Abstract
The outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a public health emergency that turns the year 2020–2021 into annus horribilis for millions of people across international boundaries. The interspecies transmission of this zoonotic virus and mutated variants are aided by exposure dynamics of infected aerosols, fomites and intermediate reservoirs. The spike in the first, second and third waves of coronavirus confirms that herd immunity is not yet reached and everyone including livestock is still vulnerable to the infection. Of serious concern are the communitarian nature of agrarians in the livestock sector, aerogenous spread of the virus and attendant cytocidal effect in permissive cells following activation of pathogen recognition receptors, replication cycles, virulent mutations, seasonal spike in infection rates, flurry of reinfections and excess mortalities that can affect animal welfare and food security. As the capacity to either resist or be susceptible to infection is influenced by numerous factors, identifying coronavirus-associated variants and correlating exposure dynamics with viral aerosols, spirometry indices, comorbidities, susceptible blood types, cellular miRNA binding sites and multisystem inflammatory syndrome remains a challenge where the lethal zoonotic infections are prevalent in the livestock industry, being the hub of dairy, fur, meat and egg production. This review provides insights into the complexity of the disease burden and recommends precision smart-farming models for upscaling biosecurity measures and adoption of digitalised technologies (robotic drones) powered by multiparametric sensors and radio modem systems for real-time tracking of infectious strains in the agro-environment and managing the transition into the new-normal realities in the livestock industry.
1. Introduction
Coronaviruses (CoVs) are fairly pleomorphic or crown-shaped, single-stranded, positive-sense RNA viruses from the Coronaviridae family. CoVs are highly pathogenic, polyadenylated in nature and primarily infect the respiratory tracts of the hosts [1,2]. These nonsegmented cis-acting viruses have the largest ribonucleic acid genomes (30–32 kb) with a 5′-cap structure and 3′-poly-A tail [3]. Genotypic and serologic tools group these viral strains into four subfamilies, namely α-, β-, γ- and δ-CoVs [4]. The recent outbreak of coronavirus infection is traceable to a cluster of isolates from pneumonia patients in the Chinese city of Wuhan in December 2019. Aetiology of the viral strain was confirmed to be β-coronavirus sequel to bronchoscopy and sequencing of bronchoalveolar lavage fluid. Comparative homological analysis shows high (~88%) genetic similarity with bat-derived severe acute respiratory syndrome (SARS)-like coronavirus (bat-SL-covzc45, bat-SL-covzxc21) and (~70%) to SARS-CoV-2, respectively [5,6]. Within 6 months, the virus (SARS-CoV-2) spread to over 213 countries and territories due to its rapid adaptive rate, high infectivity and autochthonous transmissibility across species. There are over two hundred million (211,717,079) reported cases of COVID-19 infections worldwide. Of these, victims include 17,710,881 active cases under mild condition, 109,459 active cases under serious/critical condition, 189,465,076 closed cases (for recovered/discharged patients) and 4,431,663 deaths as of 21 August 2021 [7]. Despite the administration of antiviral medications, systemic antibiotics, immune modulators and oxygen therapy and all draconian containment strategies, the world still stands at crossroads in the fight against this deadly virus.
Although knowledge about COVID-19 is evolving across the geographic landscape, there are uncertainties and conjectures on its infectivity, containment and herd immunity. SARS-CoV-2 has a genome size of 27–34 kb and incubation period of 2–14 days [8,9]. This enveloped virus contains lipid bilayer encoding the genome in spike (S), membrane (M), nucleocapsid (N), or haemagglutinin-esterase glycoproteins [10,11]. Approximately 80% of COVID-19 infections are asymptomatic or mild; 15% are severe, requiring oxygen; and 5% are critical, requiring ventilation [12]. It is transmissible symptomatically, asymptomatically or paucisymptomatically by contact with fomites, incubation carriers and respiratory droplets from persons or animals [13,14]. Oral compulsive habit of nail biting (onychophagy), reflex action of coughing or sneezing at close range and frequent touching of face or mouth with infected fingers are possible ways of contracting the virus. Infected microdroplets and aerosols may spread coronavirus during breathing, coughing or sneezing as intrafamilial spread is rising locally too. SARS-CoV-2 is transmissible through nebulising therapy, bronchoscopy, endotracheal intubation, suctioning tracheotomy or cardiopulmonary resuscitation that yields a high volume of respiratory aerosols [4,15].
1.1. Herbivory and Intricacies of Exposure Dynamics
Viruses can infect any organism in the ecosystem. Earlier scientists found plants to be the first hosts to viroids or virions [16]. Metagenomic analyses posit that plants can be latently infected by positive single-stranded RNA viruses with no symptoms but soon degenerate into acute disease condition [17,18]. Cross-species transmission of SARS-CoV-2 is not an exception where herbivory exists. Abraham (2006) [19] defines herbivory as a form of feeding by which heterotrophs feed on autotrophs or prey organisms. Herbivory exposes the nasal cavity, posterior pharynx, trachea, glottis, vocal cords and bronchopulmonary segments to ecological variables as one of the drivers of land-use dynamics. Herbivores are multicellular eukaryotic species that are anatomically and physiologically adapted to feeding on forage and fodder crops. The majority of herbivores belong to Artiodactyla (even-toed ungulates), Perissodactyla (odd-toes ungulates), Squamata (reptiles) and Testudines (chelonii) animal orders. Bats, being flying herbivores with unique aerodynamics capacity, can harbour coronaviruses. The coexistence of herbivores with sessile autotrophs in loose aggregations, complex groups or colonies creates dynamic interspecific interactions that may be mutualistic, competitive, facultative or antagonistic in nature [20]. Either by chewing or piercing–sucking mechanism, herbivorous feeding strategies are diverse and consist of algaevores (algae-feeders), folivores (leaf-eaters), granivores (seed-eaters), frugivores (fruit-eaters), nectivores (nectar-feeders), sap-feeders (sucking liquid from phloem or xylem) and leaf–stem–root chewers (Figure 1). The mode of feeding under a resource-driven system makes herbivores and transhumance pastoralists vulnerable to the vagary of microbial aerosols and infections. The permeability of the nasal cavity makes it easy for particulate matter to be filtered and trapped in the cranial fossae of the nasal septum [21].
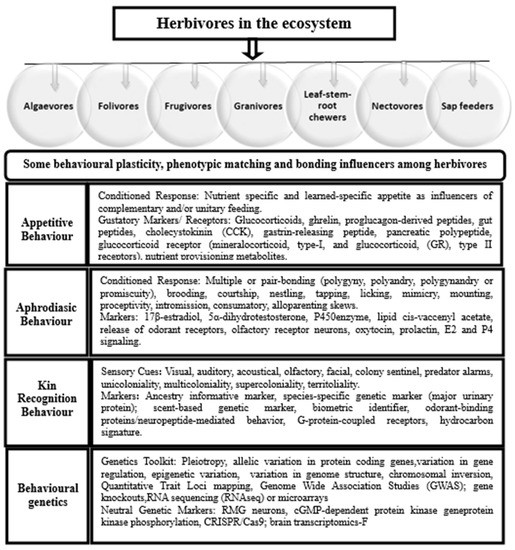
Figure 1.
Repertoire of polygenic markers influencing sociogenomics and behavioural stereotypes among vertebrates on pasture land.
By virtue of their adventurous habit, herbivores use visual and olfactory receptors with cognitive and well-coordinated instincts of sighting and selection to gather food for immediate consumption, storage or provisioning. Whilst strains of infectious diseases can be trapped by respiratory tracts, integumentary organs or natural orifices, the tonsillar crypt epithelium is probably the initial infection site for active viral replication. Certain viruses and toxins can be conveyed by axonal transport along peripheral nerves such as rabies virus (RABV), pseudorabies virus (PRV), alpha-herpesviruses (herpes simplex (HSV-1)), West Nile virus (WNV), poliovirus (PV) and Theiler’s encephalitis virus (TMEV). Exposing the conjunctiva, mucosae (nasal or larynx) and alveolar capillary network of livestock or humans to disease-carrying droplets or aerosols will affect the functionality of body systems. To ensure the welfare of free-grazing stocks and personnel in the agricultural sectors, omics-enabled tools can be optimised for sensing viral loads and make accurate theranostic predictions. Exposomics is considered a suitable option for determining the totality of nongenetic exposure dynamics and effect on system function, which is ideal for robust characterisation and validation. Thus, using robots or drones powered by biosensors and radio modem systems for viral tracking and evaluating the putative effect of exposure milieu on livestock and farmers might safeguard the livestock industry from the devastating effect of SARS-CoV-2 and its mutant variants.
