Abstract
The selection of resistant crops is an effective method for controlling geminivirus diseases. ty-5 encodes a messenger RNA surveillance factor Pelota with a single amino acid mutation (PelotaV16G), which confers effective resistance to tomato yellow leaf curl virus (TYLCV). No studies have investigated whether ty-5 confers resistance to other geminiviruses. Here, we demonstrate that the tomato ty-5 line exhibits effective resistance to various geminiviruses. It confers resistance to two representative begomoviruses, tomato yellow leaf curl China virus/tomato yellow leaf curl China betasatellite complex and tomato leaf curl Yunnan virus. The ty-5 line also exhibits partial resistance to a curtovirus beet curly top virus. Importantly, ty-5 confers resistance to TYLCV with a betasatellite. Southern blotting and quantitative polymerase chain reaction analyses showed that significantly less DNA of these geminiviruses accumulated in the ty-5 line than in the susceptible line. Moreover, knockdown of Pelota expression converted a Nicotiana benthamiana plant from a geminivirus-susceptible host to a geminivirus-resistant host. Overall, our findings suggest that ty-5 is an important resistance gene resource for crop breeding to control geminiviruses.
1. Introduction
Geminiviruses are obligate intracellular parasites that cause diseases in many economically important crops (e.g., tomato, corn, maize, cassava, and cotton); thus, they pose major threats to global food security. Geminiviruses have circular single-stranded DNA genomes that are encapsidated in twinned particles. Based on their genome organization, insect vector, and host range, geminiviruses can be classified into 14 genera, of which the genus Begomovirus is the largest [1]. Viruses in the genera Becurtovirus, Capulavirus, Citlodavirus, Curtovirus, Eragrovirus, Grablovirus, Maldovirus, Mastrevirus, Mulcrilevirus, Opunvirus, Topilevirus, Topocuvirus, and Turncurtovirus have monopartite genomes, whereas viruses in the Begomovirus genus have mono- or bipartite genomes. Monopartite begomoviruses contain six known open reading frames. The viral strand of the genome encodes the capsid protein V1 and V2, while the complementary strand encodes the C1/Rep, C2, C3, and C4 proteins [2]. Some monopartite begomoviruses are associated with satellite DNAs (α and β, each approximately 1.3–1.4 kb in length) and begomovirus/betasatellite complexes have caused numerous economically important diseases, including the earliest recorded plant viral disease [3,4,5]. Betasatellites are required for symptom expression in plants, although they depend on begomoviral DNA for replication and encapsidation [6,7].
Tomato yellow leaf curl virus (TYLCV) is a monopartite begomovirus. It is one of the most damaging and threatening viruses for tomato production worldwide. The selection of tomatoes that are resistant to the virus is an effective method for controlling disease caused by TYLCV. Currently, six TYLCV resistance genes (Ty-1 to Ty-6) are known; some are used widely for introgression breeding [8]. Ty-1, Ty-3, Ty-4, and Ty-6 are derived from the wild tomato species Solanum chilense, Ty-2 is from Solanum habrochaites, and ty-5 is from the commercial tomato cultivar Tyking [9,10,11,12,13,14,15,16,17,18,19]. Ty-1, Ty-2, Ty-3, Ty-4, and Ty-6 are dominant resistance genes, while ty-5 exhibits recessive inheritance. In recent years, Ty-1, Ty-2, Ty-3, and ty-5 have been cloned. Ty-1 and Ty-3 are allelic and encode an RNA-dependent RNA polymerase [20]. Ty-2 encodes a nucleotide-binding leucine-rich repeat protein that recognizes the Rep/C1 protein of TYLCV and induces a hypersensitive response to viral infection [21,22]. In susceptible tomatoes, ty-5 encodes the messenger RNA surveillance factor Pelota [17,19]. The ty-5 gene contains a single amino acid mutation, V16G, in Pelota (PelotaV16G); tomatoes that contain this mutation are resistant to TYLCV [17,19]. Until recently, it was unclear whether ty-5 could confer effective resistance to other geminiviruses. Ty-1 is a gene that encodes universal resistance to geminiviruses; however, it was recently reported that the resistance conferred by Ty-1 is compromised by co-infection of TYLCV with a betasatellite [9,18]. No effective resistance gene is currently available for controlling begomovirus/betasatellite complexes. There is an urgent need to screen and identify new effective resistance genes to control the emerging begomovirus/betasatellite complexes. To our knowledge, no studies have investigated whether ty-5 can confer effective resistance to begomovirus/betasatellite complexes.
In this study, we demonstrate that ty-5 confers broad-spectrum resistance to various geminiviruses. ty-5 confers effective resistance to two representative begomoviruses. The tomato ty-5 line also exhibits partial resistance to beet curly top virus (BCTV), a virus in the genus Curtovirus. Importantly, ty-5 confers resistance to infection by TYLCV with a betasatellite. Regardless of betasatellite status, significantly less DNA of these geminiviruses accumulates in the ty-5 line than in the susceptible line. Moreover, knockdown of Pelota expression converts a Nicotiana benthamiana plant from a geminivirus-susceptible host to a geminivirus-resistant host. Our findings provide important insights concerning the use of ty-5 to control geminiviruses.
