated by folate level;  : our present study’s results for rumination (with bold font if surviving correction for multiple testing); GIRK2: G protein-activated inwardly rectifying potassium channel subunit 2; 5-HT: serotonin; CREB: cAMP-response element binding protein; BDNF: brain-derived neurotrophic factor; LDL: low density lipoprotein; accum.: accumulation; phosph.: phosphorylation.
: our present study’s results for rumination (with bold font if surviving correction for multiple testing); GIRK2: G protein-activated inwardly rectifying potassium channel subunit 2; 5-HT: serotonin; CREB: cAMP-response element binding protein; BDNF: brain-derived neurotrophic factor; LDL: low density lipoprotein; accum.: accumulation; phosph.: phosphorylation.
 : biological pathways previously having been related to rumination;
: biological pathways previously having been related to rumination;  : biological pathways modulated by folate level;
: biological pathways modulated by folate level;  : our present study’s results for rumination (with bold font if surviving correction for multiple testing); GIRK2: G protein-activated inwardly rectifying potassium channel subunit 2; 5-HT: serotonin; CREB: cAMP-response element binding protein; BDNF: brain-derived neurotrophic factor; LDL: low density lipoprotein; accum.: accumulation; phosph.: phosphorylation.
: our present study’s results for rumination (with bold font if surviving correction for multiple testing); GIRK2: G protein-activated inwardly rectifying potassium channel subunit 2; 5-HT: serotonin; CREB: cAMP-response element binding protein; BDNF: brain-derived neurotrophic factor; LDL: low density lipoprotein; accum.: accumulation; phosph.: phosphorylation.
We postulated that there may be some general, umbrella-like, multifaceted biological mechanisms in the background of rumination, which affect several distinct, narrower processes and pathways. As such, ARNTL (or BMAL1) gene emerged for rumination in the whole sample. This gene is a key positive regulator of the circadian clock, showing a circadian oscillation in transcription and translation, and this rhythm then regulates the expression of more than 10% of the transcriptome [49]. ARNTL is a shared gene between rumination and several neuropsychiatric disorders [49,50,51,52,53] and obesity [54], suggesting this past-focused form of perseverative negative thinking as a potential transdiagnostic endophenotype. Another umbrella-like mechanism behind rumination, detected only in the whole sample, may be the importance of purinergic signaling. Genes of P2Y purinergic receptors, implicated in our results and in animal studies of perseverative cognition [55,56], may exert their effects on rumination through divergent biological pathways [11,12,32,57,58,59,60,61]. Purinergic effects on each of these narrower pathways are hypothesized to point to the same direction regarding the generation of rumination. These effects add up and convey a significance to the general, umbrella-like mechanism. However, external factors such as folate intake may modulate each of these distinct, narrow pathways in a different magnitude or manner [62,63,64]. Consequently, in determining the level of rumination in cases of different folate statuses, some of these pathways will become less important, while others will become more so.
4.3. Genetic Determinants of Past-Focused Perseverative Negative Thinking in Case of Suboptimal Folate Intake
In the suboptimal folate intake group, the genetics of rumination may be best determined by the specific pathways and processes most sensitive to folate deficiency. Indeed, DNA synthesis and repair [65], early prenatal development [66], prenatal brain development [67] and levels of C-C chemokines [68] and homocysteine [69] have been demonstrated to be influenced by folate levels, even in interaction with genetics [63,70,71], and have shown associations with neuropsychiatric phenotypes [63,72,73], as well. Adverse effects of genetic, intrauterine or immunometabolic factors on these phenotypes may be compensated by sufficient folate intake, although our results suggested that developmental timing is crucial in folate’s compensatory impact in case of these biological pathways and rumination, as adult folate intake has not showed association with rumination. In line with our results, cohort studies suggested an association between conception during a severe famine and increased prevalence of neurodevelopmental disorders, such as neural tube defects, as well as schizophrenia and schizoid personality [74]. Somewhat contrary to our results, independent additive effects of genetics and prenatal nutrition, not gene-by-nutrition interaction, on childhood processing speed have been suggested by a randomized, placebo-controlled, double-blind study [75].
Concurrent folate intake was not associated with an event-specific past-focused form of adults’ perseverative negative thinking in our study, perhaps because the effect of folate intake on rumination is not linear and may depend on some genetic vulnerabilities or other factors. It is important to highlight however, that folate has been suggested to impact the nervous system at all ages [15]. Higher plasma folate level has been associated with better global cognitive functioning and faster psychomotor speed as well as a reduced risk of severe white matter lesions in non-demented elderly participants [76]. Although a study did not find any associations between folate status and cognitive dysfunction or brain atrophy in Alzheimer’s patients or controls [77], a randomized, double-blind study revealed that treatment with B vitamins slowed brain atrophy [78]. Hemodynamic activity in the brain can indeed serve as an intermediate phenotype between B vitamins and cognitive functioning [79].
4.4. Genetic Determinants of Past-Focused Perseverative Negative Thinking in Case of Optimal Folate Intake
In cases of optimal folate intake, genetic vulnerabilities within numerous specific pathways may be compensated, and only narrow specific processes emerge in genetics. Hippocampal neuronal excitability [80] may be such a mechanism, which was reflected in voltage-gated K+ channel alpha subunit gene KCNH3 (BEC1 or ELK2) within our results, in line with our former rumination GWAS result KCTD12 [11]. KCTD12 acts on the same pathway between GABAB receptor and Kir3 K+ channel as G-protein subunit γ2 [81], implicated in our mapped genes. Former results were contradictory regarding folate dependence of prostaglandin D2 and E2 pathways [82,83,84] and of apolipoprotein B effects [85,86]; however, these pathways also emerged in our results in the optimal folate intake group.