1.2. Justification for the Review
Of pertinent concern is the severe impact of time-varying exposure of livestock (raised for milk, egg, meat, fur, hide and skin) and personnel involved in the food chain to the highly infectious SARS-CoV-2. The prevalence of RNA viruses in the environment is worrisome with 89% of viral species (160) being zoonotic and having a high case fatality rate [22,23]. CoVs are causative organisms of lethal zoonotic infections in avian, bovine, canine, feline, ovine and porcine species [24]. For instance, mammals and bats harbour α- and β-CoVs, mammals and avian species are often infected by γ-CoVs and birds and mammals are often infected by δ-CoVs [25]. Symptoms of severe acute respiratory syndrome (SARS) affirm that CoVs may quiescently surface from animal reservoirs and potentially elicit mild to severe respiratory disorder or septic shock in humans [24,25,26,27]. Episodic exposure during routine and periodic activities in the agro-environment can raise susceptibility to putative agents of disease infection. Unlike among humans, social distancing, face-masking and sneezing on elbow joints as means of self-protection are not practicable amidst free-ranging chickens, ruminants, bats, mink or pets due to the complexity of speciation, sociality rhythms and interconnectedness of stereotypic behavioural instincts (Figure 1). Nomadic farmers, who might not be able to afford mobile and suitable personal protection kits with adequate filtering efficiency to intercept viral particles on the farm, are also at risk. The spread of this virus in the agricultural sectors is worth serious consideration vis-a-vis the involvement of humans in food security and undue exposure to aerosols in the environment (Figure 2). Research on viral syndrome in agricultural sectors mostly adopts a reductionist approach by studying one or two stressors at a time under controlled laboratory conditions. The status quo is further worsened by the limitation of conventional approaches in producing a holistic approach of curtailing the problem of infectious diseases in the livestock industry. While it is ineffective to use a one-size-fits-all approach for addressing the multiplicity of factors linked with exposure dynamics in a lifetime, the cumulative effect of livestock and farmers’ exposure to coronavirus and its variants is life-threatening. Therefore, the aim of this review is to give insight into SARS-CoV-2 and propose the use of catch-all, smart-farming precision models and biosensor-powered digital technologies (robotic drones) for rapid-response and high-throughput screening of livestock, farmers, paddocks, poultry pens and agro-environment against coronavirus infection (Figure 3).
Figure 2.
Some spirometric indices affected by exposure to inhalable viral aerosols and microdroplets during routine or periodic activities. Key: TV: Amount of inhaled and exhaled air during a normal respiratory cycle in the lung. MRV: Amount of tidal air passing in and out of the lung per minute. ERV: Amount of air that can be expired beyond the tidal volume. RRV: Tidal volume + expiratory reserve volume; obtained by taking a deep breath (maximal breathing) and exhaling all the air from the lung or VC = by adding IRV + TV + ERV. FEVT: Timed vital capacity, which determines impairment of pulmonary function or mechanical condition of the lung, e.g., dyspnea (shortness of breath), asthma and emphysema. IC: Measures maximum voluntary ventilation or IC = IRV + TV or VC-ERV. IRV: Amount of gas that can be inhaled after normal inhalation, filling the lungs with tidal air. RV: Amount of air left after expiratory reserve volume that cannot be forcibly expelled. MIC: Measures glossopharyngeal breathing (air stacking) or maximum volume of gas that can be held within closed glottis.
Figure 3.
A compendium of environmental variables with potential to impact animal welfare and smart farming in the COVID-19 era. Livestock and other species serving as natural reservoirs for coronavirus strains (A); atmospheric aerosols (B); coronavirus strain (C); biosensors (D1,D2); robotic drone (E).
3. Viral and Host Receptor Interaction
Cellular receptors of human coronaviruses (HCoVs) belong to the membranous ectopeptidase family [4,22]. Receptors are macromolecules (glycoproteins) mediating biological processes to bind signaling molecules (ligands) intracellularly or extracellularly for initiating cellular responses. Intracellular receptors exist within the cytoplasm and bind to hydrophobic ligands roving across the plasma membrane. Cell-surface receptors (transmembrane receptors) are membrane-anchored proteins that bind to ligands on the surface of the cell to perform signal transduction functions [4,13]. Receptors have a transmembrane domain and an extracellular domain with a ligand binding (allosteric binding) site. The binding of receptors to ligands occurs at agonistic (ligand-binding) and antagonistic (ligand-blocking) sites. These include ligand-gated channels, G-protein-coupled receptors and enzyme-associated receptors. An enzyme-linked receptor is an integral membrane protein performing catalytic and receptor functions. Receptors undergo conformational changes to elicit responses by actively binding to or dissociating from ligands in cellular signaling or gated ion channels. Ligand-gated ion channels or ionotropic receptors are transmembrane ion-channel proteins that open to allow passage of Na+ K+, Ca2+ and/or Cl− ions through the membrane in response to the binding neurotransmitter. G-protein-coupled receptors such as 7T-transmembrane domain receptors, heptahelical receptors, serpentine receptors and G-protein-linked receptors detect molecules extracellularly and activate internal signal transduction pathways and cellular responses [44].
Viruses use multiple molecular species as receptors for infecting target cells. These receptors consist of sialic acid (C11H19NO9), glycosaminoglycans (negatively charged polysaccharides) and cellular adhesion molecules (intercellular adhesion molecules, vascular cell adhesion molecule-1), acetylcholine receptors, chimeric antigen receptors, interleukin-2 (IL-2) receptors, complementary receptors, growth factor receptors and neurotransmitter receptors [45,46,47]. Genome encoding occurs after viral invasion to elicit gene expression, accessory proteins and adaptation to the host. SL-CoV and SARS-CoV share similar genomic sequences and highly conserved gene products in the receptor-binding domain of the N-terminal region of S-proteins. A virus is unlikely to establish and replicate itself in the host without evasion or antagonism. As the mechanisms of attachment, intracellular trafficking, uptake and incursion into cytosol are affected by the bond between virus and receptor, the synergy between them even heightens the probability of infection occurring and activity of spike protein in the process. Whilst host cells exert defense mechanisms for rapid detection of infection or suppression of immune response, viruses have ways of evading them through pattern recognition receptors (PRRs). Upon infection, PRRs sense nucleic acids in the cytoplasm or nucleus to activate antiviral innate immunity. RNA viruses use non-primer-dependent mechanisms for replication, distortion of RNA recognition and antiviral signaling. SARS-CoV-2 exploits the angiotensin-converting enzyme-2 (ACE-2) and cellular protease TMPRSS2 receptors to penetrate the host through the ACE-2 pathway [46]. ACE-2 is a zinc metalloenzyme and transmembrane receptor expressed in the glandular cells of gastric, duodenal and rectal epithelium; pneumocytes of the lungs; enterocytes of the small intestine, heart, kidney, liver and testis; and vascular endothelium cells. ACE-2 serves as an entry point into cells for HCoV-NL63, SARS-CoV and SARS-CoV-2. It coincidentally acts as a receptor for severe acute respiratory syndrome. The density of ACE-2 in each tissue correlates with severity of disease [45,47].
3.1. Viral Tropism and Immunogenicity of Coronaviruses
By definition, viral tropism is the capacity of viral strains or isolates to cause infection in cells or tissues and induce syncytial formation as acute or chronic infections [48]. Acting as a macrophage (M), T cell or dual tropic, tropism is a major factor in pathogenesis and persistence of infection in the peripheral motor nerves and central nervous system. Viral tropism develops in stages in a receptor-dependent or receptor-independent manner. All the developmental stages culminate with the synthesis of progenies [46]. Replication of RNA viruses within the host cell depends on enzymes in the virion to synthesise its mRNA. Viral RNA also serves as its own mRNA. Most RNA viruses complete their replication in the cytoplasm, while others are transcribed in the nucleus. Several infectious pathogens go through causative and reactive pathways to reach their targets. As viral titres are highest at the early stage of infection, the viral kinetics varies between symptomatic, asymptomatic and paucisymptomatic among patients [49]; a virion with glycoprotein (GPX) enters the host and then targets the cell with GPX receptors and fuses with it to initiate reverse transcription and synthesis of viral proteins plus ribonucleic acid followed by an assemblage of viral particles to advance its tropism. Pathogenesis of coronaviruses conforms to either intestinal or respiratory infection model. Almost 80% of COVID-19 symptomatic patients develop mild conditions; 15% develop severe hypoxaemia, dyspnoea and tachypnoea conditions; and 5% become critically ill with septic shock and/or multiorgan dysfunction [50].
Another pertinent issue is the immunogenicity of coronaviruses. Coronaviruses encode accessory proteins likely involved in immune antagonism or pathogenesis. Immunogenicity is the capacity of foreign substances (antigens) to induce humoral or cell-mediated immune response in the host. Innate immune response acts as the first line of defence against pathogenic agents by detecting pathogen-associated molecular patterns [51]. Respiratory viruses thrive in a condition where the immune system is either depressed or underdeveloped. There is a wide variation in terms of immune response elicited by different viral strains. Whilst every cell has an intrinsic capacity to restrict viral infection, most respiratory viruses can suppress innate immune responses to efficiently replicate and induce it [42]. When the immune system is infected by pathogens, it concentrates in the epitopes, which allow host cells to differentiate between closely related foreign invaders. The early stage of innate response is critical in shaping the downregulation of the adaptive immune response. While some victims develop lifetime immunity against certain viral infections, others are extremely susceptible. For instance, the likelihood of reinfection of a person who has previously suffered from chicken pox is low, but reinfection with haemagglutinin-neuraminidase (H1N1) flu strain might occur after 10 years. Again, infants and adolescents who have recovered from mild COVID-19 infection soon become critically ill by reason of exaggerated immune response, known as multisystem inflammatory syndrome in children (MIS-C) [51,52,53].
