Evaluation of Different Molecular Markers for Genotyping Non-Saccharomyces Wine Yeast Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains
2.2. Cell Culture and Genomic DNA Extraction
2.3. RAPD Analysis
2.4. TRtRNA Analysis
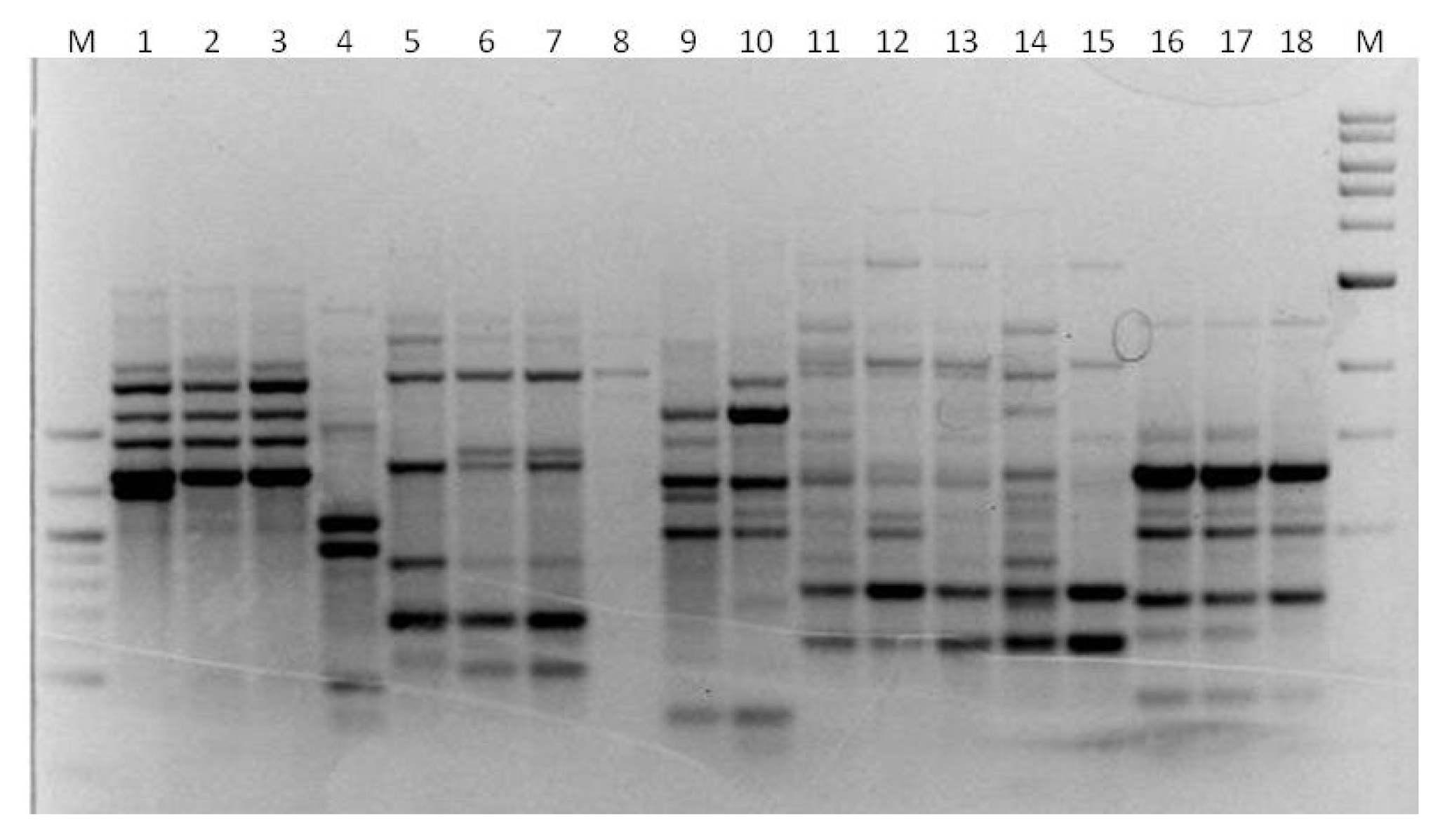
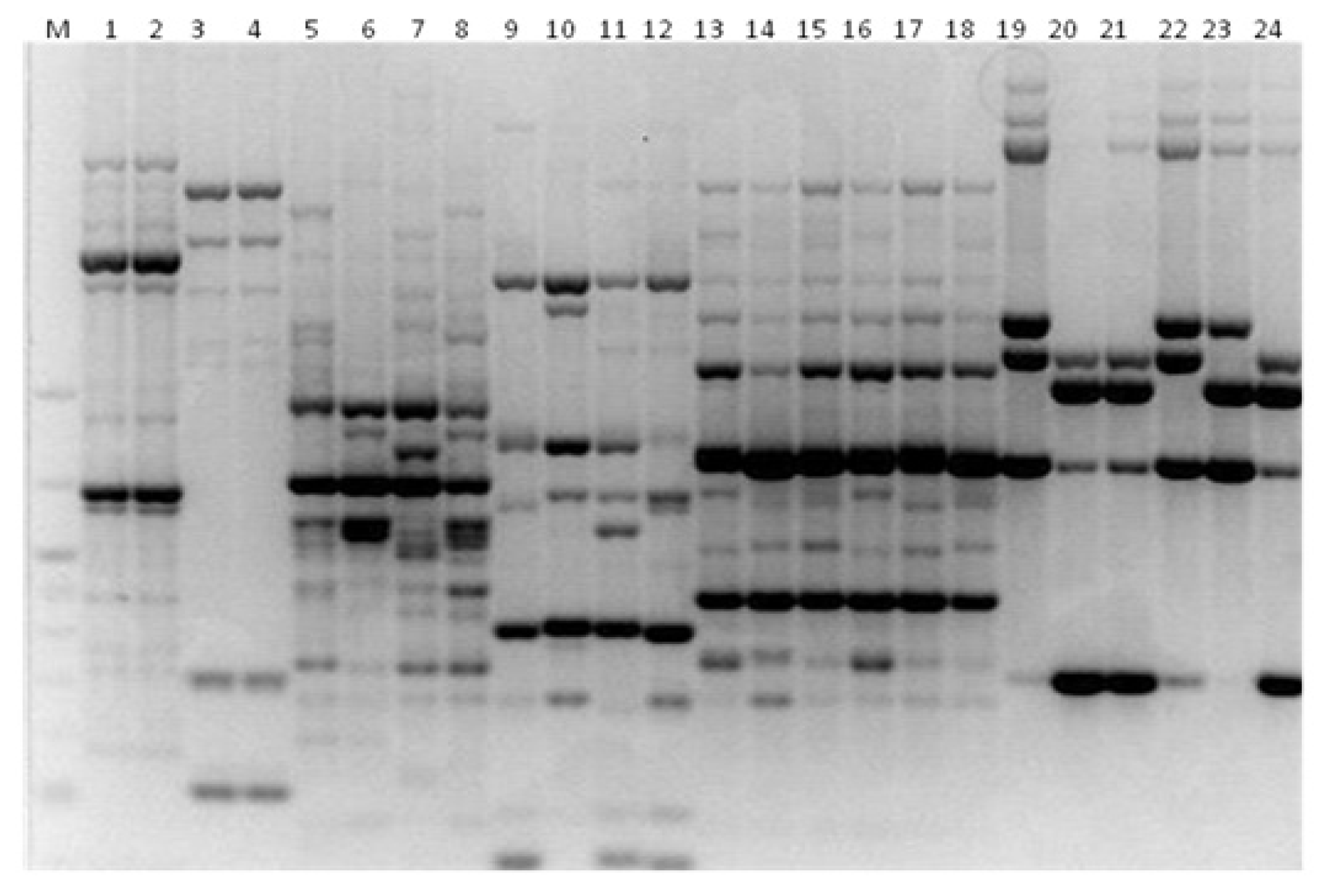
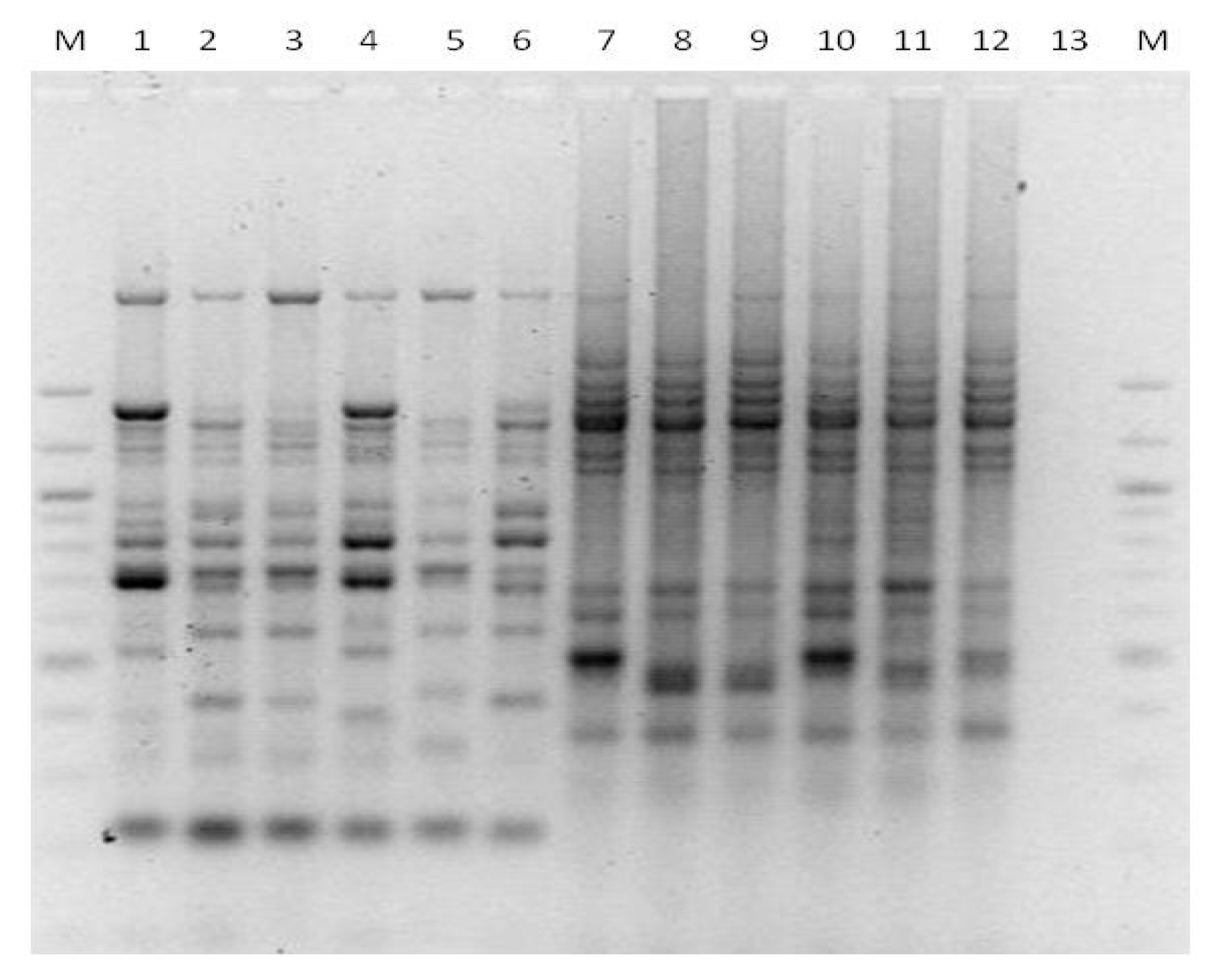
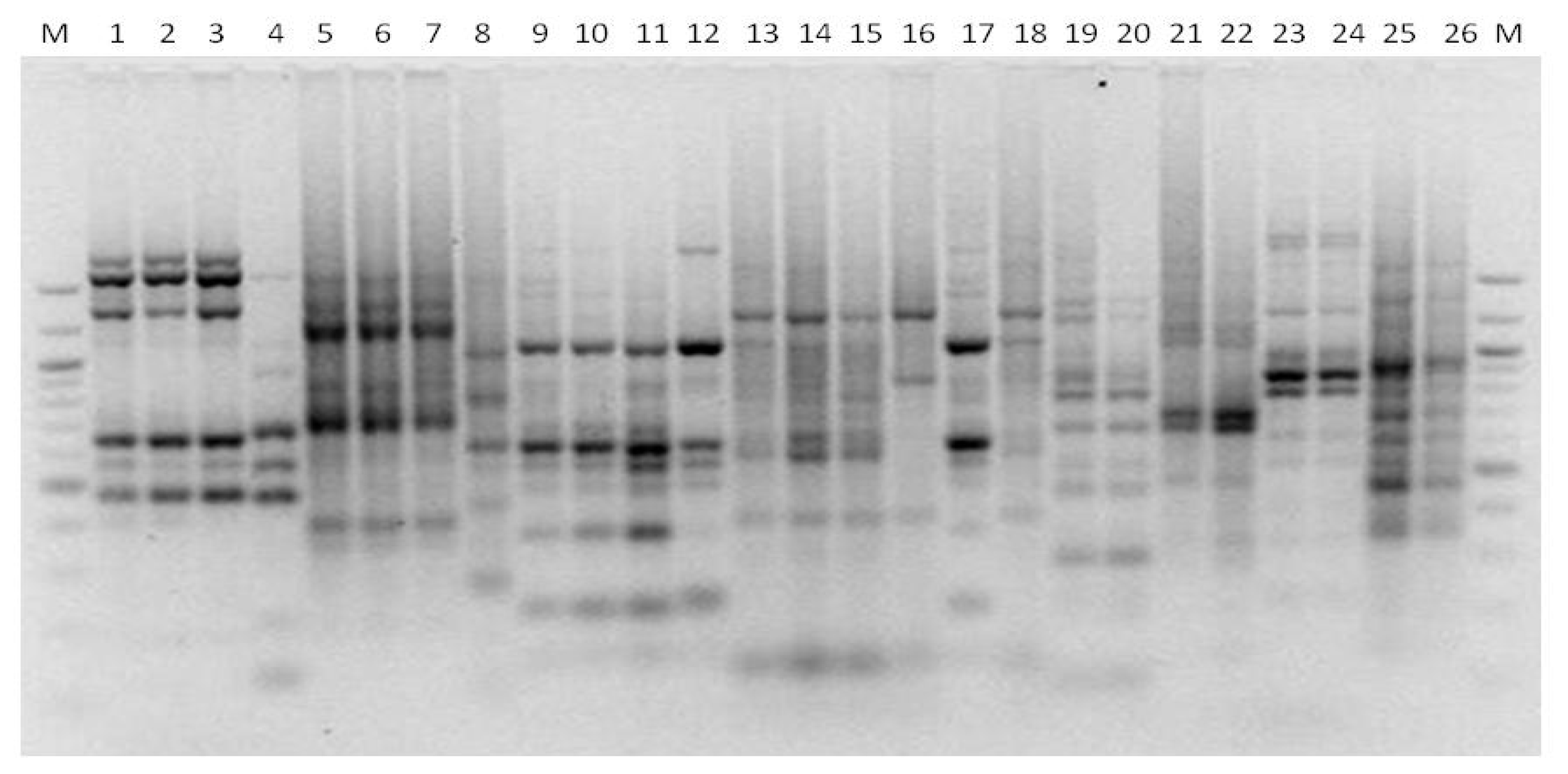
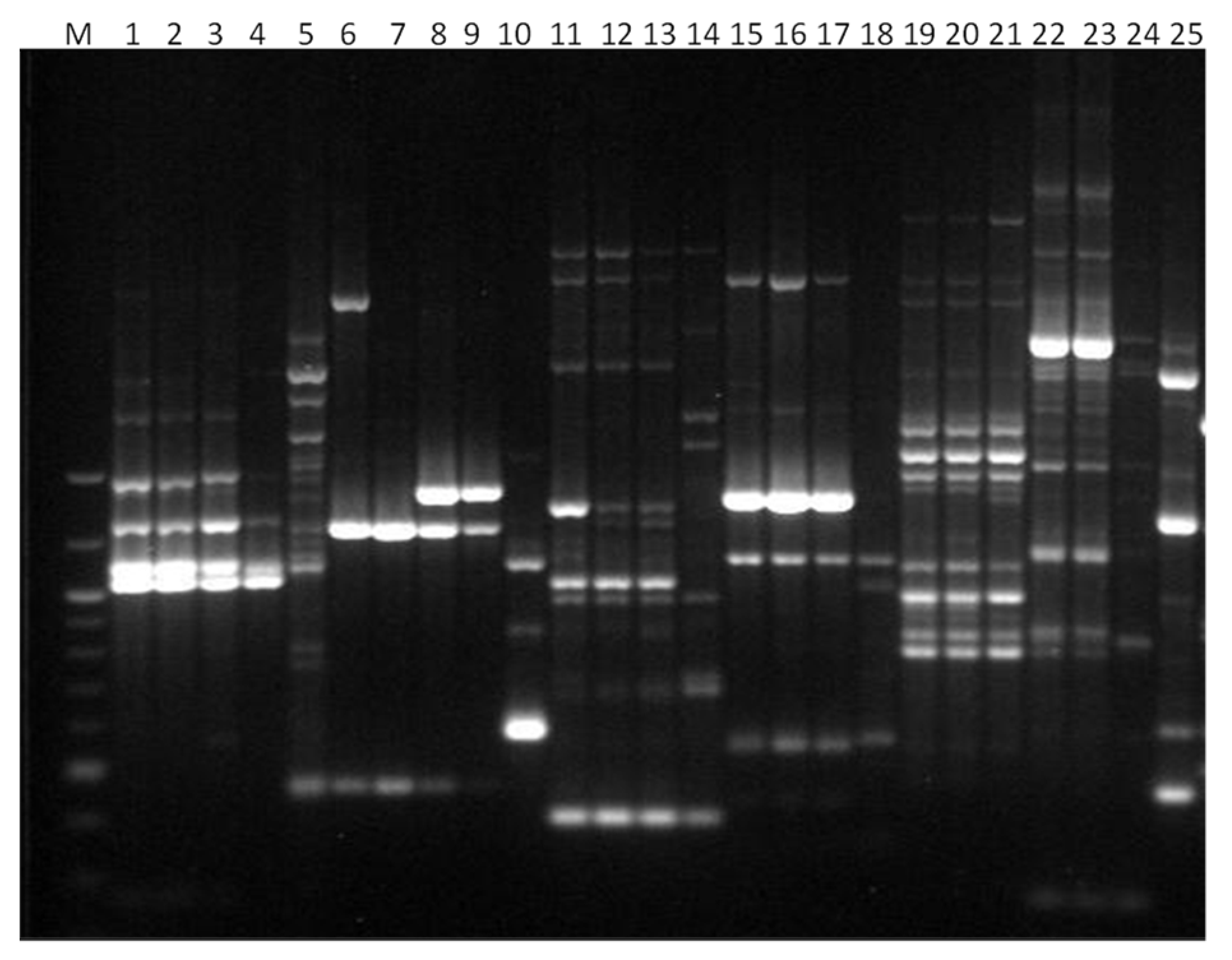
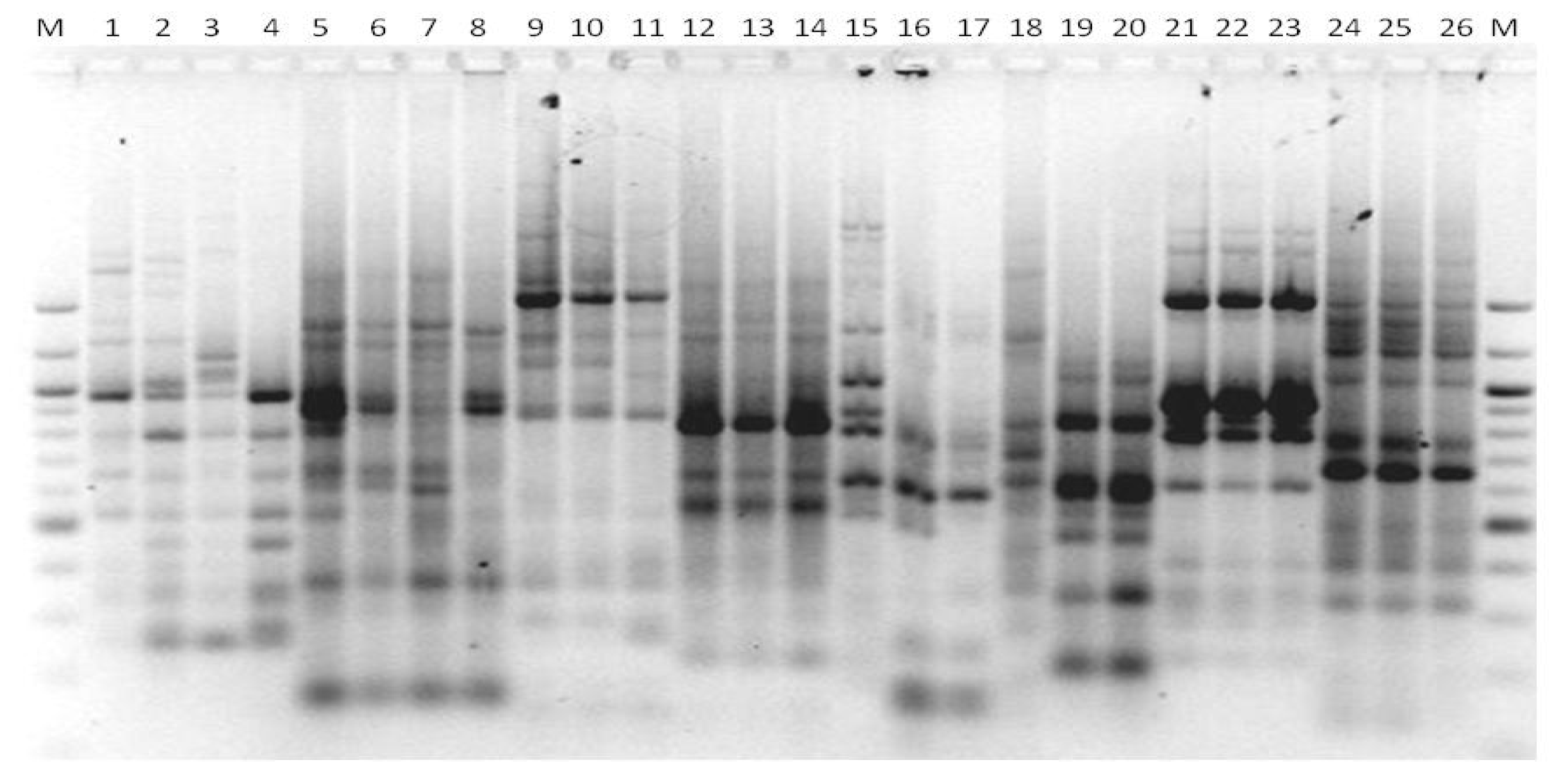
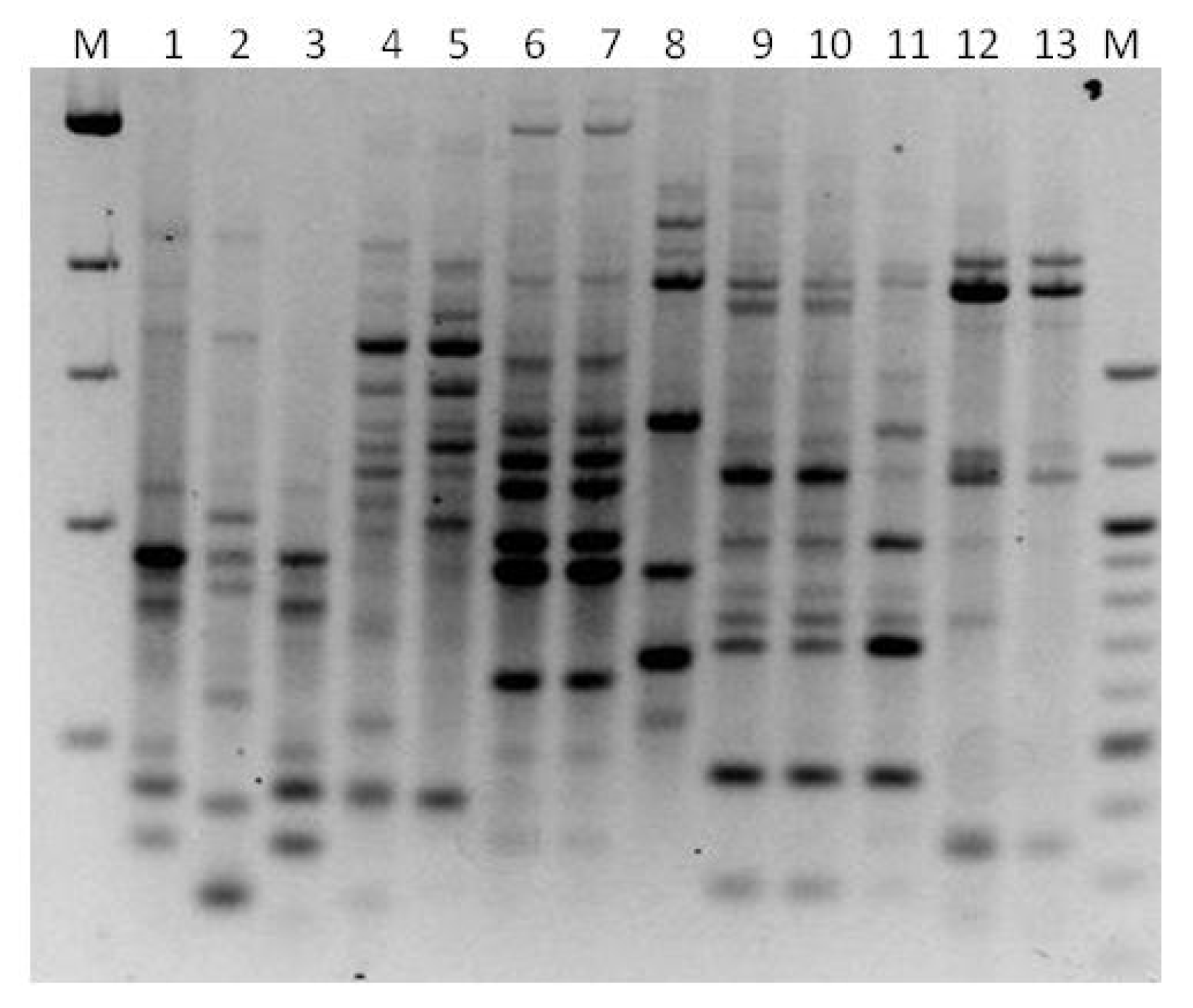
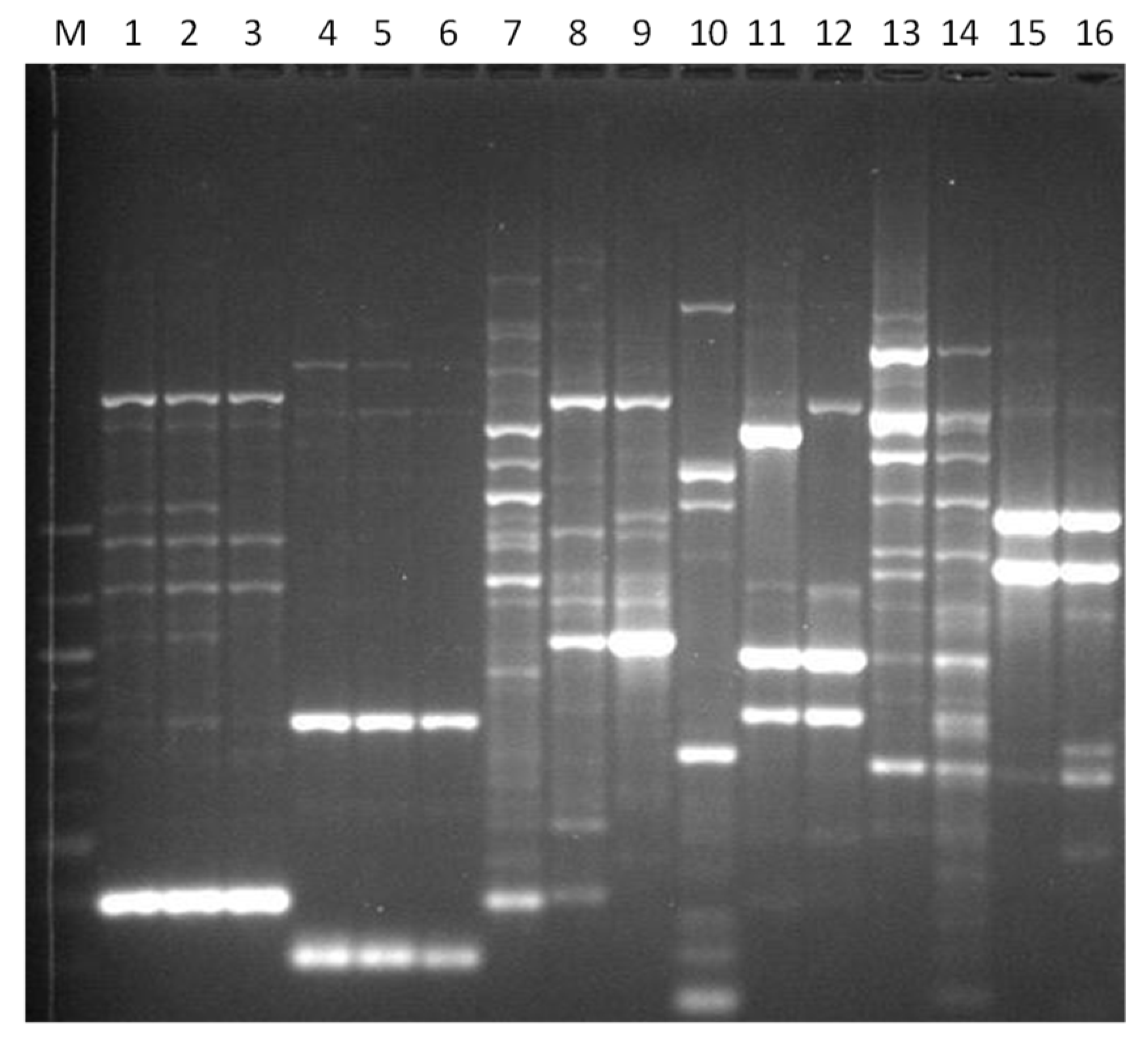
3. Results
3.1. Genotyping Lachancea therotolerans
3.2. Genotyping Starmerella bacillaris
3.3. Genotyping Metschnikowia pulcherrima
3.4. Genotyping Issatchenkia orientalis
3.5. Genotyping Torulaspora delbrueckii
3.6. Genotyping Hanseniaspora spp.
3.7. Genotyping Pichia anonala
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Martini, A.; Ciani, M.; Scorzetti, G. Direct Enumeration and Isolation of Wine Yeasts from Grape Surfaces. Am. J. Enol. Vitic. 1996, 47, 435–440. [Google Scholar]
- Fleet, G.H. Wine Yeasts for the Future. FEMS Yeast Res. 2008, 8, 979–995. [Google Scholar] [CrossRef] [PubMed]