Polygenic scores of rumination significantly explained some variance of the whole, validated rumination scale, but only if based on top risk variants within the optimal folate intake group. Although we have no information on folate intake or folate status in the target sample with the whole rumination scale, this result underlined the importance of differentiating between folate intake levels when considering genetics. Indeed, genetic background of rumination (and worry) in case of suboptimal folate intake explained less of rumination in the optimal folate intake subgroup than vice versa. This discrepancy may suggest that a high amount of genetic vulnerabilities may be compensated by an optimal folate intake. However, even the explained variance in the direction from optimal to suboptimal folate intake was around one tenth of SNP heritability, suggesting considerably different genetic underpinnings depending on folate intake level.
4.5. Limitations
Our study had some limitations. First of all, folate intake was assessed only once in 41.82%, and only as follow-up in 40.22% of all participants, with a 24 h dietary recall questionnaire with multiple pass method. However, the aim was to determine habitual dietary pattern, and validation studies suggest that this method is suitable to cost-effectively establish this information in large cohorts [87]. Second, a precise relative timing of dietary habits and perseverative cognition would be needed, serum and/or erythrocyte folate levels should be assessed, and further intervention studies should be implemented in order to validate the proposed mechanism of folate intake effects. Third, the lower sample size in the suboptimal folate intake group resulted in lower statistical power to determine genetic factors of rumination. However, the genetic pathways identified had well-known biological connections to folate metabolism, supporting their validity. Fourth, to validate our PRS results, a target sample with reliably measured folate intake and/or folate status will be required. Fifth, enrichment tests of mapped genes did not correct for LD between close genes. Therefore, our results with this methodology, though plausible, need future replication.
5. Conclusions
Our results point to a more specific genetic background of past-focused perseverative negative thinking, in contrast to the more general “Are you a worrier?” item. Importance of the specificity of past-focus was also underlined by the shared genetics between this past-focused perseverative negative thinking item and the widely used RRS rumination scale across two different populations. Rumination can thus be regarded as a good candidate for a transdiagnostic endophenotype. Furthermore, rumination as a potential endophenotype could differentiate between different risk biological pathways in different folate intake groups. This is especially interesting because rumination did not show any association with folate intake itself, in contrast to the negative association between worry and folate intake. These results point to the importance of simultaneously taking into account genetic and environmental factors to determine personalized intervention in polygenic and multifactorial disorders. Inconsistent benefits of folate supplementation in depression [17,18,19,20], and a potential benefit of folate in anxiety disorders could also be reframed in light of our results.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/nu13124396/s1, Supplementary Files S1–S8. In Supplementary File S1: Supplementary Tables S1–S4 and Supplementary Figures S1–S13. Table S1: Descriptive statistics for the whole sample and separate subgroups of the investigated UK Biobank sample (Application number 1602). S.E.M.: standard error of mean. Supplementary Table S2: Relationships between the two perseverative negative thinking items and the other variables, within the whole sample and separate subgroups of the investigated UK Biobank sample (Application number 1602). All relationships are significant, except for that of “rumination” and folate intake. χ2: Pearson chi-square, t: t-value, df: degrees of freedom, p: p-value. Supplementary Table S3: Descriptive statistics for NewMood. Range of possible values of scale scores are also indicated. S.E.M.: standard error of mean, BSI: Brief Symptom Inventory. Supplementary Table S4: Relationships between scale scores and other variables in NewMood. t: t-value, df: degrees of freedom, r: Pearson correlation coefficient, p: p-value. Supplementary Figure S1: Manhattan plots of SNP-based tests for “rumination” and “worry” items in the whole study sample (1a and 1b), in the suboptimal folate intake group (1c and 1d), and in the optimal folate intake group (1e and 1f). −log10 of p-value is displayed in function of genomic position. Red line denotes significance threshold corrected only within one analysis. SNP: single-nucleotide polymorphism. Supplementary Figure S2: QQ plots of SNP-based tests for “rumination” and “worry” items in the whole study sample (2a and 2b), in the suboptimal folate intake group (2c and 2d), and in the optimal folate intake group (2e and 2f). Observed −log10 p-values are displayed in function of their expected values. SNP: single-nucleotide polymorphism. Supplementary Figure S3: Manhattan plots of gene-based tests for “rumination” and “worry” items in the whole study sample (3a and 3b), in the suboptimal folate intake group (3c and 3d), and in the optimal folate intake group (3e and 3f). −log10 of p-value is displayed in function of genomic position. Red line denotes significance threshold corrected only within one analysis. Supplementary Figure S4: QQ plots of gene-based tests for “rumination” and “worry” items in the whole study sample (4a and 4b), in the suboptimal folate intake group (4c and 4d), and in the optimal folate intake group (4e and 4f). Observed −log10 p-values are displayed in function of their expected values. Supplementary Figure S5: Association of brain developmental stage-specific gene expression level with gene-based effect for “rumination” and “worry” items in the whole study sample (5a and 5b), in the suboptimal folate intake group (5c and 5d), and in the optimal folate intake group (5e and 5f). −log10 p-value is displayed for