As shown in Table 1 and Table 2, some nonhuman outbreaks such as bovine coronavirus (BCoV) from cattle, infectious bronchitis virus (IBV) from chickens, porcine respiratory coronavirus (PRCoV) plus porcine haemagglutinating encephalitis virus (PHEV) from pigs, feline coronavirus (FCoV) from cats, canine respiratory coronavirus (CRCoV) from carnivores and bat-SL-covzc45/bat-SL-covzxc21 from bats exhibit respiratory tropism with varying complications or outcomes [26,54]. The chain of infection across species suggests that foraging animals and nomadic farmers in an agrarian environment are vulnerable to COVID-19 infection. In response to viral infection, most cell types produce interferons for release into extracellular fluid in the host. Interferons (IFN) are naturally occurring glycoproteins from the helical cytokine family. IFNs have molecular weights of 16,776–22,093 dalton. Interferon is obtainable from leukocytes (IFN-α), fibroblasts (IFN-β) or lymphocytes (IFN-γ) of human cells. The IFN system is a major frontline defence against viral infection. IFNs are type I (IFNα and IFNβ) and type III (IFNλ) cytokines. They are secreted in response to viral infections and biological inductions. Early control of viral replication by type I interferons, complement proteins and innate immune mediators limits the spread of viruses in the early phase of infection. Expression of IFN genes occurs downstream of the double-stranded RNA-sensing by host RIG-I-like receptors for coronavirus infection.

Table 1.
Characterisation of genome and natural reservoirs of coronaviruses among avian species.

Table 2.
Characterisation of natural reservoirs of coronaviruses among selected mammalian species.
By interacting with their specific heterodimeric receptors on the cell surface, interferons initiate an array of signals that induce cellular antiviral activities, modulate inflammatory responses, inhibit or stimulate cell growth and apoptosis and modulate other components of immune system [52]. Stimulation of interferon (IFN)-dependent antiviral response at the early stage of infection is vital for monitoring immune response. Host sensing of viral double-stranded RNA triggers the activity of IFNs [55]. IFNs initiate innate immune responses by exerting direct antiviral effects on interferon-stimulated genes [56]. IFN invasive strategies involve avoidance, suppression of IFN induction and signaling. By avoidance, a virus masks itself or by-products from being recognised by the host sensors that activate the IFN system. Viruses can suppress IFN induction by inhibiting sensors in the host or downstream signaling molecules to prevent initiation. By suppressive means, gene products in the viral gene block signaling events or downregulation of type I IFN receptors to inhibit the activation of an antiviral state in the infected cell or enhancement of IFN response by activating late type I IFN genes [57].
Of five groups of immunoglobulins (IgG, IgA, IgM, IgD and IgE) with diverse amino acid sequences, antibodies exist in soluble and membrane-bound forms. Antibodies and antigens interact by spatial complementarity of lock-and-key mechanism. High specificity and affinity is the most striking feature of antigen–antibody interaction. Antibodies are secreted by β-cells in the adaptive immune system or plasma cells. Each antibody unit has a minimum of two antigen-binding sites bound bivalently or multivalently. Bonding of an antibody with an antigen forms an immune complex functioning as an entity. The immune system releases protein messengers for regulating the immune defence of host cells. The collective term for these messengers is cytokines. Cytokines are glycoproteins acting nonenzymatically on target cells of picomolar to nanomolar concentrations. The physiology of cytokines is complex and diverse. Cytokine secretion can stimulate the release of different cytokines to express the same functions [58]. It mediates the actions of interleukins, interferons and tumour necrosis and growth factors. The majority are secreted by more than one type of immune system, fibroblasts and endothelial cells. Cytokine interleukin-2 influences the function of virtually every cell in the immune system. Over-secretion of immune cells may trigger cytokine release syndrome or cytokine storm [59]. Under multiple respiratory viral infections, cytokine storm causes virus-induced tissue destruction, extreme inflammation and mortality. Several complications at the advanced stages of severe COVID-19 infection have been linked to cytokine release syndrome. Evidence has shown that the immune response of COVID-19 patients under critical condition may be as vicious as the virus responsible for the illness due to comorbidities and hyperinflammatory immune response [60].
Typically, the capacity of the body to either resist infection or be susceptible is influenced by a number of factors such as comorbidities, blood type, nutrient cycling and intactness of immune system coupled with a repertoire of polygenic makers influencing sociogenomics and behavioural stereotypes across species (Figure 1). Most of the elderly people that died of COVID-19 infection had underlying conditions and, presumably, shortened telomeres [61]. Rapid immune response occurring in the infected host during acute viral infection indicates the effect of telomere shortening on immune depression. Symptoms of telomere syndromes vary but depend on the patient’s telomere length. While some patients have few or no symptoms, others exhibit bone marrow failure; pulmonary fibrosis; liver cirrhosis; and gastrointestinal, skin and mucosal abnormalities. Again, it is not clear if the body of victims can synthesise inhibitory antibodies against viral epitopes and establish antigen-binding sites against SARS-CoV-2 molecules. As cases of reinfection with SARS-CoV-2 and its mutant variants are emerging, novel biosensors with robust sero-surveillance strategy can be exploited as a requisite standard for identifying group risks, estimating antibody levels and trends of population immunity and predicting response to outbreaks.
3.2. Evolution and Mutability of SARS-CoV-2
RNA viruses have short generation times and fast evolutionary rates. The fastest evolution corresponds to single-stranded RNA and reverse-transcribing viruses, followed by double-stranded RNA and single-stranded DNA viruses, whereas double-stranded DNA viruses evolve more slowly on average. Viral evolutions are mostly identified from open reading frames. A reading frame is a set of nonoverlapping triplets of three consecutive nucleotides. An open reading frame (ORF) is a section of the reading frame with no stop codon which is a sequence of three adjacent DNA or RNA nucleotides corresponding with amino acid during protein synthesis [62]. It should be noted that proteins cannot be synthesised if RNA transcription stops before reaching the stop codon. A gene encoding a protein has an ORF that can be translated into an amino acid sequence (AAS). To start with, the AAS or nucleotide sequence (NTS) must be obtained from the same DNA sequence. Since the AAS is the final product under evolutionary trend, multiple alignments of protein-coding genes use AAS during alignment with NTS. Coronaviruses contain specific genes in the ORF downstream regions that encode proteins for replication, nucleocapsid and spike formation. These viruses have overlapping ORFs (ORF1a, ORF1b) which are a continuous stretch of codons for gene prediction to identify regions of genomic DNA encoding protein, RNA genes and regulatory regions. The ORF7a gene creates an accessory protein for viral infection and replication in the host. If the domain of the ORF is known, the rates of mutation can be modified as wobble base pairing can permit higher mutation rates in the third nucleotide of a given codon without affecting the genetic code [63].
Genomes in RNA viruses have the highest mutation rates to circumvent the immune system of the hosts [64]. This is premised against the circulation of mutant spectra and replication by viral quasispecies. A minor alteration in NTS or AAS can annul the activity of a receptor due to errors during replication, deletion or insertion of DNA segments. The term mutation rate (MR) refers to the frequency of new variants created per site per genome replication or average number of errors produced in genomes of progeny per base per replication cycle (mut/nuc/rep) [63,64,65]. MR is affected by base composition, environment, size of gene and position in the genome. RNA viruses use encoded RNA-dependent RNA polymerase for genome replication to exploit mutation and escape antibody and cytotoxic T lymphocyte responses. The mutation rate is a critical parameter for understanding viral evolution [66]. Mutations might be induced via random errors during replication or genetic shuffling (recombination) among an infected population with weaker immunity. Mutation influencing spike protein alters amino acid (614) by substitution from aspartic acid (D) to glycine (G) at a genome domain encoding the spike protein (D614G). MR of RNA viruses occurs at rates of six orders of magnitude above that of the hosts. Mutability boosts the adaptation of RNA viruses to varied environments. MR can be calculated as follows:
where r1 = observed number of mutants at time point 1; r2 = observed number of mutants at the next time point; and N1 and N2 = numbers of cells at time points 1 and 2, respectively.