- Nisiotou, A.A.; Spiropoulos, A.E.; Nychas, G.J.E. Yeast Community Structures and Dynamics in Healthy and Botrytis-Affected Grape Must Fermentations. Appl. Environ. Microbiol. 2007, 73, 6705–6713. [Google Scholar] [CrossRef] [PubMed]
- Jolly, N.P.; Varela, C.; Pretorius, I.S. Not Your Ordinary Yeast: Non-Saccharomyces Yeasts in Wine Production Uncovered. FEMS Yeast Res. 2014, 14, 215–237. [Google Scholar] [CrossRef]
- Sgouros, G.; Chalvantzi, I.; Mallouchos, A.; Paraskevopoulos, Y.; Banilas, G.; Nisiotou, A. Biodiversity and Enological Potential of Non-Saccharomyces Yeasts from Nemean Vineyards. Fermentation 2018, 4, 32. [Google Scholar] [CrossRef]
- Nisiotou, A.; Mallouchos, A.; Tassou, C.; Banilas, G. Indigenous Yeast Interactions in Dual-Starter Fermentations May Improve the Varietal Expression of Moschofilero Wine. Front. Microbiol. 2019, 10, 1712. [Google Scholar] [CrossRef]
- Ciani, M.; Comitini, F.; Mannazzu, I.; Domizio, P. Controlled Mixed Culture Fermentation: A New Perspective on the Use of Non-Saccharomyces Yeasts in Winemaking. FEMS Yeast Res. 2010, 10, 123–133. [Google Scholar] [CrossRef]
- Jolly, N.P.; Augustyn, O.P.H.; Pretorius, I.S. The Role and Use of Non-Saccharomyces Yeasts in Wine Production. S. Afr. J. Enol. Vitic. 2006, 27, 15–39. [Google Scholar] [CrossRef]