2. Materials and Methods
2.1. Plant Materials and Virus Sources
Tomato (Solanum lycopersicum) and N. benthamiana plants were grown in an insect-free growth chamber at 25℃ under a 16-h light/8-h dark cycle. The ty-5 tomato line, AVTO1227, was introduced from the World Vegetable Center in 2013 [17]; tomato Moneymaker was the susceptible line for TYLCV. Infectious clones of TYLCV Beijing isolate (MN432609) [23], BCTV (U02311.1) [24], and tomato leaf curl Yunnan virus Y194 isolate (TbLCYnV, AJ971265; Y194 refers to TbLCYnV) [25], as well as Y10 isolates of tomato yellow leaf curl China virus (TYLCCNV, AJ319675; Y10A refers to TYLCCNV DNA-A) and tomato yellow leaf curl China betasatellite (TYLCCNB, AJ421621; Y10β refers to TYLCCNB) [7], were previously described and have been maintained in our laboratory.
2.2. Agrobacterium-Mediated Inoculation and Disease Symptom Assessment
S. lycopersicum and N. benthamiana plants were agroinfected with the geminiviruses via Agrobacterium tumefaciens-mediated infiltration. A. tumefaciens EHA105 cultures were adjusted to an optical density of OD600 = 2.0 before infiltration into S. lycopersicum plants. For agroinfiltration of N. benthamiana plants, A. tumefaciens cultures were adjusted to an optical density of OD600 = 0.2. For viral infection analysis, the numbers of virus-infected plants with different disease symptom grades were counted and converted to percentages. Grades I to IV referred to plants that were asymptomatic, showed mild leaf curling symptoms, severe leaf curling symptoms, and severely curly leaves and stunting, respectively.
2.3. Total DNA Extraction, Southern Blotting, and Quantitative Polymerase Chain Reaction
Total DNA was isolated from infected young leaves of plants using the cetyltrimethylammonium bromide (CTAB) method, separated by 1% agarose gel electrophoresis, and then transferred to nylon membranes (Hybond N+; GE Healthcare, Pittsburgh, PA, USA). The membranes were hybridized at 55℃ with digoxigenin-labeled probes that had been prepared using a commercial kit (DIG High Prime DNA Labeling and Detection Starter Kit; Roche Diagnostics, Rotkreuz, Switzerland). The agarose gels were stained (Gel Stain; TransGen Biotech, Beijing, China) and used to confirm equal sample loading. Viral DNA accumulation was also detected by quantitative polymerase chain reaction (qPCR) using TB Green Premix Ex Taq II with 40 rounds of amplification (Takara, Japan). Viral DNA accumulation was normalized to the expression of 25S rRNA using the comparative Ct method (2−ΔΔCt), as previously described [26]. Primers used in this study are listed in Table S1.
2.4. Plasmid Construction
For construction of a hairpin-based RNAi vector containing NbPelota, a partial fragment of NbPelota cDNA was amplified by PCR using the corresponding primers and cloned into the RNAi vector [27] by infusion; the reverse NbPelota fragment was cloned into the resulting vector using the restriction enzyme MluI and SalI. The primers used in this study are listed in Table S1.
3. Results
3.1. Sequence Comparison of Pelota, the Candidate ty-5 Gene, among Tomato Lines
The tomato line AVTO1227 exhibits effective resistance to TYLCV and the line Moneymaker is susceptible to TYLCV [17]. To identify nucleotide polymorphisms in the Pelota gene within the AVTO1227 resistant (R) line and susceptible (S) line, we cloned and sequenced the Pelota genes from both R and S lines. Pelota gene sequence comparisons revealed only one nucleotide difference, T47G, between the two lines. This mutation results in a single amino acid mutation, V16G, in the Pelota gene in the ty-5 R line (Figure 1). We also examined this amino acid site in the Pelota protein in S. chilense, S. peruvianum, S. pimpinellifolium, and S. pennellii, and none of these wild-type tomatoes contained the V16G mutation (Figure 1). Thus, the sequencing results confirmed that the ty-5 line of AVTO1227 specifically carries a V16G mutation in Pelota.

Figure 1.
Alignment of the predicted amino acid sequences of Pelota genes from the susceptible tomato line Moneymaker, the resistant tomato line AVTO1227, Solanum chilense, Solanum peruvianum, Solanum pimpinellifolium, and Solanum pennellii. The difference between resistant and susceptible lines (valine vs. glycine at amino acid 16) is indicated with a red box. Residues that differ from Moneymaker are indicated in solid deep red.