μ = [(r2/N2) − (r1/N1)] × ln (N2/N1) =(f1 − f2) × ln (N2/N1)],
4. Exposure Dynamics and Organ Functionality
Another caveat of importance is the effect of exposure to inhalable viral aerosols or microdroplets on lung capacity and functionality of body organs during routine or periodic activities on-farm and along the food production supply chain in the livestock industry. This scenario is presented in Figure 2. Aerosol originates from natural or anthropogenic sources and traverses the stratospheric and tropospheric layers of the atmosphere. Being a complex mixture of suspended particulate matter, aerosol varies in chemical composition, microphysical transformation, particle size (0.002–100 µm) and surface-to-volume ratio [67]. Natural aerosols consist of forest exudates, microbial spores, water droplets, geyser steam, sea salt, fog, mist, dust or pollen. Mining dust, vehicular smoke, ashes and smoke are typical aerosols from anthropogenic sources, with a litre of air containing nearly 109 particles [68]. Aerosols can spread from host cells to the adjoining environment through primary and secondary processes of aerosolisation. With varying aerodynamic diameters of >5–10 μm for respiratory droplets or <5 μm for nuclei droplets, aerosols can transmit infectious viruses via nasal cavity, oronasopharynx or mucus membranes. Although coalescence of aerosols promotes uniform (monodispersity) or nonuniform (polydispersity) size distribution, the in vivo deposition of aerosols is largely influenced by the morphology of the respiratory tract and the mode and pattern of respiration. Inhalable fractions of aerosol particles can pass through the tracheobronchial and nasopharyngeal airways with an effective diameter of ≤2.5 μm for permeation to the lungs and ≤10 μm for permeation to cilia before reaching the alveoli. Pulmonary diseases occur when air flow is obstructed due to infection, inflammation or mucus accumulation [69]. The combined effect of natural or anthropogenic activities presents a congenial environment for accumulating aerosols where livestock husbandry takes place.
Characteristically, coronaviruses can infect linings of the airways, throat, lungs and blood vessels to elicit cytocidal effects that culminate in the malfunctioning of multiple organs. The virus can stay infective in aerosols for 3 h and on surfaces for 72 h under laboratory conditions and 3–8 h postaerosolisation [70,71]. In the livestock industry, a seasonal spike in the transmission of single-stranded RNA viruses of animal origin from the Picornaviridae family occur in polygastric kraals, piggery units and poultry farms where virus-laden airborne particles are harboured in their natural reservoirs. With sketchy evidence that respiratory coronaviruses peak mostly in winter, there are still claims of their circulation year-round at lower frequency. Aerosols and infectious agents enhance the seasonal spread of human strains of coronaviruses (NL63, OC43, 229E or HKU1) causing common cold in immunocompetent persons [71,72]. Ambient temperature, relative humidity, pH and ultraviolet radiation affect their viability in aerosols and droplets. While most viruses survive best under low (≤40%) and high (≥90%) relative humidity, coronaviruses survive for shorter periods under higher temperatures and relative humidity than in cooler or dryer environments [72]. High concentrations of ultrafine aerosol particles (<100 nm) promote viral transmission; comorbidities; and unwanted clinical outcomes in form of acute myocardial injury, cardiovascular damage, low lymphocytes and abnormal clotting and renal function due to complications at early and advanced stages of COVID-19 infection [71,72]. Although case studies are scare, there is a high probability of livestock and nomadic farmers contracting the virus due to routine exposure to infected aerosols, droplets or fomites on pasturelands which can distort the mechanical properties of the pulmonary system by permeating the epithelia layer and replicating the genome to foster viral viability and infectivity. So, using appropriate farming models and biocompatible sensors for screening livestock farms and agro-processing units will boost productivity, ensure biosecurity measures and save the livestock industry from serious fatality.
4.1. Blood Types and Susceptibility to Coronavirus Variants
With high transmissibility from natural reservoirs or fomites eliciting tissue tropism in permissive hosts, the burgeoning spike in the first, second and third waves of coronavirus indicates that herd immunity is not yet attained in many countries and humans and livestock are still vulnerable to the infection. The unevenness in the pathogenicity index of the virus implies that some groups are more susceptible than others due to complex interaction with innate or environmental factors influencing viral susceptibility. A number of anthropogenic activities including exposure to infected pasture, feeds, water or fomites during livestock grazing, wildlife poaching and preslaughter handling are transmission routes to permissive hosts. Forni et al. [73] posits that wildlife and domestic animals are intermediate hosts for human coronaviruses such as severe acute respiratory syndrome (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), human coronavirus NL63 (HCoV-NL63) and human coronavirus 229E (HCoV-229E).
RNA viruses from animal origins are known to mutate more rapidly at the highest frequency. New cases of mutated SARS-CoV-2 variants have been identified based on isolates from newly sequenced viral genomes from COVID-19 patients. While some of the mutations are neutral, others are more contagious and of strong biological significance in the receptor-binding domain (RBD). These variants include H69/V70, 501.V2, P681H, E484K and D614G (Table 3). The H69/V70 or SARS-CoV-2 VUI 202012/01 is the “Variant Under Investigation, year 2020, month 12, variant 01”. It is otherwise referred to as S: N501Y to prove that it is in the spike glycoprotein of SARS-CoV-2 strains. The 501.V2 variant was isolated in South Africa while tracking the phylogenetics of the SARS-CoV-2 as one that alters the receptor-binding domain of the virus and carries other mutations, including a double deletion (positions 69 and 70). The D614G variant emerges from an ancestral D residue found in the glycosylated region of the viral spike protein or RBD of the spike protein of SARS-CoV-2. These strains are defined by multiple spike protein mutations (deletion 69–70, deletion 144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H) with an estimated potential to increase the reproductive number (R) to ≥ 0.4 and transmissibility to the high level of 70% (ECDC/WHO, 2020). Activities of these variants are attributed to the presence of 14 mutations leading to three deletions and alteration of amino acid sequence or RNA viral pathogenesis. Phylogenetic clustering of mutated strains further shows a degree of variation by 29 nucleotide substitutions from the first wave of viral strain from Wuhan city in China [74]. In comparison with the strain that constituted the first wave from Wuhan city, the mutated variants have higher viral load, transmissibility of 70% and high receptor binding capacity to evade host immune response by altering the surface structures recognised by antibodies from SARS-CoV-2.

Table 3.
Nomenclature and description of mutated strains of COVID-19 identified and reported in 2020.
Globally, coronavirus invasion and lethality are causing huge economic losses in the livestock and meat industries, with emergency mass culling and excess mortalities of meat species and fur producing mink in North America, Nordic countries and regions seriously hit by the virus. Susceptibility to infectious diseases has been linked with the ABO blood phenotypes [83]. The ABO blood group, in particular, plays a functional role in viral infections. Intriguingly, patients with non-O blood group have a higher risk for COVID-19 infection when compared to O blood groups, and the ABO blood group can influence coagulation factor VIII (FVIII) [84,85]. Blood type consists of antigenic factors of the red blood cells which represent inherited polymorphic traits among individuals. The allelic products of a given genetic locus are the blood group system. In other words, blood types are inherited traits of the red blood cells (RBCs) termed positive (A+) or negative (A−) and determined by polymorphic and antigenic components of the RBC membrane [86]. The presence or absence of species-specific antigenic molecules on the surface of red blood cells (erythrocytes) determines the blood types. A, B, C, F, G, H, I, K, L and M are symbols used for designation. When antigens exist as protein molecules or oligosaccharide sugars above the cell surface, they induce immune-mediated reactions. While no disease results from lack of expression of ABO blood group antigens, a number of illnesses may change an individual’s ABO phenotype. Animal erythrocytes have cell surface antigens that go through a polymorphic process and give rise to blood types. The systems are identified by a letter: A, B, C, D, E, H, I, J, K, L, N, P and R. For instance, bovine species such as cattle have 11 major blood group systems: A, B, C, F, J, L, M, R, S, T and Z. Avian species such as domestic chickens have 13 alloantigen systems in the nucleated red blood cells and other cell types. In the context of the coronavirus pandemic, sampling or validating blood types of various avian and mammalian species, sequencing antigenic factors and correlating such findings with herbivory and heritable susceptibility to COVID-19 infection is a knowledge gap that has not been addressed to confirm susceptibility indices of various livestock breeds exposed to different environmental dynamics. Again, apart from omics tools for syndromic detection, there is no evidence of multiple pleiotropic expressions, multiplicity of infection, autoimmune mechanisms and blood typing markers coding for susceptibility to SARS-CoV-2 or its mutated strains across avian, bovine, ovine, porcine or semiaquatic breeds and neonates. While seeking answers to these issues, using a triage colour-coding system and modifying livestock stocking density (which is a way of mimicking social distancing in humans) for contact tracing of confirmed vs. probable cases among livestock looks promising for determining positivity index in paddocks or poultry pens to raise biosecurity and safeguard against viral outbreaks on livestock farms (Figure 3 and Figure 4).
Figure 4.
Triage coding and syndromic surveillance mapping of livestock susceptibility to coronavirus in feedlots and agro-processing units.