- Ferraro, L.; Fatichenti, F.; Ciani, M. Pilot Scale Vinification Process Using Immobilized Candida stellata Cells and Saccharomyces cerevisiae. Process Biochem. 2000, 35, 1125–1129. [Google Scholar] [CrossRef]
- Nisiotou, A.; Sgouros, G.; Mallouchos, A.; Nisiotis, C.S.; Michaelidis, C.; Tassou, C.; Banilas, G. The Use of Indigenous Saccharomyces cerevisiae and Starmerella bacillaris Strains as a Tool to Create Chemical Complexity in Local Wines. Food Res. Int. 2018, 111, 498–508. [Google Scholar] [CrossRef]
- Bely, M.; Stoeckle, P.; Masneuf-Pomarède, I.; Dubourdieu, D. Impact of Mixed Torulaspora delbruecki–Saccharomyces cerevisiae Culture on High-Sugar Fermentation. Int. J. Food Microbiol. 2008, 122, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Jolly, N.P.; Augustyn, O.P.H.; Pretorius, I.S. The Effect of Non-Saccharomyces Yeasts on Fermentation and Wine Quality. S. Afr. J. Enol. Vitic. 2003, 24, 8–10. [Google Scholar] [CrossRef]
- Nisiotou, A.; Parlapani, F.; Kormas, K.; Boziaris, I. Old Targets, New Weapons: Food Microbial Communities Revealed with Molecular Tools. In Novel Food Preservation and Microbial Assessment Techniques; CRC Press: Boca Raton, FL, USA, 2014; pp. 277–312. [Google Scholar]