each of the 11 general developmental stages of BrainSpan’s brain development samples. Supplementary Figure S6: Association of tissue-specific gene expression level with gene-based effect for “rumination” and “worry” items in the whole study sample (6a and 6b), in the suboptimal folate intake group (6c and 6d), and in the optimal folate intake group (6e and 6f). −log10 p-value is displayed for each of the 30 general tissue types of GTEx v8. Supplementary Figure S7: Significantly enriched gene sets for the “rumination” item in the whole study sample. CGP: chemical and genetic perturbations, CP: canonical pathways, BP: biological processes, MF: molecular function. Supplementary Figure S8: Significantly enriched gene sets for the “rumination” item in the suboptimal folate intake group. CGP: chemical and genetic perturbations, CP: canonical pathways, BP: biological processes, MF: molecular function. Supplementary Figure S9: Significantly enriched gene sets for the “rumination” item in the optimal folate intake group. CGP: chemical and genetic perturbations, BP: biological processes. Supplementary Figure S10: Significantly enriched gene set for the “worry” item in the whole study sample. It emerged in the “canonical pathways” subcategory of the C2 curated gene set collection. Supplementary Figure S11: Significantly enriched gene sets for the “worry” item in the suboptimal folate intake group. They emerged in the “chemical and genetic perturbations” subcategory of the C2 curated gene set collection. Supplementary Figure S12: Significantly enriched gene set for the “worry” item in the optimal folate intake group. It emerged in the “chemical and genetic perturbations” subcategory of the C2 curated gene set collection. Supplementary Figure S13: Explanatory value of “rumination” risk SNPs within UK Biobank’s optimal folate intake group, for NewMood’s rumination scale. Explained variance (R2) in NewMood is displayed in function of p-value inclusion threshold in UK Biobank. Colors denote −log10 p-value of the model that regresses rumination scale on PRS, suggesting the highest explanatory value in case of the most significant few SNPs, and pointing to some shared genetics between “rumination” item and a widely used rumination scale across two different populations. PRS: polygenic risk score. Supplementary File S2: Full results for “rumination” in the whole sample. Supplementary File S3: Full results for “worry” in the whole sample. Supplementary File S4: Full results for “rumination” in the suboptimal folate intake group. Supplementary File S5: Full results for “worry” in the suboptimal folate intake group. Supplementary File S6: Full results for “rumination” in the optimal folate intake group. Supplementary File S7: Full results for “worry” in the optimal folate intake group. Supplementary File S8: eQTL mapping results for “worry” in the whole sample.
Author Contributions
Conceptualization, G.J. and N.E.; methodology, N.E. and A.M.; software, B.B., A.M., G.H. and P.P.; validation, B.B., N.E. and P.P.; formal analysis, N.E. and A.M.; investigation, G.J., X.G., G.B. (Gerome Breen), G.B. (Gyorgy Bagdy) and J.F.W.D.; resources, G.J., P.A., X.G., G.B. (Gerome Breen), G.B. (Gyorgy Bagdy) and J.F.W.D.; data curation, A.M., G.H., B.B. and P.P.; writing—original draft preparation, N.E., G.J. and G.B. (Gyorgy Bagdy); writing—review and editing, N.E., G.J., X.G., P.A., G.B. (Gyorgy Bagdy) and J.F.W.D.; visualization, N.E. and G.J.; supervision, G.J., P.A., G.B. (Gyorgy Bagdy) and J.F.W.D.; project administration, G.J., G.B. (Gyorgy Bagdy) and P.A.; funding acquisition, N.E., G.J., G.B. (Gyorgy Bagdy), P.A. and J.F.W.D. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by the UK Biobank Resource under Application Number 1602; by the Sixth Framework Program of the European Union (NewMood, LSHM-CT-2004-503474); by the Hungarian Brain Research Program (Grants KTIA_13_NAPA-II/14 and 2017-1.2.1-NKP-2017-00002), and the National Development Agency (Grant KTIA_NAP_13-1-2013-0001); by the Hungarian Academy of Sciences, Hungarian National Development Agency, Semmelweis University and the Hungarian Brain Research Program (Grant KTIA_NAP_13-2-2015-0001, MTA-SE-NAP B Genetic Brain Imaging Migraine Research Group); by the Hungarian Academy of Sciences (MTA-SE Neuropsychopharmacology and Neurochemistry Research Group); by the National Institute for Health Research Manchester Biomedical Research Centre; by the NRDI Fund based on the charter of bolster issued by the NRDI Office under the auspices of the Ministry for Innovation and Technology (BME NC TKP2020, BME IE-BIO TKP2020, Artificial Intelligence National Laboratory Programme); by the New National Excellence Program of the Ministry for Innovation and Technology from the source of the National Research, Development and Innovation Fund (ÚNKP-20-4-II-SE-9; ÚNKP-20-5-BME-92; ÚNKP-21-4-II-SE-1; ÚNKP-21-5-BME-362); by the National Research, Development and Innovation Office, Hungary (2019-2.1.7-ERA-NET-2020-00005), under the frame of ERA PerMed (ERAPERMED2019-108); and by the Thematic Excellence Programme (2020-4.1.1.-TKP2020) of the Ministry for Innovation and Technology in Hungary, within the framework of the Neurology and Translational Biotechnology thematic programmes of the Semmelweis University. G.H. and X.G. are recipients of the Janos Bolyai Research Scholarship of the Hungarian Academy of Sciences. P.P. is an international Research Fellow of the Japan Society for the Promotion of Science (Postdoctoral Fellowships for Research in Japan (Standard), P20809). None of the sponsors had any role in designing the study, collecting, analyzing or interpreting data, or in preparing or submitting the paper for publication.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the National Research Ethics Service Committee North West–Haydock (REC reference number: 21/NW/0157, dated 29 June 2021), the North Manchester Local Research Ethics Committee, Manchester, United Kingdom (REC reference number: 05/Q1406/26), and of the Scientific and Research Ethics Committee of the Medical Research Council, Budapest, Hungary (ad.225/KO/2005; ad.323-60/2005-1018EKU and ad.226/KO/2005; ad.323-61/2005-1018 EKU).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to ethical considerations.