4.2. Herd Immunity Versus Eco-Exposure to Viral Infections
Since December 2019 when the outbreak of COVID-19 was first reported in China, the viral spread has not ceased to be an endemic despite the promotion of personal protection equipment, self-isolation, social distancing, vaccination and other health surveillance strategies. Exposure to infections often elicits debilitating effects that impact general welfare vis-a-vis a range of complex factors including genetic heritability, exposomics and phylodynamics. Varying exposure levels to ambient conditions influencing viral infectivity across species and mechanisms of action make it is tricky to identify disease reservoirs or fomites where COVID-19 virions can be spotted. It is even a Herculean task to estimate the totality of dietary and pathogenic exposures for free-range herbivores, by reason of variable dietary preferences and underlying health conditions. To date, breaking the transmission chain or obtaining relative protection of population groups from infectious agents is still a mirage as the transmission of SARS-CoV-2 and its mutated strains are still traversing international boundaries with cases of corona-storms and reinfection emerging. The overstretching of primary healthcare facilities, disruption of normality in society and high mortality rate on a scale that has never been witnessed make SARS-CoV-2 a deadly virus against humanity. Ostensibly, the herd immunity threshold has not been reached for SARS-CoV-2 and its mutant strains in many countries. This natural phenomenon occurs when a large proportion of the pathogen-dependent population becomes immune to infection naturally or by vaccination. The absence of exponential growth or decline in additional infections describes this threshold point or endemic steady state in relation to antigenic drift or reassortment of separate viral genome segments for initiating changes in the viral epitope [87]. While disease eradication may not happen instantly, an overshoot can still occur. Assuming each person in a homogeneous population is susceptible to endemic infections, herd immunity threshold will be determined from effective reproduction number (Rn) and expressed as RoS = 1, where Ro is “basic reproduction number of infection” and S is “truly susceptible population or measure of contagiousness”. Further, S = (1 − p), where p is the proportion of the immune population. Paul et al. [88] posit that if S equals reciprocal of Ro (i.e., 1/Ro), the average number of transmissions per case (Rn) will be 1 and remain constant with time (t).
4.3. Biosensing and Smart Precision Farming Models in COVID-19 Era
The conventional systems of arable farming and livestock husbandry still dominate the current food system [89]. Of the 71% habitable land area, approximately 80% (40 million km2) of terrestrial space used for animal agriculture encompasses different forms of livestock ranching, stock keeping and pastoralism. In the context of highly polarised views of animal products among vegans, vegetarians and meat consumers, the livestock industry remains one of the main drivers of land-use dynamics, contributing to 25–50% of agricultural gross domestic product. As the livestock density increases and becomes closely intertwined with wildlife and humans, there is a growing concern about outbreaks of infectious diseases, with 66% of the annual emerging ones being traceable to animal origins; such outbreaks carry the potential of decreasing the production efficiency of livestock by almost 33%. Genomic evidence for SARS-CoV-2 suggests that it has a zoonotic source [32,90,91]. To proactively transform the livestock industry into a sustainable sector that conforms with the new-normal realities in the COVID-19 era, integrating precision smart livestock farming requires reliable networks of digital technologies and smart sensors for real-time tracking of infectious diseases and animal welfare (Table 4). The livestock industry is under immense pressure to curtail food insecurity by producing more by-products such as eggs, meat, milk, wool and fur using precision agriculture, digital technologies and high-value nutrient sources. With artificial intelligence, computer-based algorithms, cloud computing and big data analytics, digital animal monitoring devices such as wearables are gaining ground in cattle monitoring, enabling real-time information on individual cows.

Table 4.
Selected viral genomes, serotypes, receptors and biosensors with applications in virology.
Whilst the rates of infection and mortality rates continue to soar with no permanent solution to hasten herd immunity or eradicate the ill-fated coronavirus and its mutated variants, the use of robotic drones powered with multianalyte biosensors is proposed for eco-sensing and tracking of the natural reservoir of viral infections in livestock farms and agro-processing units (Figure 4). Biosensors are analytical devices with receptors, transducers and detectors of measurable signals [92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111]. These devices allow multitarget analyses and automation of analytes under in vivo or in vitro systems with the aid of receptors (antibodies, whole cells, enzymes, nucleic acids or aptamers); transducers (semiconductor or nanomaterial); and signal amplifiers, processors or display units for detection. The use of biosensors makes it easy to measure antigen–antibody interactions momentarily and allow accurate determination of binding kinetics and affinity constants. They have multiple applications for detecting compounds by electrical, thermal or optical signals in food, water and the environment [100,103]. Biosensors can be designed for real-time monitoring of multiple parameters at each production step or multiple time points during the automation of industrial facilities (Table 3). As a miniaturised device, a biomedical micro-electromechanical system (Bio-MEMS), synonymous with lab-on-a-chip and micro total analysis system (μTAS), is suitable for biological applications of molecular diagnostics, point-of-care diagnostics, tissue engineering, single-cell analysis and implantable microdevices. Agro-compatible, scalable, sensitive, specific, miniaturised, affordable and user-friendly biosensors can potentially boost biosecurity measures in the livestock industry and food processing units. Identification of target analytes following selection of suitable bioreceptors; transducer design that translates binding reaction into measurable signal; miniaturisation for integration into lab-on-chip tools, automation for real-time analysis, on-site testing or point-of-care detection; enhancement of sensitivity and linearity and minimisation of interference; and configuration for efficient real-time sensing are the main strengths of biosensors [100,101,102,103,104,105,106,107,108,109]. In addition to the use of biosensors for eco-sensing, their use can be extended for tracking fomites or reservoirs of infections and multiparametric measures for determining the welfare of livestock in the agro-environment (Table 5).

Table 5.
Multiparametric measures for sensing lethality of coronavirus from putative organs, aerosols and animal by-products.
As the security of animal products largely depends on eco-exposure to several variables where open grazing occurs, the choice of biosensor is based on its comparative advantage over conventional diagnostic assays for detecting nanosized virions on-farm and along food production supply chains. Precision livestock farming (PLF) is one of the novel technologies with broad spectra of applications. Agriculturists in developed nations adopt PLF principles to provide accurate information for daily operations, real-time monitoring of animal welfare and fast interventions for optimising farm management systems. Different biosensors are used in PLF in the form of eating/grazing sensors for eating, ruminating, resting and active behaviours (Agis Automatisering SensOor); neck-mounted sensors for monitoring grazing and animal position; eating sensor validation; collar-mounted sensors; and rumen sensors. Within livestock sectors, the PLF is smartly designed to monitor microclimate or agro-environment and routinely record feed utilisation, flock performance and general welfare. Landscape tracking of livestock and occupancy-based monitoring among other livestock precision farming smart technologies are advantageous in the livestock industry for improving farm labour efficiency, record-keeping and real-time decision making about routine and periodic activities. Other things to sense include the livestock storage tanks, silos, paddocks, poultry pens, milking parlours, feeds, chewing period, feeding equipment, milking equipment, feeding behaviour, water, critical transition periods (pregnancy, gestation, lactation cycle, incubation period), body temperature, rumination, pH, methane emission, spirometry indices, oestrus cycles and carbon footprints. Innovative wearable sensors can be developed to monitor animal welfare, detect the presence of infectious disease and monitor physiological parameters to allow rapid confirmatory diagnosis of suspected cases. The equations below in Figure 5 can be applied in integrated monitoring of exposure dynamics and transmission of SARS-CoV-2 variants.
with the initial condition (S(0), E(0), I(0), Q(0), R(0)) = (S0, E0, I0, Q0, R0).
N(total population) = S (t) + E (t) + I (t) + Q (t) + R (t)
Figure 5.
On-farm compartmental model for exposure dynamics and transmission of SARS-CoV-2 variants.
As shown above (Figure 5), the modified SEIR compartmental mathematical model, which we could call the SEIQR model for exposure dynamics and transmission of SARS-CoV-2 variants, implies that any animal in a population belongs to one of five states: susceptible (S), exposed (E), infectious (I), quarantine (Q) and recovered (R). The animals in the susceptible states are assumed to be healthy but have no immunity against the virus, and thus they can contract the virus at any time. The animals in the exposed state are vulnerable to the virus. The infected ones can harbour and transmit the virus to any organism they come in contact with. Any animal that is infected either recovers naturally from the disease or will be moved to the quarantine state. Some animals can also recover from the quarantine state. It is assumed that a recovered animal has a low (but not zero) chance of reinfection. The susceptible animals move to the exposed compartment (𝐸) at a rate 𝛽1 (𝐼/𝑁). Animals in the exposed compartment move to the infectious compartment (𝐼) at a rate 𝛽2. Natural death occurs at a rate 𝜇 in all the five compartments. The rate of death due to COVID-19 is α. The rate of recovery (R) for infectious animals is 𝛽3. The rates at which the infectious animals enter the quarantine compartment (Q) and return to the infectious compartment are 𝛽4 and 𝛽5, respectively. The compartmental model culminates in a five-dimensional system of ordinary differential equations (ODEs) as seen in Equations (2)–(6). Figure 6 shows the drone powered by biosensors and radio modem system to provide daily data on animal welfare including the Ultra high frequency (UHF) receiver that uses radio waves, the robot which contains an accelerometer sensor and GPS system network. This is an automatic system that responds to ambient temperature fluctuations and triggers at a threshold with an accurate precision of location of grazing livestock and time. The cloud provides a petabyte of data processing and storage system. The output is decision-based information that is available in real time. The entire system can be effectively utilized for biosensing the outbreak of infectious diseases in livestock industry.