- Martorell, P.; Barata, A.; Malfeito-Ferreira, M.; Fernández-Espinar, M.T.; Loureiro, V.; Querol, A. Molecular Typing of the Yeast Species Dekkera bruxellensis and Pichia guilliermondii Recovered from Wine Related Sources. Int. J. Food Microbiol. 2006, 106, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Baleiras Couto, M.M.; van der Vossen, J.M.B.M.; Hofstra, H.; Huis in ’t Veld, J.H.J. RAPD Analysis: A Rapid Technique for Differentiation of Spoilage Yeasts. Int. J. Food Microbiol. 1994, 24, 249–260. [Google Scholar] [CrossRef]
- Capece, A.; Fiore, C.; Maraz, A.; Romano, P. Molecular and Technological Approaches to Evaluate Strain Biodiversity in Hanseniaspora uvarum of Wine Origin. J. Appl. Microbiol. 2005, 98, 136–144. [Google Scholar] [CrossRef] [PubMed]
- De Benedictis, M.; Bleve, G.; Tristezza, M.; Tufariello, M.; Grieco, F. An Optimized Procedure for the Enological Selection of Non-Saccharomyces Starter Cultures. Antonie Van Leeuwenhoek 2011, 99, 189–200. [Google Scholar] [CrossRef]
- Agnolucci, M.; Vigentini, I.; Capurso, G.; Merico, A.; Tirelli, A.; Compagno, C.; Foschino, R.; Nuti, M. Genetic Diversity and Physiological Traits of Brettanomyces bruxellensis Strains Isolated from Tuscan Sangiovese Wines. Int. J. Food Microbiol. 2009, 130, 238–244. [Google Scholar] [CrossRef]
