Abstract
Our knowledge of the diversity and evolution of the virosphere will likely increase dramatically with the study of microbial eukaryotes, including the microalgae within which few RNA viruses have been documented. By combining total RNA sequencing with sequence and structural-based homology detection, we identified 18 novel RNA viruses in cultured samples from two major groups of microbial algae: the chlorophytes and the chlorarachniophytes. Most of the RNA viruses identified in the green algae class Ulvophyceae were related to the Tombusviridae and Amalgaviridae viral families commonly associated with land plants. This suggests that the evolutionary history of these viruses extends to divergence events between algae and land plants. Seven Ostreobium sp-associated viruses exhibited sequence similarity to the mitoviruses most commonly found in fungi, compatible with horizontal virus transfer between algae and fungi. We also document, for the first time, RNA viruses associated with chlorarachniophytes, including the first negative-sense (bunya-like) RNA virus in microalgae, as well as a distant homolog of the plant virus Virgaviridae, potentially signifying viral inheritance from the secondary chloroplast endosymbiosis that marked the origin of the chlorarachniophytes. More broadly, these data suggest that the scarcity of RNA viruses in algae results from limited investigation rather than their absence.
1. Introduction
Viruses are likely to infect every cellular organism and play fundamental roles in biosphere diversity, evolution, and ecology. Those studies of the global virosphere performed to date have revealed marked heterogeneities in virus composition. For example, while RNA viruses are commonplace in eukaryotes, they are less often found in bacteria and are yet to be conclusively identified in archaea. Rather, both the bacteria and archaea are dominated by DNA viruses [1,2]. It is unclear, however, whether such highly skewed virus distributions reflect fundamental biological, cellular or ecological factors of the hosts in question, or because RNA viruses in bacteria and archaea are often so divergent in sequence that they are difficult to detect using primary sequence comparisons alone.
The advent of “omics” technologies has fueled more intensive efforts to assess global viral diversity, especially in marine environments [3,4,5,6,7]. However, despite the substantial increase in virus sampling, our picture of the virosphere remains largely restricted to bacteria, some animal lineages and plants [8,9,10,11,12]. Clearly, such a sampling bias also impacts our understanding of the fundamental patterns and processes of virus evolution. A good example of this major knowledge bias are the unicellular eukaryotes, grouped under the term “protists”, and particularly the microalgae. Despite recent efforts to document the RNA virome of marine micro-organisms [2,13,14], to date only 61 viruses have been formally recognized in microalgae [15] comprising just 82 viral sequences [16]. This represents only 0.6% of the total 14,679 viral sequences listed on the Viral-Host database (release April 2020).
Since the first cultivation in 1979 [17,18], the isolation and characterization of algal viruses (phycoviruses) has largely focused on those with DNA genomes [15,19] (including 55 of the 82 algal virus sequences available at VirusHostdb, release April 2020). These include the well-known giant viruses, the majority of which (53%) have been described in the green algae (Chlorophyta). The DNA-dominated virome of green algae contrasts with those of their sister-group, the land plants [20], for which 60% of the 3590 viral reference entries are RNA viruses (i.e., the “Riboviria”; VirusHostdb, April 2020 release). Most (85%) of the 27 RNA viruses described from algae to date have been identified in diatoms [21]. Although limited in number, the algal RNA viruses characterized thus far display impressive diversity, belonging to the families Totiviridae, Reoviridae, Marnaviridae, Endornaviridae, Flaviviridae, Narnaviridae and Alvernaviridae. It is currently unclear, however, whether the seemingly differing distributions of DNA and RNA viruses reflect a major switch in virus composition that occurred during the expansion of land plants or is indicative of the inherent limitations in sampling and cultivation of both algae and their viruses, or the difficulties in identifying highly-divergent RNA viruses [22]. Indeed, because RNA viruses are the fastest evolving entities described [23], phylogenetic signal is rapidly lost over evolutionary time. Hence, it is possible that the low number of algal RNA viruses detected to date simply reflects the fact that they are highly divergent in sequence, even in the canonical RNA-dependent RNA polymerase (RdRp), and hence refractory to detection using primary sequence similarity. Importantly, protein structures are expected to be an order of magnitude more conserved than amino acid sequences [24]. As a consequence, the study of conserved secondary or tertiary structures could help identify distant homologies among RNA viruses [25,26], including novel viruses within the microalgae.