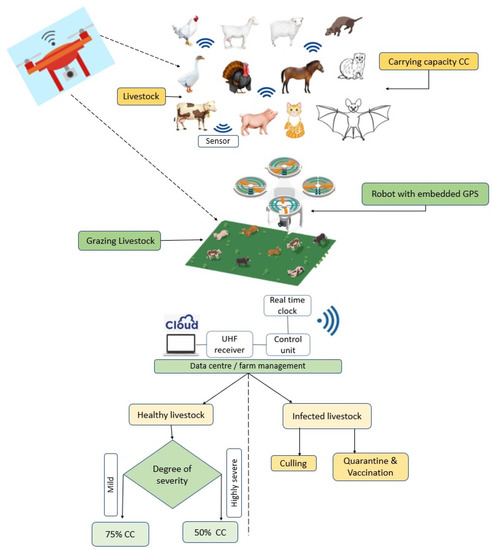
Figure 6.
Robot-embedded model with drone powered by biosensors and radio modem system to provide daily data on animal welfare.
5. Conclusions
The outbreak of SARS-CoV-2 adds to the scale of disease burden and challenges the livestock industry is facing vis-a-vis the menace of infectious diseases. Due to recurrent surges in viral waves across agro-ecological zones, it is uncertain what the devastating effects of these infections will be in the nearest future. Another worrisome issue for the livestock sector is the potential of a wide range of mammalian and avian species being natural reservoirs for different strains of infectious zoonotic diseases such as coronavirus with its highly transmissible variants. Knowing that herd immunity is not yet attained in many climes and that livelihood partly depends on food security from animal products, indiscriminate exposure of farmers and livestock during routine and periodic activities to environmental variables harbouring coronaviruses may pose a serious threat to public health and food production. Thus, neglecting the impacts of viral aerosols on agrarian communities will unavoidably have devastating effects on animal welfare and food security. Therefore, effective use of smart precision livestock models, biosensor-powered digital technology and Internet of Things (IoT) may enhance catch-all and efficient monoplex or multiplex platforms for rapid-response and high-throughput screening of livestock and the agro-environment to limit transmissibility of zoonotic infections along food chains. A combination of digitalised technologies can be introduced into communal farming systems and rebranded with a modernised sense of “cooperative smart livestock farming” to help the resource-poor farmers mitigate transitioning into the new-normal realities in the livestock industry for realising sustainable and high production efficiency of animal products.
Author Contributions
Conceptualisation, P.O.F. and O.E.F.; writing—original draft preparation, P.O.F. and O.E.F.; writing—review and editing, P.O.F.; visualisation, P.O.F., O.E.F., L.O.J. and M.G.O.; supervision, P.O.F. and O.E.F.; project administration, P.O.F., O.E.F., L.O.J. and M.G.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
Not Applicable.
Acknowledgments
Authors are grateful to the North-West University, MaSIM and the National Research Foundation of South Africa (Thutuka) for financial support with grant holder UID 117009.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hui, D.S.; IAzhar, E.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McIntosh, A.; Bachmann, M.; Siedner, M.J.; Gareta, D.; Seeley, J.; Herbst, K. Effect of COVID-19 lockdown on hospital admissions and mortality in rural KwaZulu-Natal, South Africa: Interrupted time series analysis. BMJ Open 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Di Gennaro, F.; Pizzol, D.; Marotta, C.; Antunes, M.; Racalbuto, V.; Veronese, N.; Smith, L. Coronavirus Diseases (COVID-19) Current Status and Future Perspectives: A Narrative Review. Int. J. Environ. Res. Public Health 2020, 17, 1-11. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.R.; Cao, Q.D.; Hong, Z.S.; Tan, Y.Y.; Chen, S.D.; Jin, H.J.; Tan, K.S.; Wang, D.Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—An update on the status. Mil. Med Res. 2020, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019-novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zheng, Y.; Gou, X.; Pu, K.; Chen, Z.; Guo, Q.; Ji, R.; Wang, H.; Wang, Y.; Zhou, Y. Prevalence of comorbidities in the novel Wuhan coronavirus (COVID-19) infection: A systematic review and meta-analysis. Int. J. Infect. Dis. 2020, 94, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Worldometers, 2021. COVID-19 Coronavirus Pandemic. Available online: worldometers.info/coronavirus/?zarsrc=130 (accessed on 21 August 2021).
- Jin, Y.; Yang, H.; Ji, W.; Wu, W.; Chen, S.; Zhang, W.; Duan, G. Virology, Epidemiology, Pathogenesis, and Control of COVID-19. Viruses 2021, 12, 1-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahin, A.R.; Erdogan, A.; Agaoglu, P.M.; Dineri, Y.; Cakirci, A.Y.; Senel, M.E.; Okyay, R.A.; Tasdogan, A.M. Novel Coronavirus (COVID-19) Outbreak: A review of the current literature. EJMO 2020, 4, 1–7. [Google Scholar]
- Peiris, J.S.M.; Chu, C.M.; Cheng, V.C.C.; Chan, K.S.; Hung, I.F.N.; Poon, L.M.; Law, K.I.; Tang, B.S.F.; Hon TY, W.; Chan, C.S.; et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: A prospective study. Lancet 2003, 361, 1767–1772. [Google Scholar] [CrossRef] [Green Version]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- World Health Organization. WHO Statement Regarding Cluster of Pneumonia Cases in Wuhan, China Geneva (updated 9–14 January 2020). Available online: https://www.who.int/china/news/detail/09-01-2020-who-statement-regarding-cluster-of-pneumoniacases-in-wuhan-china (accessed on 21 August 2021).
- Burrell, C.J.; Howard, C.R.; Murphy, F.A. Coronaviruses. In Fenner and White’s Medical Virology; Elsevier: Philadelphia, PA, USA, 2017; pp. 437–446. [Google Scholar]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Beijerinck, M.W. Concerning a contagium vivum fluidum as cause of the spot disease of tobacco leaves. American Phytopathological Society, Johnson, J., Ed. Classics 1898, 7, 33–52. [Google Scholar]
- Takahashi, H.; Fukuhara, T.; Kitazawa, H.; Kormelink, R. Virus latency and the impact on plants. Front. Microbiol. 2019, 10, 1–18. [Google Scholar] [CrossRef]
- Zhang, Y.-Z.; Shi, M.; Holmes, E.C. Using metagenomics to characterize an expanding virosphere. Cell 2018, 172, 1168–1172. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.A.A. (Ed.) Defining Principles; Sustainability Science and Engineerings Series; Elsevier: Amsterdam, The Netherlands, 2006; Volume 1, pp. 513–518. [Google Scholar]
- Schowalter, T.; Pandeya, M.; Presley, S.J.; Willig, M.R.; Zimmerman, J.K. Arthropods are not declining but are responsive to disturbance in the Luquillo Experimental Forest, Puerto Rico. Proc. Nat. Acad. Sci. USA 2021, 118, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hefner, J.T.; Linde, K.C. Nasal Aperture shape: Functional Morphology. In Atlas of Human Cranial Macromorphoscopic Traits, 1st ed.; Elsevier: Philadelphia, PA, USA, 2018; pp. 73–88. [Google Scholar]
- Woolhouse, M.E.J.; Adair, K.; Brierley, L. RNA viruses: A case study of the biology of emerging infectious diseases. Microbiol. Spectr. 2013, 1, 1–16. [Google Scholar] [CrossRef]
- Ashour, H.M.; Elkhatib, W.F.; Rahman, M.M.; Elshabrawy, H.A. Insights into the Recent 2019 Novel Coronavirus (SARS-CoV-2) in Light of Past Human Coronavirus Outbreaks. Pathogens 2020, 9, 1-15. [Google Scholar] [CrossRef] [Green Version]
- Al-Ahmadi, A.; Alahmadi, M.; Al-Zahrani, A. Spatial association between primary Middle East respiratory syndrome coronavirus infection and exposure to dromedary camels in Saudi Arabia Zoo. Public Health 2020, 67, 382–390. [Google Scholar]
- Cossart, P.; Helenius, A. Endocytosis of viruses and bacteria. Cold Spring Habor Perspect. Biol. 2014, 4, 1–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maclachlan, N.J.; Edward, J.; Dubovi, E.J. Coronaviridae. In Fenner’s Veterinary Virology, 5h ed.; Elsevier: London, UK, 2017; pp. 435–461. [Google Scholar]
- Almendros, A. Can companion animals become infected with Covid-19? Vet. Record 2020, 186, 388–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.I.; Kim, S.G.; Kim, S.M.; Kim, E.H.; Park, S.J.; Yu, K.M.; Chang, J.H.; Kim, E.J.; Lee, S.; Casel, M.A.B.; et al. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 2020, 27, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Principi, N. School Closure during the Coronavirus Disease 2019 (COVID-19) Pandemic: An Effective Intervention at the Global Level? JAMA Pediatr. 2021, 174, 921–922. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult in patients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Wei, M.; Yuan, J.; Liu, Y.; Fu, T.; Yu, X.; Zhang, Z.J. Novel Coronavirus Infection in Hospitalized Infants Under 1 Year of Age in China. JAMA 2020, 323, 1313–1314. [Google Scholar] [CrossRef]