- Tofalo, R.; Schirone, M.; Torriani, S.; Rantsiou, K.; Cocolin, L.; Perpetuini, G.; Suzzi, G. Diversity of Candida zemplinina Strains from Grapes and Italian Wines. Food Microbiol. 2012, 29, 18–26. [Google Scholar] [CrossRef]
- Barquet, M.; Martín, V.; Medina, K.; Pérez, G.; Carrau, F.; Gaggero, C. Tandem Repeat-TRNA (TRtRNA) PCR Method for the Molecular Typing of Non-Saccharomyces Subspecies. Appl. Microbiol. Biotechnol. 2012, 93, 807–814. [Google Scholar] [CrossRef]
- Chalvantzi, I.; Banilas, G.; Tassou, C.; Nisiotou, A. Biogeographical Regionalization of Wine Yeast Communities in Greece and Environmental Drivers of Species Distribution at a Local Scale. Front. Microbiol. 2021, 12, 705001. [Google Scholar] [CrossRef]
- Senses-Ergul, S.; Ágoston, R.; Belák, Á.; Deák, T. Characterization of Some Yeasts Isolated from Foods by Traditional and Molecular Tests. Int. J. Food Microbiol. 2006, 108, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Martín, B.; Garriga, M.; Hugas, M.; Bover-Cid, S.; Veciana-Nogués, M.T.; Aymerich, T. Molecular, Technological and Safety Characterization of Gram-Positive Catalase-Positive Cocci from Slightly Fermented Sausages. Int. J. Food Microbiol. 2006, 107, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Vicente, J.; Ruiz, J.; Belda, I.; Benito-Vázquez, I.; Marquina, D.; Calderón, F.; Santos, A.; Benito, S. The Genus Metschnikowia in Enology. Microorganisms 2020, 8, 1038. [Google Scholar] [CrossRef] [PubMed]