3.2. ty-5 Confers Resistance to Two Representative Begomoviruses in China
To determine the resistance spectrum of ty-5, we used two representative begomoviruses from China (TYLCCNV/TYLCCNB and TbLCYnV). The tomato AVTO1227 line carrying the ty-5 gene (hereafter referred to as the ty-5 line) was inoculated with infectious clones of TYLCCNV/TYLCCNB or TbLCYnV by Agrobacterium-mediated infiltration. Plants agroinfiltrated with the pBinPLUS empty vector were used as mock controls. The tomato Moneymaker line (hereafter referred to as the WT line), in which TYLCV can induce strong disease symptoms, was also inoculated with the abovementioned infectious clones and used as controls. The disease symptoms of inoculated tomatoes were monitored at 3–7 weeks post-inoculation. Compared with mock controls, the Moneymaker WT line showed severe disease symptoms, including yellowing and leaf curling, at 3–7 weeks after inoculation with TYLCCNV/TYLCCNB (Y10Aβ) or TbLCYnV (Y194) (Figure 2A). However, the ty-5 line inoculated with these two viruses remained symptomless. We then classified the disease symptoms from grade I (no symptoms) to grade IV (very severe symptoms; Figure S1). All inoculated plants in the WT line exhibited grade III symptoms, whereas no plant in the ty-5 line exhibited any obvious disease symptoms (grade I; Figure 2B). Southern blotting analysis showed that the ty-5 line accumulated significantly less genomic DNA from TYLCCNV/TYLCCNB (Y10Aβ) or TbLCYnV (Y194), compared with the WT line (Figure 2C). qPCR assays confirmed that ty-5 inhibited the viral accumulation of these two begomoviruses in the resistant line (Figure 2D). These results suggest that ty-5 confers resistance to the two representative begomoviruses.
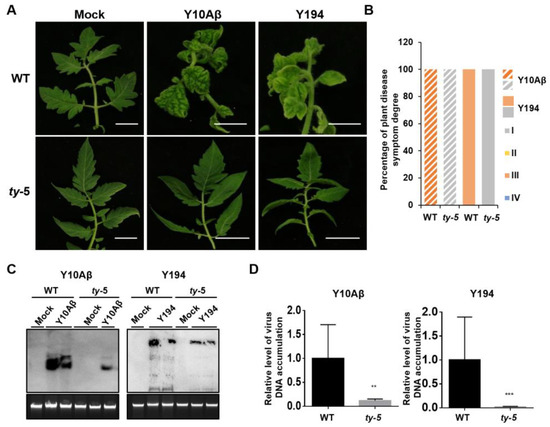
Figure 2.
The ty-5 gene confers resistance to two representative begomoviruses from China. (A) Viral symptoms in the tomato ty-5 line with a PelotaV16G mutation and the wild-type (WT) line, agroinfected with either the pBinPLUS empty vector (mock), an infectious clone of tomato yellow leaf curl China virus (TYLCCNV)/tomato yellow leaf curl China betasatellite (TYLCCNB) (Y10Aβ), or tomato leaf curl Yunnan virus (TbLCYnV, Y194). The inoculated plants were photographed at 7 weeks post-inoculation (wpi). (B) The percentage and degree of plant disease symptoms in WT and ty-5 plants infected by TYLCCNV/TYLCCNB (Y10Aβ) or TbLCYnV (Y194) at 7 wpi. In total, 15 plants from each line were used for the assays. Disease symptoms were classified from grade I (no symptoms) to grade IV (very severe symptoms). (C) Southern blotting analysis of viral DNA accumulation in WT and ty-5 lines infected with TYLCCNV/TYLCCNB (Y10Aβ) or TbLCYnV (Y194) at 7 wpi. DNA fragments of TYLCCNV or TbLCYnV capsid protein were used as probes for detection of genomic DNA from TYLCCNV and TbLCYnV, respectively. (D) qPCR analysis of viral DNA accumulation in WT and ty-5 lines infected with TYLCCNV/TYLCCNB (Y10Aβ) and TbLCYnV (Y194) at 7 wpi. Specific primers for TYLCCNV or TbLCYnV capsid protein were used to quantify the accumulation of viral DNA; 25S rRNA was used as the internal control. Asterisks denote statistically significant differences evaluated with Student’s t test, ** p < 0.01, *** p < 0.001.
3.3. ty-5 Confers Resistance to Curtovirus
To further characterize the resistance spectrum of ty-5, we tested its resistance to the curtovirus BCTV. Both the ty-5 and WT lines were inoculated with the infectious clone of BCTV or the pBinPLUS empty vector (mock control) by agroinfiltration. Compared to plants that were inoculated with the mock control, the WT line showed severe stunting at 18–30 days after BCTV infection (Figure 3A,B). However, the ty-5 line inoculated with BCTV showed mild leaf curl symptoms and no stunting was observed at 18–30 days post-inoculation. We also classified the disease symptoms from BCTV into grades I (none) to IV (very severe). All inoculated plants in the WT line exhibited grade IV disease symptoms, while most plants in the ty-5 line exhibited grade II disease symptoms (Figure 3C). Southern blotting analysis showed that significantly less BCTV genomic DNA accumulated in the ty-5 line than in the WT line (Figure 3D). This finding was confirmed by qPCR (Figure 3E). Thus, the results suggest that ty-5 confers resistance not only to begomoviruses, but also to a curtovirus.