- Desforges, M.; Coupanec, A.L.; Dubeau, P.; Bourgouin, A.; Lajoie, L.; Dube, M.; Talbot, P.J. Human. Coronaviruses opportunistic pathogens of the central nervous system? Viruses 2019, 12, 1-28. [Google Scholar] [CrossRef] [Green Version]
- Chow, E.J.; Schwartz, N.G.; Tobolowsky, F.A.; Zacks, R.L.; Huntington-Frazier, M.; Reddy, S.C.; Rao, A.K. Symptom screening at illness onset of health care personnel with SARS-CoV-2 Infection in King County, Washington. JAMA 2020, 323, 2087–2089. [Google Scholar] [CrossRef]
- Ogoina, D. Fever, fever patterns and diseases called ‘fever’—A review. J. Infec. Public Health 2011, 4, 108–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartung, T. Pyrogen testing revisited on occasion of the 25th anniversary of the whole blood monocyte activation test. ALTEX 2021, 38, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Pranitha, N.; Sthira, S.S.; Mohanan, P.V. Pyrogens, a polypeptide produces fever by metabolic changes in hypothalamus: Mechanisms and detections. Immunol. Lett. 2018, 204, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020, 92, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, Y.; Helenius, A. Virus entry at a glance. J. Cell Sci. 2013, 126, 1289–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.; She, G.; Wu, T.; Xue, C.; Cao, Y. PEDV enters cells through clathrin, caveolae and lipid raft-mediated endocytosis and trafcs via the endo-lysosome pathway. Vet. Res. 2020, 51, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Carrasco-Hernandez, R.; Jácome, R.; LópezVidal, Y.; Ponce de León, S. Are RNA viruses candidate agents for the Next Global Pandemic? A Review. ILAR J. 2017, 58, 343–358. [Google Scholar] [CrossRef] [Green Version]
- Beachboard, D.C.; Horner, S.M. Innate immune evasion strategies of DNA and RNA viruses. Curr. Opin. Microbiol. 2016, 32, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Han, S.; Wang, L.F.; Shi, Z. Immunogenicity difference between the SARS coronavirus and the bat SARS-like coronavirus spike(S) proteins. Biochem. Biophy. Res. Com. 2009, 387, 326–329. [Google Scholar] [CrossRef] [PubMed]
- Baig, A.M.; Khaleeq, A.; Ali, U.; Syeda, H. Evidence of the COVID-19 virus targeting the CNS: Tissue distribution, host-virus interaction and proposed neurotropic mechanisms. ACS Chem. Neurosci. 2020, 11, 995–998. [Google Scholar] [CrossRef] [Green Version]
- Hamming, I.; Timens, W.; Bulthuis, M.L.; Lely, A.T.; Navis, G.; van Goor, H. Tissue distribution of ACE-2 protein, the functional receptor for SARS coronavirus: A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade long structural studies of SARS. J. Virol. 2020, 94, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Zhong, L.; Deng, J.; Peng, J.; Dan, H.; Zeng, X.; Li, T.; Chen, Q. High expression of ACE-2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020, 12, 1–5. [Google Scholar] [CrossRef]
- Nomaguchi, M.; Fujita, M.; Miyazaki YAdachi, A. Viral tropism. Front. Microbiol. 2012, 3, 281. [Google Scholar] [CrossRef] [Green Version]
- Lescure, F.-X.; Bouadma, L.; Nguyen, D.; Parisey, M.; Wicky, P.-H.; Behillil, S.; Gaymard, A.; Bouscambert-Duchamp, M.; Donati, F.; Le Hingrat, Q.; et al. Clinical and virological data of the first cases of COVID-19 in Europe: A case series. Lancet Infect. Dis. 2020, 20, 697–706. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; McGoogan, J.M. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) outbreak in China: Summary of a report of 72314 cases from the Chinese Centre for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef]
- Abdul Amir, A.S.; Hafidh, R.R. The possible immunological pathways for the variable immunopathogenesis of COVID-19 Infections among healthy adults, elderly and children. Electron. J. Gen. Med. 2020, 17, 1–4. [Google Scholar]
- Katze, M.G.; He, Y.; Gale, M. Viruses and interferon: A fight for supremacy. Nat. Rev. Immunol. 2002, 2, 675–687. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, Q.; Chen, J.; Xiang, R.; Song, H.; Shu, S.; Chen, L.; Liang, L.; Zhou, J.; You, L.; et al. Detection of Covid-19 in children in early January 2020 in Wuhan, China. N. Engl. J. Med. 2020, 382, 1370–1371. [Google Scholar] [CrossRef] [PubMed]
- Saif, L.J. Animal coronaviruses: What can they teach us about the severe acute respiratory syndrome? Rev. Sci. Technol. 2004, 23, 643–660. [Google Scholar] [CrossRef]
- Goubau, D.; Deddouche, S.; Reis e Sousa, C. Cytosolic sensing of viruses. Immunity 2013, 38, 855–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maverakis, E.; Kim, K.; Shimoda, M.; Gershwin, M.E.; Patel, F.; Wilken, R.; Raychaudhuri, S.; Ruhaak, L.R.; Lebrilla, C.B. Glycans in the immune system and altered glycan theory of autoimmunity: A critical review. J. Autoimmun. 2015, 57, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kumagai, I.; Tsumoto, K. Antigen-Antibody binding. Encycl. Life Sci. 2016, 1–8. [Google Scholar] [CrossRef]
- Teijaro, J.R.; Walsh, K.B.; Rice, S.; Rosen, H.; Oldstone, M.B.A. Mapping the innate signaling cascade essential for cytokine storm during influenza virus infection. Proc. Natl. Acad. Sci. USA 2014, 111, 3799–3804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.J.; Su, I.J.; Theron, M.; Wu, Y.C.; Lai, S.K.; Liu, C.C.; Lei, H.Y. An interferon-gamma-related cytokine storm in SARS patients. J. Med. Virol. 2005, 75, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Konig, M.F.; Powell, M.A.; Staedtke, V.; Bai, R.-Y.; Thomas, D.L.; Fischer, N.M.; Huq, S.; Khalafallah, A.M.; Koenecke, A.; Xiong, R.; et al. Preventing cytokine storm syndrome in COVID-19 using α-1 adrenergic receptor antagonists. J. Clin. Investig. 2020, 130, 3345–3347. [Google Scholar] [CrossRef]
- Effros, R.B. Telomerase induction in T cells: A cure for aging and disease? Exp. Geront. 2007, 42, 416–420. [Google Scholar] [CrossRef] [Green Version]
- Hung, C.L.; Lin, C.Y. Open reading frame phylogenetic analysis on the cloud. Int. J. Genom. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Rodgers, K.; McVey, M. Error-prone repair of DNA-double-strand breaks. J. Cell. Physiol. 2016, 231, 15–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanjuan, R.; Nebot, M.R.; Chirico, N.; Mansky, L.M.; Belshaw, R. Viral mutation rates. J. Virol. 2010, 84, 9733–9748. [Google Scholar] [CrossRef] [Green Version]
- Stern, A.; Andino, R. Viral evolution: It is all about mutations. In Viral Pathogenesis: From Basics to System Biology, 3rd ed; Elsevier: Amsterdam, The Netherlands, 2016; pp. 233–240. [Google Scholar]
- Duffy, S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018, 16, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrault, D.; Moineau, S.; Duchaine, C. Methods for sampling of airborne viruses. Microbiol. Mol. Biol. Rev. 2008, 72, 413–444. [Google Scholar]
- Aliabadi, A.A.; Rogak, S.N.; Bartlett, K.H.; Green, S.I. Preventing airborne disease transmission: Review of methods for ventilation design in health care facilities. Adv. Prev. Med. 2011, 2011, 124064. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical characteristics of 138 hospitalized patients with 2019 Novel Coronavirus-infected pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Leung, N.H.; Chu, D.K.; Shiu, E.Y.; Chan, K.H.; McDevitt, J.J.; Hau, B.J.; Yen, H.L.; Li, Y.; Ip, D.K.; Peiris, J.M.; et al. Respiratory virus shedding in exhaled breath and efficacy of face masks. Nat. Med. 2020, 26, 676–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaman, J.; Kohn, M. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc. Natl. Acad. Sci. USA 2009, 106, 3243–3248. [Google Scholar] [CrossRef] [Green Version]