Given the phylogenetic diversity and array of genomic features characterized to date in algal RNA viruses (with linear, circular, segmented, nonsegmented, single-strand and double-strand genomes), as well as their wide range of habitats and ubiquity, it is reasonable to expect that microalgae will harbour an abundance of RNA viruses. In addition, 72,500 species of microalgae have been identified to date distributed across diverse branches of the eukaryotic phylogeny (in the TSAR (Telonemia, Stramenopiles, Alveolates, Rhizaria), Archaeplastida, Haptista, Cryptista and “excavates” supergroups [27,28,29]), with estimates of the true number of species in excess of 300,000 [30,31]. Microalgae constitute a primary food source in the marine and freshwater food chain. Together with the ancient (ca. 1.8 billion years) nature of eukaryotic algae and their involvement in secondary plastid endosymbiosis events involving many branches of the eukaryotic phylogeny, it has been proposed that algal viruses played a key role in the early evolution of eukaryote viruses [15,32]. Revealing the nature of RNA virus diversity in algae may therefore have major consequences for understanding the processes that have shaped long-term virus biodiversity and evolution.
We aimed to reveal more of the RNA virosphere in cultured samples of two clades of microalgae: (i) the green algae (Chlorophyta), that are part of the Archaeplastida eukaryotic supergroup, and (ii) the chlorarachniophytes, a lineage of Rhizaria that obtained a chloroplast through secondary endosymbiosis of a green alga [33]. To this end we performed an unbiased (i.e., bulk RNA-sequencing) meta-transcriptomic analysis, with an emphasis on detecting remote signals of homology in the RdRp, the gene hallmark of RNA viruses, using protein-profile based approaches. The comparison of the viromes of these two distant groups enabled us to address a number of fundamental evolutionary questions: (i) is the deep divergence between the two algae taxa also reflected in their RNA virome compositions? (ii) does their RNA virome provide evidence for complex evolutionary histories, including horizontal transfer events? (iii) is the RNA/DNA virus bias between algae and green plants an artefact of sampling or reflect a more fundamental biological division? More generally, we aimed to broaden our understanding of the biodiversity of the algal virosphere as this may have implications for understanding and managing the roles played by algae in global element cycling, climate forcing and biotechnology, and as reservoirs for genetic novelty [34,35].
2. Materials and Methods
2.1. Algal Cultures
Algal strains were isolated from marine sand (Microrhizoidea pickettheapsiorum [36]; Kraftionema allantoideum [37]; Chlorarachnion reptans and Lotharella sp. from the Wye River, Victoria, Australia) or coral skeletons (Ostreobium sp. HV05007, Kavieng, Papua New Guinea), or obtained from the NIVA Culture Collection of Algae (Dolichomastix tenuilepis, SCCPA strain K-0587). Cultures were maintained in K-enriched seawater medium (transferring every other week) at either 26 °C (Ostreobium) or 16 °C (all others) under cool white LED lights at 1 Photosynthetic active radiation (PAR) (Ostreobium) or 15 PAR (all others). Cultures were pelleted by centrifugation in falcon tubes and stored in RNAlater at −80 °C until RNA extraction.
2.2. Total RNA Extractions
For total RNA extractions, RNAlater was removed by low centrifugation, algal cells were disrupted using thaw/freezing cycles and bead beating (0.1–0.5 mm), and total RNA was extracted using the Qiagen ® RNeasy Plant mini kit following the manufacturer’s instructions.
For the initial meta-transcriptomic screening, RNAs were pooled into three groups: (i) the chlorophyta Dolichomastix tenuilepis and Microrhizoidea pickettheapsiorum (Mamiellophyceae) and Kraftionema allantoideum (Ulvophyceae) were pooled into meta-transcriptome ‘ALG_1′; (ii) the ulvophyte Ostreobium sp. comprised ‘ALG_2′; and (iii) the two chlorarachniophytes Chlorarachnion reptans and Lotharella sp. were pooled into ‘ALG_3′ (Table 1).

Table 1.
Sample and library description.
2.3. Total RNA Sequencing
RNA quality was assessed and TruSeq stranded libraries were synthetized by the Australian Genome Research Facility (AGRF), using either (i) TruSeq stranded with a eukaryotic rRNA depletion step (RiboZero Gold kit, Illumina) for ALG_2, or (ii) the SMARTer Stranded Total RNA-Seq Kit v2—Pico Input Mammalian libraries (Takara Bio, Mountain View, CA, USA) for ALG_1 and ALG_3 (Table S1), due to the low amount of input RNAs in these libraries. The resulting libraries were sequenced on an Illumina HiSeq2500 (paired-end, 100bp) at the AGRF. Library descriptions and RNA-seq statistics are summarized in Table S1.