Acknowledgments
We thank Charles Curtis for his assistance in genotyping (King’s College London, SGDP Centre, Institute of Psychiatry), and Anna Petschner for her help in figure preparation.
Conflicts of Interest
G.B. (Gerome Breen) has received honoraria, research or conference grants and consulting fees from Illumina, Otsuka, and COMPASS Pathfinder Ltd. G.B. (Gyorgy Bagdy) is a member of the Board of directors at Gedeon Richter. J.F.W.D. has share options in P1vital, and he has performed research, consultancy, and speaking engagements for AstraZeneca, Autifony, Bristol-Myers Squibb, Eli Lilly, Janssen-Cilag, P1vital, Schering Plough, and Servier, for which all fees have been paid to the University of Manchester to reimburse them for the time taken. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. The remaining authors declare no conflict of interest.
References
- Denovan, A.; Dagnall, N.; Lofthouse, G. Neuroticism and somatic complaints: Concomitant effects of rumination and worry. Behav. Cogn. Psychother. 2019, 47, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Merino, H.; Ferreiro, F.; Senra, C. Cognitive vulnerability to emotional symptoms: Reconsidering the role of worry and rumination. J. Psychopathol. Behav. Assess. 2014, 36, 136–142. [Google Scholar] [CrossRef]
- Broeren, S.; Muris, P.; Bouwmeester, S.; van der Heijden, K.B.; Abee, A. The role of repetitive negative thoughts in the vulnerability for emotional problems in non-clinical children. J. Child Fam. Stud. 2011, 20, 135–148. [Google Scholar] [CrossRef]
- Merino, H.; Senra, C.; Ferreiro, F. Are worry and rumination specific pathways linking neuroticism and symptoms of anxiety and depression in patients with generalized anxiety disorder, major depressive disorder and mixed anxiety-depressive disorder? PLoS ONE 2016, 11, e0156169. [Google Scholar] [CrossRef] [PubMed]
- Roelofs, J.; Huibers, M.; Peeters, F.; Arntz, A.; van Os, J. Rumination and worrying as possible mediators in the relation between neuroticism and symptoms of depression and anxiety in clinically depressed individuals. Behav. Res. Ther. 2008, 46, 1283–1289. [Google Scholar] [CrossRef]
- Brosschot, J.F.; Gerin, W.; Thayer, J.F. The perseverative cognition hypothesis: A review of worry, prolonged stress-related physiological activation, and health. J. Psychosom. Res. 2006, 60, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M.; Alloy, L.B. A roadmap to rumination: A review of the definition, assessment, and conceptualization of this multifaceted construct. Clin. Psychol. Rev. 2009, 29, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Ottaviani, C. Brain-heart interaction in perseverative cognition. Psychophysiology 2018, 55, 1–14. [Google Scholar] [CrossRef]
- Topper, M.; Molenaar, D.; Emmelkamp, P.M.; Ehring, T. Are rumination and worry two sides of the same coin? A structural equation modelling approach. J. Exp. Psychopathol. 2014, 5, 363–381. [Google Scholar] [CrossRef]
- du Pont, A.; Rhee, S.H.; Corley, R.P.; Hewitt, J.K.; Friedman, N.P. Are rumination and neuroticism genetically or environmentally distinct risk factors for psychopathology? J. Abnorm. Psychol. 2019, 128, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Eszlari, N.; Millinghoffer, A.; Petschner, P.; Gonda, X.; Baksa, D.; Pulay, A.J.; Rethelyi, J.M.; Breen, G.; Deakin, J.F.W.; Antal, P.; et al. Genome-wide association analysis reveals KCTD12 and miR-383-binding genes in the background of rumination. Transl. Psychiatry 2019, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Eszlari, N.; Petschner, P.; Gonda, X.; Baksa, D.; Elliott, R.; Anderson, I.M.; Deakin, J.F.W.; Bagdy, G.; Juhasz, G. Childhood adversity moderates the effects of HTR2A epigenetic regulatory polymorphisms on rumination. Front. Psychiatry 2019, 10, 394. [Google Scholar] [CrossRef] [PubMed]
- Shaw, Z.A.; Hilt, L.M.; Starr, L.R. The developmental origins of ruminative response style: An integrative review. Clin. Psychol. Rev. 2019, 74, 101780. [Google Scholar] [CrossRef] [PubMed]
- Eszlari, N.; Kovacs, D.; Petschner, P.; Pap, D.; Gonda, X.; Elliott, R.; Anderson, I.M.; Deakin, J.F.W.; Bagdy, G.; Juhasz, G. Distinct effects of folate pathway genes MTHFR and MTHFD1L on ruminative response style: A potential risk mechanism for depression. Transl. Psychiatry 2016, 6, e745. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, E.H. Folic acid, ageing, depression, and dementia. BMJ 2002, 324, 1512–1515. [Google Scholar] [CrossRef] [PubMed]