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017, 25, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.; Lau, E.; Wong, J.Y.; et al. Early Transmission dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Fan, Q.; Zhang, W.; Li, B.; Li, D.J.; Zhang, J.; Zhao, F. Association Between ABO Blood Group System and COVID-19 Susceptibility in Wuhan. Front. Cell. Inf. Microbiol. 2020, 10, 1–7. [Google Scholar] [CrossRef]
- Rowe, E.; Dawkins, M.S.; Gebhardt-Henrich, S.G. A Systematic Review of Precision Livestock Farming in the Poultry Sector: Is Technology Focused on Improving Bird Welfare? Animals 2019, 9, 1-18. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [Google Scholar] [CrossRef] [Green Version]
- Del Valle, D.M.; Schulze, S.K.; Huang, H.H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef]
- Kikkert, M. Innate Immune Evasion by Human Respiratory RNA Viruses. J. Innate Immun. 2020, 12, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Yu, H.; Yang, H.; Xue, F.; Wu, Z.; Shen, W.; Li, J.; Zhou, Z.; Ding, Y.; Zhao, Q.; et al. Structures of two coronavirus main proteases: Implications for substrate binding and antiviral drug design. J. Virol. 2008, 82, 2515–2527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, K.V. SARS coronavirus: A new challenge for prevention and therapy. J. Clin. Investig. 2009, 111, 1605–1609. [Google Scholar] [CrossRef] [PubMed]
- Di Maria, E.; Latini, A.; Borgiani, P.; Novelli, G. Genetic variants of the human host influencing the coronavirus-associated phenotypes (SARS, MERS and COVID-19): Rapid systematic review and field synopsis. Hum. Genom. 2020, 14, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 2020, 184, 64–75. [Google Scholar] [CrossRef]
- Tasneem, A.A.; Abdulhamd, M.A. ABO blood groups among Coronavirus disease 2019 patients. Ibero Am. J. Med. 2020, 2, 268–274. [Google Scholar]
- Song, J.; Chen, F.; Campos, M.; Bolgiano, D.; Houck, K.; Chambless, L.E.; Wu, K.K.; Folsom, A.R.; Couper, D.; Boerwinkle, E.; et al. Quantitative Influence of ABO Blood Groups on Factor VIII and Its Ratio to von Willebrand Factor, Novel Observations from an ARIC Study of 11,673 Subjects. PLoS ONE 2015, 10, e0132626. [Google Scholar] [CrossRef] [Green Version]
- Reid, M.E.; Lomas-Francis, C. The Blood Group Antigen Facts Book, 2nd ed.; Elsevier Academic Press: New York, NY, USA, 2004. [Google Scholar]
- Walls, A.; Park, Y.; Tortorici, M.; Wall, A.; McGuire, A.; Veesler, D.A. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Paul, E.M.; Fine, K.M.; Scott, J.A.; Edmunds, W.J. Community Protection. Plotkin’s Vaccines: USA, 7th ed.; Elsevier: Philadelphia, PA, USA, 2018; pp. 1512–1531. [Google Scholar]
- Alobid, M.; Derardja, B.; Szucs, I. Food Gap Optimization for Sustainability Concerns, the Case of Egypt. Sustainability 2021, 13, 1-17. [Google Scholar] [CrossRef]
- Van Boheemen, S.; de Graaf, M.; Lauber, C.; Bestebroer, T.M.; Raj, V.S.; Zaki, A.M. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio 2012, 3, 473–512. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- Lin, J.; Wang, R.; Jiao, P.; Li, Y.; Liao, M. An impedance immunosensor based on low-cost microelectrodes and specific monoclonal antibodies for rapid detection of avian influenza virus H5N1 in chicken swabs. Biosens. Bioelectron. 2015, 67, 546–552. [Google Scholar] [CrossRef] [PubMed]
- Callaway, Z.; Wang, Y.; Zhang, B.; Zhang, T.; Costello, T.A.; Slavik, M.F.; Li, Y. A portable impedance biosensing system for rapid detection of avian influenza virus. Sensors 2016, 17, 1–15. [Google Scholar]
- Fu, Y.; Callaway, Z.; Lum, J.; Wang, R.; Lin, J.; Li, Y. Exploiting enzyme catalysis in ultra-low ion strength media for impedance biosensing of avian influenza virus using a bare interdigitated electrode. Anal. Chem. 2014, 86, 1965–1971. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Das, A.; Dhawane, A.N.; Sweeney, J.; Zhang, X.; Chivukula, V.; Iyer, S.S. Highly specific and rapid glycan-based amperometric detection of influenza viruses. Chem. Sci. 2017, 8, 3628–3634. [Google Scholar] [CrossRef] [Green Version]
- Aydinlik, S.; Ozkan-Ariksoysal, D.; Kara, P.; Ayiner, A.A.; Ozsoz, M.S. A nucleic acid-based electrochemical biosensor for the detection of influenza B virus from PCR samples using gold nanoparticle-adsorbed disposable graphite electrode and Meldola’s blue as an intercalator. Anal. Methods 2011, 3, 1607–1614. [Google Scholar] [CrossRef]
- Tian, J.; Wang, D.; Zheng, Y.T.; Jing, A. High Sensitive Electrochemical Avian Influenza Virus H7 Biosensor Based on CNTs/MoSx Aerogel. Int. J. Electrochem. Sci. 2017, 12, 2658–2668. [Google Scholar] [CrossRef]
- Lam-Dai, T.; Binh Hai, N.; Nguyen Van, H.; Hoang Vinh, T.; Huy Le, N.; Phuc Xuan, N. Electrochemical detection of short HIV sequences on chitosan/Fe3O4 nanoparticle based screen printed electrodes. Mater. Sci. Eng. C 2011, 31, 477–485. [Google Scholar]
- Qureshi, A.; Kang, W.P.; Davidson, J.L.; Gurbuz, Y. Review on carbon- derived, solid-state, micro and nano sensors for electrochemical sensing applications. Diam. Rel. Mat. 2009, 18, 1401–1420. [Google Scholar] [CrossRef] [Green Version]
- Braustein, H.E.; Braustein, I.E. Real time diagnostic point of care by amperometric immuno-biosensor kit by flow technology. ECS Trans. 2014, 58, 1–17. [Google Scholar] [CrossRef]
- Mashhadizadeh, H.M.; Talemi, R.P. A highly sensitive and selective hepatitis B DNA biosensor using gold nanoparticle electrodeposition on an Au electrode and mercaptobenzaldehyde. J. Anal. Methods 2014, 6, 8956–8964. [Google Scholar] [CrossRef]
- Malecka, K.; Grabowska, I.; Radecki, J.; Stachyra, A.; Góra-Sochacka, A.; Sirko, A.; Radecka, H. Electrochemical detection of avian influenza virus genotype using amino-ssDNA probe modified gold electrodes. Electroanalysis 2013, 25, 1871–1878. [Google Scholar] [CrossRef]
- Kiilerich-Pedersen, K.; Dapra, J.; Cherre SRozlosnik, N. High sensitivity point-of-care device for direct virus diagnostics. Biosens. Bioelectron. 2003, 49, 374–379. [Google Scholar] [CrossRef]
- Jarocka, U.; Sawicka, R.; Gora-Sochacka, A.; Sirko, A.; Zagórski-Ostoja, W.; Radecki, J.; Radecka, H. An immunosensor based on antibody binding fragments attached to gold nanoparticles for the detection of peptides derived from avian influenza hemagglutinin H5. Sensors 2014, 14, 15714–15728. [Google Scholar] [CrossRef] [Green Version]
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for highly accurate Severe Acute Respiratory Syndrome Coronavirus-2 Detection. ACS Nano 2020, 14, 5268–5277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dincer, C.; Kling, A.; Urban, G.A.; Bruch, R. Single-Channel Multianalyte Biosensor. Patent WO 2019/134741 A1, 2019. [Google Scholar]
- Kaur, H.; Shorie, M. Nanomaterial based aptasensors for clinical and environmental diagnostic applications. Nanoscale Adv. 2019, 1, 2123–2138. [Google Scholar] [CrossRef] [Green Version]
- Bora, U.; Sett, A.; Singh, D. Nucleic Acid Based Biosensors for Clinical Applications. Biosens. J. 2013, 2, 1–8. [Google Scholar] [CrossRef]
- Slütter, B.; Jiskoot, W. Sizing the optimal dimensions of a vaccine delivery system: A particulate matter. Expert Opin. Drug Deliv. 2016, 13, 167–170. [Google Scholar] [CrossRef]
- Mehrotra, P. Biosensors and their applications—A review. J. Oral Biol. Craniofacial Res. 2016, 6, 153–159. [Google Scholar] [CrossRef] [Green Version]
- Fayemi, O.E.; Adekunle, S.A.; Ebenso, E.E. A Sensor for the Determination of Lindane Using PANI/Zn, Fe(III) Oxides and Nylon 6,6/MWCNT/Zn, Fe(III) Oxides Nanofibers Modified Glassy Carbon Electrode. J. Nanomater. 2016, 1, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Stringer, R.C.; Schommer, S.; Hoehn, D.; Grant, S.A. Development of an optical biosensor using gold nanoparticles and quantum dots for the detection of Porcine Reproductive and Respiratory Syndrome Virus. Sens. Actuators B Chem. 2008, 134, 427–431. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).