2.4. In Silico Processing of Meta-Transcriptomic Data
2.4.1. Read Depletion and Contig Assembly
The RNA-seq data were first subjected to low-quality read and Illumina adapter filtering using the Trimmomatic v0.36 program [38]. Ribosomal RNA was depleted with the SortmeRNA v3.0.3 program [39] using the SILVA v32 database [40], which removed between 86 and 94% of the total unfiltered reads (Figure S1A/2). Read-depleted libraries were then de novo assembled using the Trinity v2.5.1 program [41] and contigs shorter than 200 nt were removed (the average length of contig assembly is shown in Figure S1B/2). Contig abundances were calculated from the RNA-seq data using the Expectation–Maximization (RSEM) v1.3.1 software [42] and expressed as the expected read counts. An analysis of the assembly quality was attempted by estimating the proportion of full-length transcripts in each library using the “analyze_blastPlus_topHit_coverage.pl” script available in Trinity package. Briefly, this analysis consists in aligning all the transcripts obtained after each de novo assembly against the SwissProt/UniProt database using BLASTx and extracting the number of proteins aligned depending on their level of coverage (percentage of the top-hit sequence).
2.4.2. RNA Virus Detection Using BLASTx and BLASTn
The similarity of contigs to the current NCBI nucleotide (nt) and protein (nr) databases was determined using the BLASTn v2.2.30 and Diamond BLASTx v0.9.32 programs [43], respectively, employing 10−10 and 10−05 as e-value cut-offs and the more sensitive option in BLASTx. RNA virus-like sequences were also identified using BLASTx against all RdRp protein sequences available on NCBI/GenBank. False-positive signals for RNA viruses were removed by BLASTing RdRp-like sequences against the nr database and discarding sequences displaying a nonviral sequence as the best hit, based on BLASTx scores.
2.4.3. RNA Virus Profile-Based Homology Detection
To detect especially diverse RdRp-based sequences, orphan contigs (i.e., those with no match in either the nr and nt databases) were compared to the Pfam RdRp-protein profiles ‘MitoVir_RdRp’ (PF05919), ‘Birna_RdRp’ (PF04197), ‘Viral_RdRp_C’ (PF17501), ‘RdRP_1′ (PF00680), ‘RdRP_2′ (PF00978), ‘RdRP_3′ (PF00998) and ‘RdRP_4′ (PF02123), as well as to the entire VOG profile database (http://vogdb.org) [44] using the HMMer3 v3.3 program [45]. To check for false-positive signals, these orphan sequences were submitted to the entire Pfam database using the same HMMer version and default parameters.
2.4.4. D Protein Structure Prediction of RdRp-Like Contigs
To infer a structural model for the distant RdRp signals detected using profiles, sequences displaying a RdRp-like signal were subjected to the normal mode search of the Protein Homology/analogY Recognition Engine v 2.0 (Phyre2) web portal [46]. Briefly, this program first compares the submitted amino acid sequence to a curated nonredundant nr20 data set using HHblits [47]. It then converts the conserved secondary structure information as a query against known 3D-structures using HHsearch [48]. A final structural modeling step based on identified structural homologies is performed as described previously [46].
2.4.5. RNA Virus Sequence Analysis and Annotation
Total non-rRNA reads were mapped onto RNA virus-like contigs using Bowtie2 v2.3.3.1 and heterogeneous coverage and potential mis-assemblies were manually resolved using Geneious v11.1.4 [49]. Open reading frames (ORFs) were first predicted using getORF from EMBOSS v6.6.0, in which ORFs were defined as regions that are free of stop codons (−find 0 option), although partial sequences (i.e., missing start or stop codons) were retained for analysis. Protein domains were annotated using the InterProscan software package from EMBL-EBI, using the InterPro consortium databases PROSITEpatterns v2019_11, PROSITEprofiles v2019_11, PRINTS v42.0, Pfam v33.1, PIRSF v3.10, TIGRFAM v15.0, SuperFamily, CDD v3.17 and PANTHER v14.1 (https://github.com/ebi-pf-team/interproscan).
2.4.6. Revealing Host-Virus Associations
A challenge faced by all metagenomic studies is confidently assigning each viral sequence to a particular host in a given sample. We used algal cultures to minimize the number of potential additional cellular hosts. These cultures were, however, nonaxenic (i.e., cultures not purified from other contaminating organisms), with mainly bacteria present. To evaluate the possibility of additional microeukaryotic cells in the sample, we obtained taxonomic identification for contigs in the meta-transcriptome by aligning them to the NCBI nt database using the KMA aligner v1.2.11a and the CCMetagen program [50,51] v1.1.3. Contigs matching an entry in the nt database were displayed as Krona plots and classified based on their taxonomy (using high taxonomic levels for clarity).