- Sanhueza, C.; Ryan, L.; Foxcroft, D.R. Diet and the risk of unipolar depression in adults: Systematic review of cohort studies. J. Hum. Nutr. Diet. 2013, 26, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Bedson, E.; Bell, D.; Carr, D.; Carter, B.; Hughes, D.; Jorgensen, A.; Lewis, H.; Lloyd, K.; McCaddon, A.; Moat, S.; et al. Folate augmentation of treatment—Evaluation for depression (FolATED): Randomised trial and economic evaluation. Health Technol. Assess. 2014, 18, vii–viii. [Google Scholar] [CrossRef] [PubMed]
- Papakostas, G.I.; Shelton, R.C.; Zajecka, J.M.; Etemad, B.; Rickels, K.; Clain, A.; Baer, L.; Dalton, E.D.; Sacco, G.R.; Schoenfeld, D.; et al. L-methylfolate as adjunctive therapy for SSRI-resistant major depression: Results of two randomized, double-blind, parallel-sequential trials. Am. J. Psychiatry 2012, 169, 1267–1274. [Google Scholar] [CrossRef]
- Taylor, M.J.; Carney, S.M.; Goodwin, G.M.; Geddes, J.R. Folate for depressive disorders: Systematic review and meta-analysis of randomized controlled trials. J. Psychopharmacol. 2004, 18, 251–256. [Google Scholar] [CrossRef]
- Roberts, E.; Carter, B.; Young, A.H. Caveat emptor: Folate in unipolar depressive illness, a systematic review and meta-analysis. J. Psychopharmacol. 2018, 32, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Durga, J.; van Boxtel, M.P.; Schouten, E.G.; Bots, M.L.; Kok, F.J.; Verhoef, P. Folate and the methylenetetrahydrofolate reductase 677C-->T mutation correlate with cognitive performance. Neurobiol. Aging 2006, 27, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Nicholl, B.I.; Cullen, B.; Martin, D.; Ul-Haq, Z.; Evans, J.; Gill, J.M.; Roberts, B.; Gallacher, J.; Mackay, D.; et al. Prevalence and characteristics of probable major depression and bipolar disorder within UK biobank: Cross-sectional study of 172,751 participants. PLoS ONE 2013, 8, e75362. [Google Scholar] [CrossRef] [PubMed]
- Nagel, M.; Watanabe, K.; Stringer, S.; Posthuma, D.; van der Sluis, S. Item-level analyses reveal genetic heterogeneity in neuroticism. Nat. Commun. 2018, 9, 905. [Google Scholar] [CrossRef]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Nagel, M.; Jansen, P.R.; Stringer, S.; Watanabe, K.; de Leeuw, C.A.; Bryois, J.; Savage, J.E.; Hammerschlag, A.R.; Skene, N.G.; Munoz-Manchado, A.B.; et al. Meta-analysis of genome-wide association studies for neuroticism in 449, 484 individuals identifies novel genetic loci and pathways. Nat. Genet. 2018, 50, 920–927. [Google Scholar] [CrossRef]
- Hullam, G.; Antal, P.; Petschner, P.; Gonda, X.; Bagdy, G.; Deakin, B.; Juhasz, G. The UKB envirome of depression: From interactions to synergistic effects. Sci. Rep. 2019, 9, 9723. [Google Scholar] [CrossRef]
- Liu, B.; Young, H.; Crowe, F.L.; Benson, V.S.; Spencer, E.A.; Key, T.J.; Appleby, P.N.; Beral, V. Development and evaluation of the Oxford WebQ, a low-cost, web-based method for assessment of previous 24 h dietary intakes in large-scale prospective studies. Public Health Nutr. 2011, 14, 1998–2005. [Google Scholar] [CrossRef]
- Galante, J.; Adamska, L.; Young, A.; Young, H.; Littlejohns, T.J.; Gallacher, J.; Allen, N. The acceptability of repeat Internet-based hybrid diet assessment of previous 24-h dietary intake: Administration of the Oxford WebQ in UK Biobank. Br. J. Nutr. 2016, 115, 681–686. [Google Scholar] [CrossRef] [PubMed]
- John, O.P.; Srivastava, S. Handbook of Personality: Theory and Research; John, O.P., Ed.; Guilford Press: New York, NY, USA, 1999; Volume 2, pp. 102–139. [Google Scholar]
- Treynor, W.; Gonzalez, R.; Nolen-Hoeksema, S. Rumination reconsidered: A psychometric analysis. Cognit. Ther. Res. 2003, 27, 247–259. [Google Scholar] [CrossRef]
- Derogatis, L.R. BSI: Brief Symptom Inventory: Administration, Scoring, and Procedures Manual; National Computer Systems Pearson, Inc.: Minneapolis, MN, USA, 1993. [Google Scholar]
- Juhasz, G.; Dunham, J.S.; McKie, S.; Thomas, E.; Downey, D.; Chase, D.; Lloyd-Williams, K.; Toth, Z.G.; Platt, H.; Mekli, K.; et al. The CREB1-BDNF-NTRK2 pathway in depression: Multiple gene-cognition-environment interactions. Biol. Psychiatry 2011, 69, 762–771. [Google Scholar] [CrossRef]