- du Toit, M.; Pretorius, I.S. Microbial Spoilage and Preservation of Wine: Using Weapons from Nature’s Own Arsenal-A Review. S. Afr. J. Enol. Vitic. 2019, 21, 74–96. [Google Scholar] [CrossRef]
- Howell, K.; Varela, C.A.; Grieco, F.; Tristezza, M.; Tufariello, M.; Capozzi, V.; Spano, G.; Mita, G. The Oenological Potential of Hanseniaspora uvarum in Simultaneous and Sequential Co-Fermentation with Saccharomyces cerevisiae for Industrial Wine Production. Front. Microbiol. 2016, 1, 670. [Google Scholar] [CrossRef]
- Caruso, M.; Capece, A.; Salzano, G.; Romano, P. Typing of Saccharomyces cerevisiae and Kloeckera apiculata Strains from Aglianico Wine. Lett. Appl. Microbiol. 2002, 34, 323–328. [Google Scholar] [CrossRef]
- Gobbi, M.; Comitini, F.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Lachancea thermotolerans and Saccharomyces cerevisiae in Simultaneous and Sequential Co-Fermentation: A Strategy to Enhance Acidity and Improve the Overall Quality of Wine. Food Microbiol. 2013, 33, 271–281. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Beisert, B.; Navascués, E.; Marquina, D.; Calderón, F.; Rauhut, D.; Benito, S.; Santos, A. Influence of Torulaspora delbrueckii in Varietal Thiol (3-SH and 4-MSP) Release in Wine Sequential Fermentations. Int. J. Food Microbiol. 2017, 257, 183–191. [Google Scholar] [CrossRef]
- Banilas, G.; Sgouros, G.; Nisiotou, A. Development of Microsatellite Markers for Lachancea thermotolerans Typing and Population Structure of Wine-Associated Isolates. Microbiol. Res. 2016, 193, 1–10. [Google Scholar] [CrossRef]
- Benito, S. The Impact of Torulaspora delbrueckii Yeast in Winemaking. Appl. Microbiol. Biotechnol. 2018, 102, 3081–3094. [Google Scholar] [CrossRef]
- Ramírez, M.; Velázquez, R. The Yeast Torulaspora Delbrueckii: An Interesting but Difficult-to-Use Tool for Winemaking. Fermentation 2018, 4, 94. [Google Scholar] [CrossRef]
- Renault, P.; Miot-Sertier, C.; Marullo, P.; Hernández-Orte, P.; Lagarrigue, L.; Lonvaud-Funel, A.; Bely, M. Genetic Characterization and Phenotypic Variability in Torulaspora delbrueckii Species: Potential Applications in the Wine Industry. Int. J. Food Microbiol. 2009, 134, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Canonico, L.; Comitini, F.; Ciani, M. TdPIR Minisatellite Fingerprinting as a Useful New Tool for Torulaspora delbrueckii Molecular Typing. Int. J. Food Microbiol. 