2.4.7. Phylogenetic Analysis
RdRp amino acid sequences were aligned using the L-INS-I algorithm and default parameters in the MAFFT program v7.402 [52] and trimmed with TrimAI v1.4.1 (automated1 model). Maximum likelihood phylogenetic trees were then estimated using IQ-TREE v2.0-rc1, employing ModelFinder to obtain the best-fit model of amino acid substitution in each case, with nodal support assessed using 1000 bootstrap replicates and 1000 replicates of SH-like approximate likelihood ratio test (SH-aLRT) [53,54]. For each tree, reference genomes and corresponding RdRp sequences were retrieved from the NCBI viral genome resource (https://www.ncbi.nlm.nih.gov/genome/viruses/). To depict the evolutionary relationships of the newly discovered viruses as meaningfully as possible, the closest unclassified BLASTx homologs were used in the phylogenetic analysis. This resulted in alignments of 237, 76, 198, 558 and 325 RdRp protein sequences for Amalga-like, Mito-like, Tombus-like, Virga-like and Bunya-like virus groups, respectively.
2.5. RT-PCR Validation
Viral contigs were validated experimentally and associated to individual algal sample using Reverse Transcription (SuperScript ™ IV reverse transcriptase – Invitrogen, Carlsbad, CA, USA ™) followed by PCR (Platinum ™ SuperFi ™ DNA polymerase – Invitrogen ™), with specific primer sets for each contig. The rbcL and tufA marker genes were used as PCR positive controls using sets of primers designed in [55]. All primers and PCR conditions used in this study are described in Table S2.
2.6. Data Availability
The libraries sequenced here are available at the Sequence Read Archive (SRA) under BioProject PRJNA668187. The consensus sequences of the all novel viruses identified here have been submitted to GenBank and assigned accession numbers MW086576-MW086593.
3. Results
3.1. The RNA Viromes of Two Divergent Groups of Microalgae
Our aim was to determine the RNA viromes of six microalgae cultures from six different algal species classified into two highly phylogenetically distinct algal clades: the chlorarachniophytes (Rhizaria) and the green algae (Chlorophyta, Archaeplastida) (Figure 1A,B). To the best of our knowledge, this is the first identification of viruses in samples from the Chlorarachniophyceae (Figure 1C) [15].
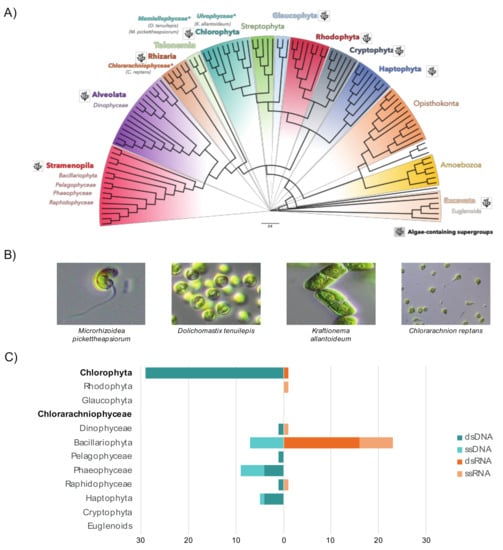
Figure 1.
The enormous diversity of algae contrasts with their poorly characterized viromes. (A) Representation of algae supergroups among the diversity of eukaryotes (latest eukaryotic classification retrieved from [29]). The phylogenetic tree was adapted from [56]. Pictures illustrate some of the samples used in this study and corresponding clades are marked with “*”. (B) Pictures of algae cultures used in this study. (C) The current extent of the microalgae virosphere. The viral sequence counts for each virus class (DNA or RNA, single-stranded or double-stranded) were retrieved from VirusHostdb [16] according to 11 major eukaryotic algae lineages. Microalgal lineages investigated in this study are highlighted in bold.
While our limited understanding of chlorarachniophyte viruses can be explained by the small number of species characterized to date (only 15 in Algaebase database), the Ulvophyceae is an abundant and diverse algal lineage in existence since the late Proterozoic and comprises at least 1933 species [57]. It contains a wide range of morphologies from unicellular benthic algae to large seaweeds [58] and its representatives commonly occur in marine, terrestrial and freshwater habitats [20]. We therefore performed RNA sequencing (meta-transcriptomics) on six microalgal species belonging to both the green algae (classes Ulvophyceae and Mamiellophyceae) and chlorarachniophytes (Table 1).