- Bulik-Sullivan, B.K.; Loh, P.-R.; Finucane, H.K.; Ripke, S.; Yang, J.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Patterson, N.; Daly, M.J.; Price, A.L.; Neale, B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015, 47, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Galinsky, K.J.; Bhatia, G.; Loh, P.R.; Georgiev, S.; Mukherjee, S.; Patterson, N.J.; Price, A.L. Fast principal-component analysis reveals convergent evolution of ADH1B in Europe and east Asia. Am. J. Hum. Genet. 2016, 98, 456–472. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, s13742-015. [Google Scholar] [CrossRef]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLOS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Human Genomics. The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Kawasawa, Y.I.; Cheng, F.; Zhu, Y.; Xu, X.; Li, M.; Sousa, A.M.; Pletikos, M.; Meyer, K.A.; Sedmak, G.; et al. Spatio-temporal transcriptome of the human brain. Nature 2011, 478, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; O’Reilly, P.F. PRSice-2: Polygenic risk score software for biobank-scale data. Gigascience 2019, 8, giz082. [Google Scholar] [CrossRef]
- Zetsche, U.; Burkner, P.C.; Schulze, L. Shedding light on the association between repetitive negative thinking and deficits in cognitive control—A meta-analysis. Clin. Psychol. Rev. 2018, 63, 56–65. [Google Scholar] [CrossRef]
- Gottesman, I.I.; Gould, T.D. The endophenotype concept in psychiatry: Etymology and strategic intentions. Am. J. Psychiatry 2003, 160, 636–645. [Google Scholar] [CrossRef]
- Atmaca, M.; Tezcan, E.; Kuloglu, M.; Kirtas, O.; Ustundag, B. Serum folate and homocysteine levels in patients with obsessive-compulsive disorder. Psychiatry Clin. Neurosci. 2005, 59, 616–620. [Google Scholar] [CrossRef]
- Esnafoğlu, E.; Yaman, E. Vitamin B12, folic acid, homocysteine and vitamin D levels in children and adolescents with obsessive compulsive disorder. Psychiatry Res. 2017, 254, 232–237. [Google Scholar] [CrossRef]
- Saraswathy, K.N.; Ansari, S.N.; Kaur, G.; Joshi, P.C.; Chandel, S. Association of vitamin B12 mediated hyperhomocysteinemia with depression and anxiety disorder: A cross-sectional study among Bhil indigenous population of India. Clin. Nutr. ESPEN 2019, 30, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Bjelland, I.; Tell, G.S.; Vollset, S.E.; Refsum, H.; Ueland, P.M. Folate, vitamin B12, homocysteine, and the MTHFR 677C->T polymorphism in anxiety and depression: The Hordaland homocysteine study. Arch. Gen. Psychiatry 2003, 60, 618–626. [Google Scholar] [CrossRef]
- Rainka, M.; Aladeen, T.; Westphal, E.; Meaney, J.; Gengo, F.; Greger, J.; Capote, H. L-methylfolate calcium supplementation in adolescents and children: A retrospective analysis. J. Psychiatr. Pract. 2019, 25, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Tural, Ü.; Çorapçıoğlu, A.; Boşgelmez, Ş.; Köroğlu, G.; Ünver, H.; Duman, C.; Önder, E. Double blind controlled study of adding folic acid to fluoxetine in the treatment of OCD. Psychiatr. Danub. 2019, 31, 69–77. [Google Scholar] [CrossRef]
- Cronin, P.; McCarthy, M.J.; Lim, A.S.P.; Salmon, D.P.; Galasko, D.; Masliah, E.; De Jager, P.L.; Bennett, D.A.; Desplats, P. Circadian alterations during early stages of Alzheimer’s disease are associated with aberrant cycles of DNA methylation in BMAL1. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2017, 13, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Baird, A.L.; Coogan, A.N.; Siddiqui, A.; Donev, R.M.; Thome, J. Adult attention-deficit hyperactivity disorder is associated with alterations in circadian rhythms at the behavioural, endocrine and molecular levels. Mol. Psychiatry 2012, 17, 988–995. [Google Scholar] [CrossRef]
- Borglum, A.D.; Demontis, D.; Grove, J.; Pallesen, J.; Hollegaard, M.V.; Pedersen, C.B.; Hedemand, A.; Mattheisen, M.; Uitterlinden, A.; Nyegaard, M.; et al. Genome-wide study of association and interaction with maternal cytomegalovirus infection suggests new schizophrenia loci. Mol. Psychiatry 2014, 19, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, R.; Gonzalez, S.; Villa, E.; Ramirez, M.; Zavala, J.; Armas, R.; Contreras, J.; Dassori, A.; Leach, R.J.; Flores, D.; et al. Identification of circadian gene variants in bipolar disorder in Latino populations. J. Affect. Disord. 2015, 186, 367–375. [Google Scholar] [CrossRef][Green Version]
- Partonen, T.; Treutlein, J.; Alpman, A.; Frank, J.; Johansson, C.; Depner, M.; Aron, L.; Rietschel, M.; Wellek, S.; Soronen, P.; et al. Three circadian clock genes Per2, Arntl, and Npas2 contribute to winter depression. Ann. Med. 2007, 39, 229–238. [Google Scholar] [CrossRef]
- Milagro, F.I.; Gomez-Abellan, P.; Campion, J.; Martinez, J.A.; Ordovas, J.M.; Garaulet, M. CLOCK, PER2 and BMAL1 DNA methylation: Association with obesity and metabolic syndrome characteristics and monounsaturated fat intake. Chronobiol. Int. 2012, 29, 1180–1194. [Google Scholar] [CrossRef]