2015, 200, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Pfliegler, W.P.; Horváth, E.; Kállai, Z.; Sipiczki, M. Diversity of Candida zemplinina Isolates Inferred from RAPD, Micro/Minisatellite and Physiological Analysis. Microbiol. Res. 2014, 169, 402–410. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nisiotou, A.; Gyftogianni, E.; Banilas, G. Evaluation of Different Molecular Markers for Genotyping Non-Saccharomyces Wine Yeast Species. Microbiol. Res. 2022, 13, 643-654. https://doi.org/10.3390/microbiolres13030046
Nisiotou A, Gyftogianni E, Banilas G. Evaluation of Different Molecular Markers for Genotyping Non-Saccharomyces Wine Yeast Species. Microbiology Research. 2022; 13(3):643-654. https://doi.org/10.3390/microbiolres13030046
Chicago/Turabian StyleNisiotou, Aspasia, Emanouela Gyftogianni, and Georgios Banilas. 2022. "Evaluation of Different Molecular Markers for Genotyping Non-Saccharomyces Wine Yeast Species" Microbiology Research 13, no. 3: 643-654. https://doi.org/10.3390/microbiolres13030046
APA StyleNisiotou, A., Gyftogianni, E., & Banilas, G. (2022). Evaluation of Different Molecular Markers for Genotyping Non-Saccharomyces Wine Yeast Species. Microbiology Research, 13(3), 643-654. https://doi.org/10.3390/microbiolres13030046