Because of variable RNA quality and quantity, fewer nonrRNA reads were obtained for the ALG_1 and ALG_3 libraries. This may in large part explain the limited length of contigs, the reduced number of estimated full-length transcripts, and ultimately the lower number of viral sequences, compared to ALG_2 (Figure S3).
In total, we identified 18 new putative viral sequences using a standard sequence similarity search among the three libraries. These largely comprised viruses with double-stranded (ds) RNA or single-stranded positive-sense (ssRNA(+)) genomes (Table 2). However, a divergent bunya-like partial sequence was also retrieved from Chlorarachnion reptans and may constitute the first negative-sense (ssRNA(−−−)) virus identified in microalgae. Importantly, the presence of these viruses was validated by RT-PCR on each total extracted RNA (Figure S1). In each case these viruses exhibited very low levels of sequence similarity to existing RdRp amino acid sequences, with sequence identities ranging from only 27 to 38%. With the exception of Virga-like bellevilovirus, Bunya-like bridouvirus and Amalga-like boulavirus for which partial sequences have been retrieved, the length and genomic organization (ORF numbers, predicted protein length, etc.) of all of the new viruses described in this study are similar to the well-annotated full-length genomes of reference homologs. It is therefore likely that they correspond to full-length genomes. In addition, the lack of frameshifts and premature stop codons means these sequences are very likely true (exogenous) viruses rather than endogenous viral elements (EVEs) inserted into host genomes.

Table 2.
BLASTx results of the newly described virus-like sequences against nr database.
Eight of the viruses identified in Ostreobium sp. fell within the Narnaviridae or Tombusviridae. In contrast, five viral sequences from Ostreobium sp. and K. allantoideum do not fit into defined taxonomic groups and were instead related to the broad set of ‘partiti-like’ viruses that comprise the Partitiviridae, Totiviridae and Amalgaviridae. Finally, more divergent but detectable sequence similarities to the Virgaviridae (+ssRNA) were obtained for samples from the chlorarachniophyte library.
3.2. Detection of Divergent Viruses Using Protein Structural Data
An additional attempt to detect even more divergent RNA viruses was conducted was using protein structure. In particular, it is possible that highly divergent viruses are part of the unknown orphan sequences (i.e., contigs with no match in nt/nr databases, or the ‘dark matter’) that comprise between 50–60% of total contigs obtained in this study (Figure 2A).
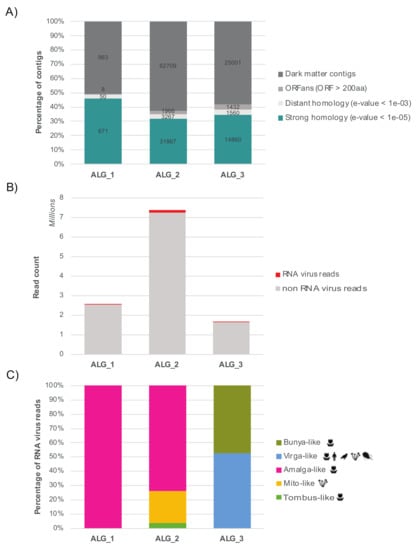
Figure 2.
Abundance of unknown and RNA virus-like contigs detected in the algal libraries. (A) Percentage of nonassigned contigs. For clarity, numbers are normalized as the percentage of total contigs (actual contig numbers are indicated in bold). Blue: number of contigs showing strong sequence similarity to the nr database (e-value < 10−05); light grey: contigs showing weak sequence similarity to the nr database (e-values 10−05 to 10−03); middle-dark grey: contigs with no sequence similarity detected by BLASTx/BLASTp but predicted to encode one or more ORFs longer than 200 amino acids (600 nt); dark grey: genomic ‘dark matter’ - contigs without any signal detected or any long ORFs encoded. (B) Total number of RNA virus reads per total number of non-rRNA reads in each library. (C) Distribution of RNA virus diversity in the three libraries and percentage of RNA virus reads associated with each viral super-clade. The host range is represented for each viral clade.
Accordingly, we attempted to detect evolutionary-conserved features of protein structural and functional motifs in orphan sequences that encode unknown ORFs, using a cut-off of 200 amino acids (600 nucleotides): we chose this size because it is shorter than most RdRps [59] yet should be long enough to make evolutionary inferences. The corresponding translated ORFs were compared to protein profiles from the PFAM RdRp clan and the VOG databases using the hidden-Markov model-based HMMer3 program. To help exclude false-positives, all positive hits were compared to the entire PFAM database. This resulted in the identification of three nonphage contigs that displayed homology to the RNA virus RdRp: ALG_2_DN19089, ALG_2_DN594 and ALG_3_DN34624 (Table 3).