- Koch, H.; Bespalov, A.; Drescher, K.; Franke, H.; Krügel, U. Impaired cognition after stimulation of P2Y1 receptors in the rat medial prefrontal cortex. Neuropsychopharmacology 2015, 40, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Krügel, U.; Köles, L.; Illés, P. Integration of neuronal and glial signalling by pyramidal cells of the rat prefrontal cortex; control of cognitive functions and addictive behaviour by purinergic mechanisms. Neuropsychopharmacol. Hung. 2013, 15, 206–213. [Google Scholar]
- Antypa, N.; Van der Does, A.J. Serotonin transporter gene, childhood emotional abuse and cognitive vulnerability to depression. Genes Brain Behav. 2010, 9, 615–620. [Google Scholar] [CrossRef]
- Beevers, C.G.; Wells, T.T.; McGeary, J.E. The BDNF Val66Met polymorphism is associated with rumination in healthy adults. Emotion 2009, 9, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Guzman, S.J.; Gerevich, Z. P2Y receptors in synaptic transmission and plasticity: Therapeutic potential in cognitive dysfunction. Neural Plast. 2016, 2016, 1207393. [Google Scholar] [CrossRef] [PubMed]
- Lazary, J.; Juhasz, G.; Anderson, I.M.; Jacob, C.P.; Nguyen, T.T.; Lesch, K.P.; Reif, A.; Deakin, J.F.; Bagdy, G. Epistatic interaction of CREB1 and KCNJ6 on rumination and negative emotionality. Eur. Neuropsychopharmacol. 2011, 21, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Zhou, Q.; Cao, Y.; Shi, H.; Wu, H.; Zhang, B.; Huang, F.; Wu, X. P2Y(12) deficiency in mouse impairs noradrenergic system in brain, and alters anxiety-like neurobehavior and memory. Genes Brain Behav. 2019, 18, e12458. [Google Scholar] [CrossRef]
- Bottiglieri, T.; Laundy, M.; Crellin, R.; Toone, B.K.; Carney, M.W.; Reynolds, E.H. Homocysteine, folate, methylation, and monoamine metabolism in depression. J. Neurol. Neurosurg. Psychiatry 2000, 69, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Kronenberg, G.; Harms, C.; Sobol, R.W.; Cardozo-Pelaez, F.; Linhart, H.; Winter, B.; Balkaya, M.; Gertz, K.; Gay, S.B.; Cox, D.; et al. Folate deficiency induces neurodegeneration and brain dysfunction in mice lacking uracil DNA glycosylase. J. Neurosci. Off. J. Soc. Neurosci. 2008, 28, 7219–7230. [Google Scholar] [CrossRef] [PubMed]
- Robinson, N.; Grabowski, P.; Rehman, I. Alzheimer’s disease pathogenesis: Is there a role for folate? Mech. Ageing Dev. 2018, 174, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Catala, G.N.; Bestwick, C.S.; Russell, W.R.; Tortora, K.; Giovannelli, L.; Moyer, M.P.; Lendoiro, E.; Duthie, S.J. Folate, genomic stability and colon cancer: The use of single cell gel electrophoresis in assessing the impact of folate in vitro, in vivo and in human biomonitoring. Mutat. Res. 2019, 843, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Van Winkle, L.J.; Ryznar, R. One-carbon metabolism regulates embryonic stem cell fate through epigenetic DNA and histone modifications: Implications for transgenerational metabolic disorders in adults. Front. Cell Dev. Biol. 2019, 7, 300. [Google Scholar] [CrossRef] [PubMed]
- Ars, C.L.; Nijs, I.M.; Marroun, H.E.; Muetzel, R.; Schmidt, M.; Steenweg-de Graaff, J.; van der Lugt, A.; Jaddoe, V.W.; Hofman, A.; Steegers, E.A.; et al. Prenatal folate, homocysteine and vitamin B(12) levels and child brain volumes, cognitive development and psychological functioning: The generation R study. Br. J. Nutr. 2019, 122, S1–S9. [Google Scholar] [CrossRef]
- Denny, K.J.; Jeanes, A.; Fathe, K.; Finnell, R.H.; Taylor, S.M.; Woodruff, T.M. Neural tube defects, folate, and immune modulation. Birth Defects Res. Part A Clin. Mol. Teratol. 2013, 97, 602–609. [Google Scholar] [CrossRef]
- Selhub, J.; Jacques, P.F.; Dallal, G.; Choumenkovitch, S.; Rogers, G. The use of blood concentrations of vitamins and their respective functional indicators to define folate and vitamin B12 status. Food Nutr. Bull. 2008, 29, S67–S73. [Google Scholar] [CrossRef]
- Hammons, A.L.; Summers, C.M.; Woodside, J.V.; McNulty, H.; Strain, J.J.; Young, I.S.; Murray, L.; Boreham, C.A.; Scott, J.M.; Mitchell, L.E.; et al. Folate/homocysteine phenotypes and MTHFR 677C>T genotypes are associated with serum levels of monocyte chemoattractant protein-1. Clin. Immunol. 2009, 133, 132–137. [Google Scholar] [CrossRef]
- Stanger, O.; Fowler, B.; Piertzik, K.; Huemer, M.; Haschke-Becher, E.; Semmler, A.; Lorenzl, S.; Linnebank, M. Homocysteine, folate and vitamin B12 in neuropsychiatric diseases: Review and treatment recommendations. Expert Rev. Neurother. 2009, 9, 1393–1412. [Google Scholar] [CrossRef]