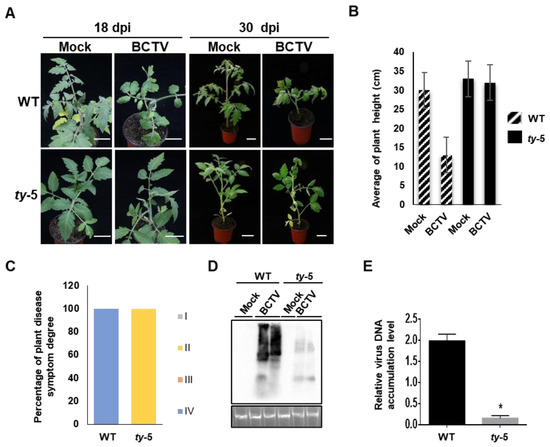
Figure 3.
ty-5 confers resistance to beet curly top virus (BCTV), a geminivirus in the genus Curtovirus. (A) Viral symptoms of the tomato ty-5 line and the wild-type (WT) line agroinfected with either the pBinPLUS empty vector (mock) or an infectious clone of BCTV. Systemically infected leaves and entire plants were photographed at 18 and 30 days post-inoculation (dpi), respectively. (B) The mean height of WT and ty-5 plants infected with BCTV at 30 dpi. (C) The percentage and degree of disease symptoms in WT and ty-5 plants infected with BCTV at 30 dpi. Fifteen plants from each of the WT and ty-5 lines were used for the assays. Disease symptoms were classified from grade I (no symptoms) to grade IV (very severe symptoms). (D) Southern blotting analysis of viral DNA accumulation in WT and ty-5 lines infected with BCTV at 30 dpi. A DNA fragment of BCTV capsid protein was used as a probe for detection of genomic DNA from BCTV. (E) qPCR analysis of viral DNA accumulation in WT and ty-5 lines infected with BCTV at 30 dpi. Specific primers for BCTV capsid protein were used to quantify the accumulation of viral DNA; 25S rRNA was used as the internal control. Asterisks denote statistically significant differences evaluated with Student’s t test, * p < 0.05.
3.4. ty-5 Confers Resistance to TYLCV with Betasatellite
It has also been recently reported that the resistance of Ty-1 to geminiviruses is compromised during co-infection by TYLCV with a betasatellite [9,18]. To test whether ty-5 could confer resistance to TYLCV during co-infection with a betasatellite, the tomato ty-5 line was agroinfiltrated with infectious clones of TYLCV alone, TYLCV/TYLCCNB, or a pBinPLUS empty vector (mock control). The WT line was also inoculated with TYLCV or TYLCV/TYLCCNB complex for comparison. Compared with plants that had been inoculated with TYLCV, the WT line showed severe disease symptoms after infection by the TYLCV/TYLCCNB (TYLCV/Y10β) complex (Figure 4A,B). TYLCV did not cause any obvious symptoms in the ty-5 line. In the presence of a betasatellite, TYLCV induced no or mild leaf curl symptoms in the ty-5 line (Figure 4A,B). Approximately 12.5% of the ty-5 plants infected by the TYLCV/Y10β complex showed mild disease symptoms, whereas all others showed no symptoms (Figure 4C). In contrast, 100% of WT plants infected either by TYLCV or the TYLCV/Y10β complex showed severe disease symptoms (Figure 4C). Southern blotting analysis showed that substantial genomic DNA from TYLCV accumulated in the WT line, regardless of TYLCCNB status. Significantly less viral DNA accumulated in the ty-5 line (Figure 4D). In the ty-5 plant with mild leaf curl symptoms caused by the TYLCV/Y10β (Figure 4D), some viral DNA accumulated (the lane before the last lane); however, this was less than in the WT line infected with the TYLCV/Y10β. In the ty-5 plant without leaf curl symptoms under infection by TYLCV/Y10β, the quantity of viral DNA (the last lane) was comparable to the quantity in the ty-5 plant that was infected with TYLCV (Figure 4D).
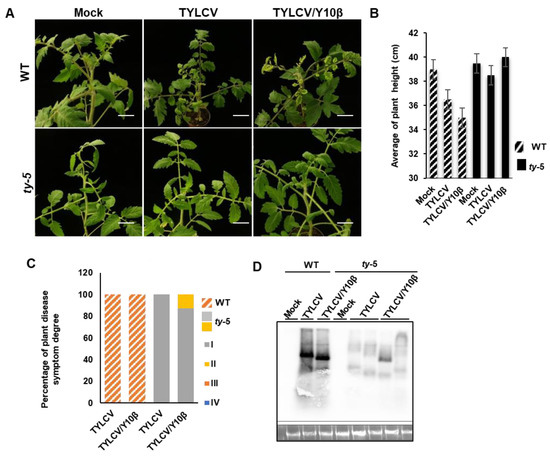
Figure 4.