Table 3.
Results of the VOGdb and PFAM HMM analysis. Light pink: phage-like sequences. Grey: nonviral sequences; Light blue: DNA virus-like sequences; Orange: RNA virus-like sequences. Abund: expected read counts estimated using the RSEM program.
To manually assess the level of confidence of the RdRp signal detected in the HMM comparisons, the protein sequences of the three RdRp candidates were aligned to amino acid sequences retrieved from the RdRp_C, RdRp_4 and RdRp_1 PFAM profiles. The ALG_2_DN594 contig displayed similarities with the RdRp_C profile that represents the C-terminal of the RdRp (Protein A) found in alphanodaviruses. Unfortunately, this C-terminal region lacks the key functional motifs usually associated with the RdRp, preventing us from definitively establishing the ALG_2_DN594 contig as a true RNA virus. Similarly, the ALG_3_DN34624 alignment with the viral RdRp sequences that comprise the PFAM RdRp_4 profile (PF02123) does not show conservation of the crucial functional residues at the A, B and C-motifs within the RdRp, particularly the canonical motif C that is normally GDD, yet GFD in ALG_3 contig (Figure S2B): strikingly, the GFD motif is absent from an alignment of 4627 viral RdRp sequences [60]. Whether this reflects a newly identified functional motif remains to be determined, but we cannot safely conclude that ALG_3_DN34624 encodes a viral RdRp. In contrast, the ALG_2_DN19089-encoded ORF shared motifs with RdRp_1 profile (PF00680), including motif A at positions 437–442, motif B at positions 507–517 and a GDD motif C at positions 557–559 of the RdRp alignment (Figure S2A). Because of the presence of these functional motifs, ALG_2_DN19089 can be confidently considered as a true RdRp-encoding contig and will be referred to here as ‘Partiti-like adriusvirus’. Interestingly, this contig also revealed significant similarity to some eukaryotic chloroplast-associated double-stranded RNA replicons (BDRC) obtained from the green algae species Bryopsis cinicola [61] (Table 2). It is therefore likely that these BDRC dsRNA Bryopsis-replicons in fact represent viral RdRp sequences [22], and we treat them as such in this study.
An additional BLASTx comparison using this divergent Partiti-like RdRp as a reference identified two other BDRC-like contigs in the Ostreobium sp. data set—ALG_2_DN19300 and ALG_2_DN19436 (Table 2). Along with the Partiti-like adriusvirus, these two additional sequences were both validated by RT-PCR (Figure S1) and are listed in the viral contig table as ‘Partiti-like lacotivirus’ and ‘Partiti-like alassinovirus’, respectively (Table 2).
The remaining hits from the RdRp-profile analysis—ALG_2_DN594_c0_g2_i1_len711 and ALG_3_DN34624_c0_g1_i1_len2077—were used in a Phyre2 protein structural analysis. However, this revealed no confident identification of a viral RdRp (i.e., the confidence levels of structural models obtained were <90%).
3.3. Relative Abundance and Prevalence of RNA Viruses in the Samples
Relative virus abundance varied between libraries and viruses of the same family in the Ostreobium sp. culture, with viral-like sequences constituting between 0.01 (considered as average abundance) and 1.2% (considered as very high abundance) of the total non-rRNA reads (Table 2, Figure 2). Each virus described was identified in only one of the cultures sequenced (Figure S1). However, intersample BLAST-comparisons revealed similarity between the partiti-like sequence identified in the K. allantoideum and Ostreobium samples (30% amino acid identity). A nonannotated ORF from a K. allantoideum contig, ALG_1_DN2506, aligned with the N-terminal ORF of Ostreobium sp. amalga-like virus contigs that potentially encode the virus coat protein, but the high level of divergence prevented us from performing any phylogenetic analysis of proteins other than the RdRp. Because of their co-occurrence in K. allantoideum (1.1 and 1.2 PCR, Figure S1) and their similar abundance levels, it is likely that ALG_1 DN2506 and ALG_1 DN2691 (referred as ‘Amalga-like boulavirus’, Table 2) contigs are part of the same genome. Unfortunately, the poor quality of RNAs and the resulting high degree of fragmentation obtained in the ALG_1 RNA-seq library did not allow us to resolve this question.