- Huber, A.K.; Giles, D.A.; Segal, B.M.; Irani, D.N. An emerging role for eotaxins in neurodegenerative disease. Clin. Immunol. 2018, 189, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, A.A.; Hewedi, D.H.; Eissa, A.M.; Frydecka, D.; Misiak, B. Homocysteine levels in schizophrenia and affective disorders-focus on cognition. Front. Behav. Neurosci. 2014, 8, 343. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Susser, E.; Hoek, H.W.; Brown, A. Neurodevelopmental disorders after prenatal famine: The story of the Dutch Famine Study. Am. J. Epidemiol. 1998, 147, 213–216. [Google Scholar] [CrossRef] [PubMed]
- Campoy, C.; Azaryah, H.; Torres-Espínola, F.J.; Martínez-Zaldívar, C.; García-Santos, J.A.; Demmelmair, H.; Haile, G.; Györei, E.; Ramírez-Tortosa, M.D.C.; Reischl, E.; et al. Long-chain polyunsaturated fatty acids, homocysteine at birth and fatty acid desaturase gene cluster polymorphisms are associated with children’s processing speed up to age 9 years. Nutrients 2020, 13, 131. [Google Scholar] [CrossRef] [PubMed]
- de Lau, L.M.; Refsum, H.; Smith, A.D.; Johnston, C.; Breteler, M.M. Plasma folate concentration and cognitive performance: Rotterdam scan study. Am. J. Clin. Nutr. 2007, 86, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Rabensteiner, J.; Hofer, E.; Fauler, G.; Fritz-Petrin, E.; Benke, T.; Dal-Bianco, P.; Ransmayr, G.; Schmidt, R.; Herrmann, M. The impact of folate and vitamin B12 status on cognitive function and brain atrophy in healthy elderly and demented Austrians, a retrospective cohort study. Aging 2020, 12, 15478–15491. [Google Scholar] [CrossRef]
- Smith, A.D.; Smith, S.M.; de Jager, C.A.; Whitbread, P.; Johnston, C.; Agacinski, G.; Oulhaj, A.; Bradley, K.M.; Jacoby, R.; Refsum, H. Homocysteine-lowering by B vitamins slows the rate of accelerated brain atrophy in mild cognitive impairment: A randomized controlled trial. PLoS ONE 2010, 5, e12244. [Google Scholar] [CrossRef]
- Cecchetti, L.; Lettieri, G.; Handjaras, G.; Leo, A.; Ricciardi, E.; Pietrini, P.; Pellegrini, S. Brain hemodynamic intermediate phenotype links vitamin B(12) to cognitive profile of healthy and mild cognitive impaired subjects. Neural Plast. 2019, 2019, 6874805. [Google Scholar] [CrossRef] [PubMed]
- Miyake, A.; Takahashi, S.; Nakamura, Y.; Inamura, K.; Matsumoto, S.; Mochizuki, S.; Katou, M. Disruption of the ether-a-go-go K+ channel gene BEC1/KCNH3 enhances cognitive function. J. Neurosci. 2009, 29, 14637–14645. [Google Scholar] [CrossRef]
- David, M.; Richer, M.; Mamarbachi, A.M.; Villeneuve, L.R.; Dupré, D.J.; Hebert, T.E. Interactions between GABA-B1 receptors and Kir 3 inwardly rectifying potassium channels. Cell. Signal. 2006, 18, 2172–2181. [Google Scholar] [CrossRef]
- Carroll, N.; Hughes, L.; McEntee, G.; Parle-McDermott, A. Investigation of the molecular response to folate metabolism inhibition. J. Nutr. Biochem. 2012, 23, 1531–1536. [Google Scholar] [CrossRef]
- Li, G.; Xia, M.; Abais, J.M.; Boini, K.; Li, P.L.; Ritter, J.K. Protective action of anandamide and its COX-2 metabolite against l-homocysteine-induced NLRP3 inflammasome activation and injury in podocytes. J. Pharmacol. Exp. Ther. 2016, 358, 61–70. [Google Scholar] [CrossRef]
- McKay, D.L.; Perrone, G.; Rasmussen, H.; Dallal, G.; Hartman, W.; Cao, G.; Prior, R.L.; Roubenoff, R.; Blumberg, J.B. The effects of a multivitamin/mineral supplement on micronutrient status, antioxidant capacity and cytokine production in healthy older adults consuming a fortified diet. J. Am. Coll. Nutr. 2000, 19, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Nakouzi, G.A.; Nadeau, J.H. Does dietary folic acid supplementation in mouse NTD models affect neural tube development or gamete preference at fertilization? BMC Genet. 2014, 15, 91. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, M.; Yuan, M.M.; Yuan, L.; Huang, L.L.; Liao, J.H.; Yu, X.L.; Su, C.; Chen, Y.H.; Yang, Y.Y.; Yu, H.; et al. Chronic folate deficiency induces glucose and lipid metabolism disorders and subsequent cognitive dysfunction in mice. PLoS ONE 2018, 13, e0202910. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, D.C.; Hardie, L.J.; Frost, G.S.; Alwan, N.A.; Bradbury, K.E.; Carter, M.; Elliott, P.; Evans, C.E.L.; Ford, H.E.; Hancock, N.; et al. Validation of the Oxford webQ online 24-hour dietary questionnaire using biomarkers. Am. J. Epidemiol. 2019, 188, 1858–1867. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).