ty-5 confers resistance to tomato yellow leaf curl virus (TYLCV) with a betasatellite. (A) Viral symptoms of the tomato ty-5 line and wild-type (WT) line agroinfected with either the pBinPLUS empty vector (mock), an infectious clone of TYLCV, or infectious clones of TYLCV/tomato yellow leaf curl China betasatellite (Y10β). The mock- and virus-inoculated plants were photographed at 7 weeks post-inoculation (wpi). (B) The mean height of WT and ty-5 plants infected with TYLCV, with and without a betasatellite, at 7 wpi. (C) The percentage of degree of disease symptoms in WT and ty-5 plants infected by TYLCV, with and without a betasatellite at 7 wpi. Fifteen plants from each of the WT and ty-5 lines were used for the assays. Disease symptoms were classified from grade I (no symptoms) to grade IV (very severe symptoms). (D) Southern blotting analysis of viral DNA accumulation in WT and ty-5 lines infected by TYLCV or TYLCV/Y10β at 7 wpi. A DNA fragment of TYLCV capsid protein was used as a probe for detection of genomic DNA from TYLCV.
3.5. Suppression of Pelota Expression in N. benthamiana Converts a Geminivirus-Susceptible Host to a Geminivirus-Resistant Host
Pelota with the V16G mutation confers resistance to geminiviruses. Thus, we tested whether knockdown of the Pelota expression of N. benthamiana plants would confer geminivirus resistance. N. benthamiana plants were treated with Agrobacterium that carries a construct expressing hairpin RNA targeting Pelota (RNAi-Pelota) or Agrobacterium that carries a control vector expressing hairpin RNA GUS gene (RNAi-GUS), and then agroinfected with TYLCV, TYLCCNV/TYLCCNB, TbLCYnV, or BCTV. Compared to the RNAi-GUS-treated control plant, the RNAi-Pelota-treated plant infected by TYLCV or TYLCCNV/TYLCCNB showed very mild or no disease symptoms (Figure 5A). Significantly less viral DNA accumulated systemically in the leaves of RNAi-Pelota-treated plants than in the leaves of RNAi-GUS control plants infected by TYLCV or TYLCCNV/TYLCCNB (Figure 5B,C). Similarly, RNAi-GUS control plants agroinfected with TbLCYnV exhibited severe disease symptoms in systemically infected leaves. In contrast, RNAi-Pelota-treated plants agroinfected with TbLCYnV showed mild disease symptoms (Figure 5A). As for BCTV, symptoms were similar between RNAi-Pelota and RNAi-GUS control plants (Figure 5A). Southern blotting analysis showed that less genomic DNA from TYLCV or TYLCCNV accumulated in RNAi-Pelota plants than in RNAi-GUS control plants, whereas no significant difference was found in terms of genomic DNA accumulation from TbLCYnV or BCTV (Figure 5C).
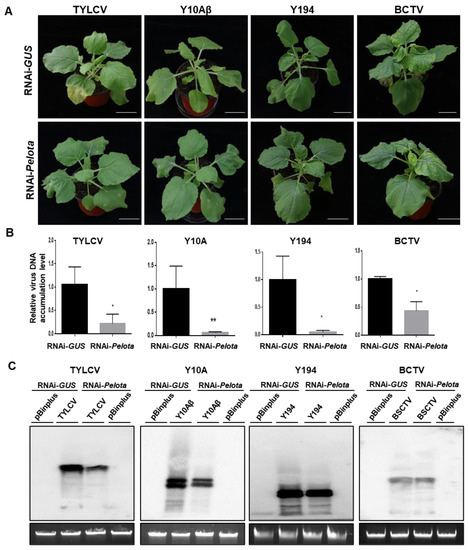
Figure 5.
Knockdown of Pelota expression converts a Nicotiana benthamiana plant from a geminivirus-susceptible host to a geminivirus-resistant host. (A) Viral symptoms of N. benthamiana plants treated with agrobacterium that contains the construct expressing hairpin RNA targeting Pelota (RNAi-Pelota) or a hairpin RNA GUS control (RNAi-GUS), and then agroinoculated with either the pBinPLUS empty vector (mock), an infectious clone of tomato yellow leaf curl virus (TYLCV), tomato yellow leaf curl China virus/tomato yellow leaf curl China betasatellite (Y10Aβ), tomato leaf curl Yunnan virus (Y194), or beet curly top virus (BCTV). (B) qPCR analysis of viral DNA accumulation in geminivirus-infected RNAi-Pelota- and RNAi-GUS-treated plants. Leaf samples were collected from TYLCV, Y10Aβ, Y194, and BCTV-infected N. benthamiana plants at 9, 5, 9, and 12 dpi, respectively. Specific primers for TYLCV, Y10, Y194, or BCTV capsid protein were used to quantify the accumulation of viral DNA; 25S rRNA was used as the internal control. Asterisks denote statistically significant differences evaluated with Student’s t test, * p < 0.05, ** p < 0.01. (C) Viral DNA accumulation in geminivirus-infected RNAi-Pelota- and RNAi-GUS-treated plants was analyzed by Southern blotting. Leaf samples were collected from TYLCV, Y10Aβ, Y194, and BCTV-infected N. benthamiana plants at 12, 8, 12, and 9 dpi, respectively. DNA fragment of TYLCV, Y10, Y194, or BCTV capsid protein was used to quantify the accumulation of viral DNA.