Notably, a large majority of the new RNA viruses reported here come from the ulvophyte Ostreobium sp. clonal culture, although this may in part result from differences in RNA quality and sequencing rather than a true biological difference in RNA virome composition and diversity (Figure S3). Difficulties in detecting highly divergent viral sequences, especially in poorly characterized and distant clades such as the chlorarachniophytes, may also contribute to the different numbers of viruses observed between libraries.
3.4. Detection of Possible Secondary Hosts
As the algal cultures analyzed here were not axenic, we assessed the diversity and relative abundance of other potential eukaryotic organisms in these samples. Indeed, algae cultures are commonly co-cultured with bacteria, fungi and other endosymbiotic algae [62]. The rRNA depletion performed during the RNA-Seq library preparation prevented us from using standard 16S/18S profiling. We therefore evaluated the presence of other eukaryotes in the samples using CCMetagen [50]. According to the Krona plots obtained for each library, cultivated algae were, as expected, the dominant organism found in the samples, representing between 79–99% of all assigned contigs (Files S1–S3). Nevertheless, a small proportion (2–8%) of contigs from ALG_1 and ALG_3 were assigned to dinoflagellates and Cyanophora algae (Files S1 and S3). Although sequences assigned to Lingulodinium polyedrum potentially result from GenBank mis-annotation and were likely of bacterial origin, the Coolia malayensis (Dinophyceae), Amphidinium sp. (Dinophyceae) and Cyanophora paradoxa (Glaucophyta) associated sequences likely constitute true assignments. We therefore suspect that these additional micro-eukaryotic transcripts may have arisen from cross-contamination with additional algae samples that were extracted and prepared at the same time and sequenced in the same run. Importantly, however, none of the viruses identified in the three libraries studied here could be detected in the transcriptomes of co-processed Dinophyceae and Glaucophyta cultures, suggesting that these low-abundance contaminants are not the hosts of the viruses reported here. In addition, a minor portion of the contigs in ALG_2 and ALG_9 was assigned to bacterial species (0.3% and 5%, respectively; Supplementary Files S2 and S3). To prevent any misinterpretation, particular care was taken to remove bacteriophage-like signals from the final virus-like sequence files.
3.5. Phylogenetic Analysis of the Newly Identified Viruses
3.5.1. Partiti-Like dsRNA Viruses
Eight of the newly described viral sequences exhibited RdRp amino acid sequence similarity to Partiti-like viruses (i.e., relatives of the Partitiviridae) and found at various levels of abundance (Table 2). Based on phylogenetic studies, five of the viral-like sequences are close to those from the Amalgaviridae (named after their mosaic status comprising both a Partitiviridae-like RdRp and the dicistronic and monopartite genomic organization of the Totiviridae [63]) and form a clade with the Bryopsis mitochondria-associated dsRNA (BDRM), although they share only 28–32% sequence identity (Table 2, Figure 3). BDRM was first described as a dsRNA associated with mitochondria in Bryopsis cinicola macroalgae [64] and later classified as a virus by the International Committee on Taxonomy of Viruses (ICTV). Like Ostreobium and Kraftionema, Bryopsis belongs to the class Ulvophyceae, and it seems likely that all these five newly identified Amalga-like viral sequences (Figure 3) form an Ulvophyceae-infecting viral clade.

Figure 3.
RdRp phylogeny of the newly identified chlorophyte viruses among the Amalgaviridae, Partitiviridae, Picobirnaviridae and Hypoviridae. Sequences identified in this study are labeled in red. Unclassified sequences from [65] are highlighted in grey. For clarity, some families and genera have been collapsed. Left, phylogenetic tree estimated using IQ-Tree with bootstrap replicates and SH-aLTR set to 1000 (values in parenthesis). Right, genomic organization of both viral genomes identified in this study (red) and representative species of each major family and genus used in the phylogeny (Cryphonectria hypovirus 2—Hypoviridae; Chicken picobirnavirus—Picobirnaviridae; Southern tomato virus—Amalgaviridae; Cryptosporidium parvum virus 1—Cryspovirus; Discula destructiva virus 1—Gammapartitivirus; Figure cryptic virus—Deltapartitivirus; Ceratocystis resinifera partitivirus—Betapartitivirus; White clover cryptic virus 1—Alphapartitivirus. The tree is mid-pointed rooted and branch lengths are scaled according to the number of amino acid substitutions per site.