4. Discussion
In the present study, we demonstrated that ty-5 confers broad-spectrum resistance to geminiviruses; it provided effective resistance to two representative begomoviruses present in China, TYLCCNV/TYLCCNB and TbLCYnV. ty-5 also conferred partial resistance to BCTV, a virus in the genus Curtovirus. Finally, ty-5 exhibited resistance to TYLCV co-infected with a betasatellite. Southern blotting and qPCR analyses showed that significantly less genomic DNA from these geminiviruses accumulated in the ty-5 line than in the susceptible one. Moreover, knockdown of Pelota expression converted an N. benthamiana plant from a geminivirus-susceptible host to a geminivirus-resistant host.
In Aedes aegypti, Pelota deficiency suppresses Drosophila C virus capsid protein synthesis and dengue replication [28,29]. In tomatoes, Pelota with a V16G mutation confers resistance to the begomovirus TYLCV [17]. It was recently reported that a single-nucleotide polymorphism (A to G) located at the splice site of the ninth intron of Pelota in BaPep-5 pepper confers resistance to pepper yellow leaf curl Indonesia virus and pepper yellow leaf curl Aceh virus [30]. A single base substitution (T556A) in the coding sequence of OsPelota confers bacterial blight resistance by activating the salicylic acid pathway [31]. A Pelota mutant also confers resistance to rice blast [32]. Here, we found that ty-5 with the V16G amino acid mutation in Pelota confers broad-spectrum resistance to various geminiviruses. Moreover, we found that knockdown of Pelota expression converted a N. benthamiana plant from a geminivirus-susceptible host to a geminivirus-resistant host. These results suggest that the Pelota gene is a good target for engineering to confer geminivirus resistance to various hosts. Geminiviruses cause diseases in many economically important crops, such as tomato, corn, maize, cassava, and cotton. Thus, ty-5 is a promising resistance gene that can be targeted in various crops. ty-5 only contains a single amino acid substitution, V16G, in Pelota. The introduction of this single amino acid change into Pelota might generate resistance lines in various crops. Although the necessary T to G gene editing cannot yet be conducted by the CRISPR/Cas9 method, future technology developments may enable sufficient editing for the conversion of geminivirus-susceptible crops to geminivirus-resistant crops.
Begomovirus/betasatellite complexes have recently emerged as causal agents for many economically important diseases. Many begomoviruses characterized in China are associated with betasatellites. We found that ty-5 conferred resistance to TYLCCNV/TYLCCNB. Furthermore, TYLCV reportedly can associate with a betasatellite [8,9,33]. Ty-1 is the most widely used resistance gene in tomato breeding. However, the resistance conferred by Ty-1 is compromised by TYLCV upon co-infection with a betasatellite. This suggests that the TYLCV/betasatellite complex is able to overcome the widely used Ty-1 resistance gene. However, our current findings indicate that the resistance conferred by ty-5 cannot be compromised by co-replication of TYLCV with a betasatellite; thus, ty-5 offers the potential to control this newly emerged TYLCV/betasatellite complex.
Currently, we are not aware of how a single mutation in Pelota confers resistance to geminiviruses. In both animals and plants, Pelota proteins are reportedly involved in mRNA surveillance [34,35]. One possible scenario is that the mutant PelotaV16G may confer greater resistance to geminiviruses. The other scenario is that WT Pelota may be required for viral replication or transcription and the mutant PelotaV16G may interfere with its ability to assist in viral replication or transcription. However, further efforts are required to dissect the role of Pelota in geminivirus infection. In conclusion, our findings suggest that ty-5 can confer effective resistance to various geminiviruses. ty-5 offers an important resistance gene resource for tomato crop breeding to control begomovirus/betasatellite complexes or other geminiviruses. Genome editing of Pelota also holds promise for the generation of geminivirus-resistant lines in other crops.
Supplementary Materials
The following supporting information can be downloaded at: www.mdpi.com/article/10.3390/v14081804/s1, Figure S1: Disease symptoms from grade I (no symptoms) to grade IV (very severe symptoms); Table S1: Primers used in this study.
Author Contributions
X.Z., X.Y. and D.L. conceived and designed the experiments; Y.R. and X.Y. performed experiments; Y.R., X.T., X.Y. and X.Z. analyzed the data and wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31720103914 and 31972245) and a project from the Yunnan Provincial Government (202205AF150047).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data used in this study are already provided in the manuscript in the required section. There are no underlying data available.
Acknowledgments
We thank Yinlei Wang (Jiangsu Academy of Agricultural Sciences) for providing tomato ty-5 seeds.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Davison, A.J.; Dempsey, D.M.; Dutilh, B.E.; García, M.L.; et al. Changes to virus taxonomy and to the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses. Arch. Virol. 2021, 166, 2633–2648. [Google Scholar] [CrossRef]
- Yang, X.; Guo, W.; Li, F.; Sunter, G.; Zhou, X. Geminivirus-associated betasatellites: Exploiting chinks in the antiviral arsenal of plants. Trends Plant Sci. 2019, 24, 519–529. [Google Scholar] [CrossRef]
- Rojas, M.R.; Macedo, M.A.; Maliano, M.R.; Soto-Aguilar, M.; Souza, J.O.; Briddon, R.W.; Kenyon, L.; Rivera Bustamante, R.F.; Zerbini, F.M.; Adkins, S.; et al. World management of geminiviruses. Annu. Rev. Phytopathol. 2018, 56, 637–677. [Google Scholar] [CrossRef]
- Hu, T.; Song, Y.; Wang, Y.; Zhou, X. Functional analysis of a novel βV1 gene identified by a geminivirus betasatellite. Sci. China Life Sci. 2020, 63, 688–696. [Google Scholar] [CrossRef]
- Rojas, M.R.; Hagen, C.; Lucas, W.J.; Gilbertson, R.L. Exploiting chinks in the plant’s armor: Evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 2005, 43, 361–394. [Google Scholar] [CrossRef]
- Zhou, X.; Xie, Y.; Tao, X.; Zhang, Z.; Li, Z.; Fauquet, C.M. Characterization of DNAβ associated with begomoviruses in China and evidence for co-evolution with their cognate viral DNA-A. J. Gen. Virol. 2003, 84, 237–247. [Google Scholar] [CrossRef]
- Cui, X.; Tao, X.; Xie, Y.; Fauquet, C.M.; Zhou, X. A DNAβ associated with tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 2004, 78, 13966–13974. [Google Scholar] [CrossRef]
- Yan, Z.; Wolters, A.A.; Navas-Castillo, J.; Bai, Y. The global dimension of tomato yellow leaf curl disease: Current status and breeding perspectives. Microorganisms 2021, 9, 740. [Google Scholar] [CrossRef] [PubMed]
- Voorburg, C.M.; Yan, Z.; Bergua-Vidal, M.; Wolters, A.A.; Bai, Y.; Kormelink, R. Ty-1, a universal resistance gene against geminiviruses that is compromised by co-replication of a betasatellite. Mol. Plant Pathol. 2020, 21, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, D. Co-dominant SCAR markers for detection of the Ty-3 and Ty-3a loci from Solanum chilense at 25 cM of chromosome 6 of tomato. Tomato. Genet Cooper 2008, 57, 25–29. [Google Scholar]
- Yang, X.; Caro, M.; Hutton, S.F.; Scott, J.W.; Guo, Y.; Wang, X.; Rashid, M.H.; Szinay, D.; Jong, H.; Visser, R. Fine mapping of the tomato yellow leaf curl virus resistance gene Ty-2 on chromosome 11 of tomato. Mol. Breed. 2014, 34, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Hutton, S.F.; Scott, J.W.; Schuster, D.J. Recessive resistance to tomato yellow leaf curl virus from the tomato cultivar Tyking is located in the same region as Ty-5 on chromosome 4. HortScience 2012, 47, 324–327. [Google Scholar] [CrossRef]
- Ji, Y.; Schuster, D.J.; Scott, J.W. Ty-3, a begomovirus resistance locus near the tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Mol. Breed. 2007, 20, 271–284. [Google Scholar] [CrossRef]
- Ji, Y.; Scott, J.W.; Schuster, D.J.; Maxwell, D.P. Molecular mapping of Ty-4, a new tomato yellow leaf curl virus resistance locus on chromosome 3 of tomato. J. Am. Soci. Hortic. Sci. 2009, 134, 281–288. [Google Scholar] [CrossRef]
- Zamir, D.; Ekstein-Michelson, I.; Zakay, Y.; Navot, N.; Zeidan, M.; Sarfatti, M.; Eshed, Y.; Harel, E.; Pleban, T.; van-Oss, H.; et al. Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, Ty-1. Theor. Appl. Genet. 1994, 88, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Hanson, P.M.; Green, S.K.; Kuo, G. Ty-2, a gene on chromosome 11 conditioning geminivirus resistance in tomato. Tomato Genet. Coop. Rep. 2006, 56, 17–18. [Google Scholar]
- Wang, Y.; Jiang, J.; Zhao, L.; Zhou, R.; Yu, W.; Zhao, T. Application of whole genome resequencing in mapping of a tomato yellow leaf curl virus resistance gene. Sci. Rep. 2018, 8, 9592. [Google Scholar] [CrossRef]
- Lapidot, M.; Karniel, U.; Gelbart, D.; Fogel, D.; Evenor, D.; Kutsher, Y.; Makhbash, Z.; Nahon, S.; Shlomo, H.; Chen, L.; et al. A novel route controlling begomovirus resistance by the messenger RNA surveillance factor pelota. PLoS. Genet. 2015, 11, e1005538. [Google Scholar] [CrossRef]
- Anbinder, I.; Reuveni, M.; Azari, R.; Paran, I.; Nahon, S.; Shlomo, H.; Chen, L.; Lapidot, M.; Levin, I. Molecular dissection of tomato leaf curl virus resistance in tomato line TY172 derived from Solanum peruvianum. Theor. Appl. Genet. 2009, 119, 519–530. [Google Scholar] [CrossRef]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.F.; Scott, J.W.; Edwards, J.D.; Bai, Y.; Mcdowell, J.M. The tomato yellow leaf curl virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA–dependent RNA polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef]
- Shen, X.; Yan, Z.; Wang, X.; Wang, Y.; Arens, M.; Du, Y.; Visser, R.G.F.; Kormelink, R.; Bai, Y.; Wolters, A.A. The NLR protein encoded by the resistance gene Ty-2 is triggered by the replication-associated protein Rep/C1 of tomato yellow leaf curl virus. Front. Plant Sci. 2020, 11, 545306. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Ohnishi, J.; Saito, A.; Ohyama, A.; Nunome, T.; Miyatake, K.; Fukuoka, H. An NB-LRR gene, TYNBS1, is responsible for resistance mediated by the Ty-2 Begomovirus resistance locus of tomato. Theor. Appl. Genet. 2018, 131, 1345–1362. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Tan, H.; Zhao, S.; Li, H.; Liu, H.; Ma, Y.; Zhang, X.; Rong, J.; Fu, X.; Lozano-Durán, R.; et al. Geminiviruses encode additional small proteins with specific subcellular localizations and virulence function. Nat. Commun. 2021, 12, 4278. [Google Scholar] [CrossRef] [PubMed]
- Stenger, D.C. Complete nucleotide sequence of the hypervirulent CFH strain of beet curly top virus. Mol. Plant Microbe Interact. 1994, 7, 154–157. [Google Scholar] [CrossRef]
- Xie, Y.; Zhao, L.; Jiao, X.; Jiang, T.; Gong, H.; Wang, B.; Briddon, R.W.; Zhou, X. A recombinant begomovirus resulting from exchange of the C4 gene. J. Gen. Virol. 2013, 94, 1896–1907. [Google Scholar] [CrossRef]
- Zhou, T.; Zhang, M.; Gong, P.; Li, F.; Zhou, X. Selective autophagic receptor NbNBR1 prevents NbRFP1-mediated UPS-dependent degradation of βC1 to promote geminivirus infection. PLoS Pathog. 2021, 17, e1009956. [Google Scholar] [CrossRef]
- Huang, C.; Xie, Y.; Zhou, X. Efficient virus-induced gene silencing in plants using a modified geminivirus DNA 1 component. Plant Biotechnol. J. 2009, 7, 254–265. [Google Scholar] [CrossRef]
- Wu, X.; He, W.T.; Tian, S.; Meng, D.; Li, Y.; Chen, W.; Li, L.; Tian, L.; Zhong, C.Q.; Han, F.; et al. Pelo is required for high efficiency viral replication. PLoS Pathog. 2014, 10, e1004034. [Google Scholar] [CrossRef]
- Asad, S.; Hussain, M.; Hugo, L.; Osei-Amo, S.; Zhang, G.; Watterson, D.; Asgari, S. Suppression of the pelo protein by Wolbachia and its effect on dengue virus in Aedes aegypti. PLoS Negl. Trop. Dis. 2018, 12, e0006405. [Google Scholar] [CrossRef]
- Koeda, S.; Onouchi, M.; Mori, N.; Pohan, N.S.; Nagano, A.J.; Kesumawati, E. A recessive gene pepy-1 encoding Pelota confers resistance to begomovirus isolates of PepYLCIV and PepYLCAV in Capsicum annuum. Theor. Appl. Genet. 2021, 134, 2947–2964. [Google Scholar] [CrossRef]
- Zhang, X.B.; Feng, B.H.; Wang, H.M.; Xu, X.; Shi, Y.F.; He, Y.; Chen, Z.; Sathe, A.P.; Shi, L.; Wu, J.L. A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J. Integr. Plant Biol. 2018, 60, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.; Fan, S.; Deng, L.; Zhong, G.; Zhang, S.; Li, M.; Chen, W.; Wang, G.; Tu, B.; Wang, Y.; et al. LML1, Encoding a conserved eukaryotic release factor 1 protein, regulates cell death and pathogen resistance by forming a conserved complex with SPL33 in rice. Plant Cell Physiol. 2018, 59, 887–902. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.F.; Rojas, M.; Kon, T.; Gamby, K.; Xoconostle-Cazares, B.; Gilbertson, R.L. A severe symptom phenotype in tomato in Mali is caused by a reassortant between a novel recombinant begomovirus (tomato yellow leaf curl Mali virus) and a betasatellite. Mol. Plant Pathol. 2009, 10, 415–430. [Google Scholar] [CrossRef]
- Szadeczky-Kardoss, I.; Csorba, T.; Auber, A.; Schamberger, A.; Nyiko, T.; Taller, J.; Orban, T.I.; Burgyan, J.; Silhavy, D. The nonstop decay and the RNA silencing systems operate cooperatively in plants. Nucleic Acids Res. 2018, 46, 4632–4648. [Google Scholar] [CrossRef] [PubMed]
- Arribere, J.A.; Fire, A.Z. Nonsense mRNA suppression via nonstop decay. eLife 2018, 7, e33292. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).