Given the level of RdRp pairwise identity between these sequences (Table S3, top) we assumed that each constituted a new species. To perform a preliminary taxonomic assessment, we used the PAirwise Sequence Comparison (PASC) tool available at the NCBI [66]. Each of these five new BDRM-like viral genome sequences were compared with the Amalgaviridae full-length genomes available. The closest matches to existing Amalgaviridae sequences were retrieved for each newly discovered virus, and the resulting pairwise identity distributions compared with those observed within and between Amalgaviridae genera (Figure 4A). While this analysis indicates that these newly identified virus sequences are not part of any existing Amalgaviridae genus (Figure 4A), whether they can be considered as a new genus within the Amalgaviridae, or even a new family, is currently unclear and will require formal taxonomic assessment by the ICTV.
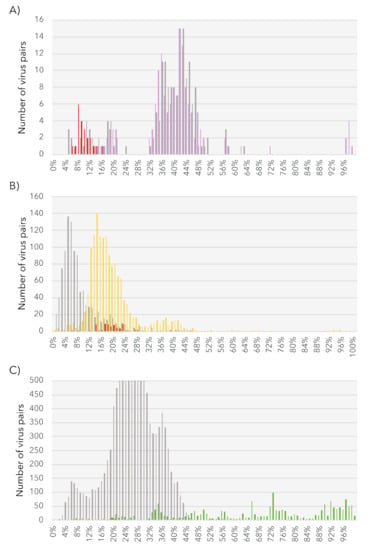
Figure 4.
Genome pairwise identity distributions of the new algal viral sequences. The level of pairwise identity between the newly identified viruses and existing members of each viral family are represented in red. (A) Intergenus (grey) and intra-genus (purple) identity levels within the Amalgaviridae. (B) Intergenus (grey) and intra-genus (yellow) identity levels within the Narnaviridae. (C) Intergenus (grey) and intra-genus (green) identity levels within the Tombusviridae.
Interestingly, if these Amalga-like sequences are translated into amino acids using the protozoan mitochondrial code they display the same genomic organization as BDRM, encoding two overlapping ORFs: the 5′ one encoding a hypothetical protein and the other encoding a replicase through a -1 ribosomal frameshift [67] (Figure S4). However, it is unclear if these sequences should be translated using the standard cytoplasmic code, and such sub-cellular localization remains to be validated. It is also notable that the two closest homologs of BDRM, the Amalga-like dominovirus and Amalga-like chaucrivirus, also contain the GGAUUUU ribosomal-1 frameshift motif and the two encoded ORFs could plausibly be translated in this manner (Figure S4).
The length and two-ORF encoding genomic structure of the BDRM-like sequences generally correspond to genomic features of the amalgaviruses (Figure 3, right). Despite a lack of detectable sequence similarity at both the sequence (BLASTx) and structural levels (i.e., the Phyre2 analysis), the second ORFs predicted in these amalgavirus-like sequences are expected to encode a CP-like protein, even if the involvement of this potential CP in encapsidation remains unclear [68].
The three BDRC-like sequences identified from Ostreobium sp. also cluster with the Partiti-like viruses (Figure 3, Table 2) and can be classified as three different species after applying the commonly-used 90% RdRp percentage identity species demarcation criteria (Table S3, Middle). Notably, the genomic organization of these BDRC-like contigs seems “inverted” compared to members of the Totiviridae and Amalgaviridae; that is, a first ORF encoding a CP protein is followed by a second that represents the RdRp (Figure 5).
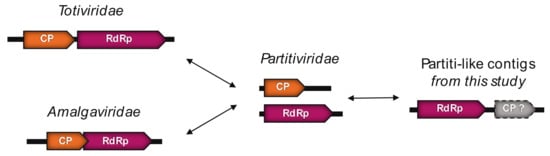
Figure 5.
Genomic organization of the Partitiviridae, Totiviridae and Amalgaviridae. Possible evolutionary scenarios for the BDRC-like contigs observed in Ostreobium sp.
Indeed, the RdRp encoded by the Partiti-like ALG_2 contigs is close to the 5′ extremity, followed by a second ORF. This second ORF could potentially encode a CP, although functional annotation could not be achieved due to the high level of sequence divergence.
3.5.2. Mitovirus-Like ssRNA(+) Viruses
Seven viral sequences from Ostreobium sp. clustered in the Narnaviridae, forming a clade within the genus Mitovirus (Figure 6). With their single ORF likely encoding an RdRp, and a genome length of ~3000 nt that is typical of the Narnaviridae (Figure 6, right), and the relatively high levels of abundances (Table 2), these viral sequences very likely constitute replicating viruses and thus represent a newly-described clade of protist-associated mitoviruses potentially restricted to green microalgae. The closest relative virus identified, Shahe Narna-like virus 6, was isolated from freshwater small planktonic crustaceans (Daphnia magna, Daphnia carinata and Moina macrocopa) belonging to the order Cladocera [65]. These “grazer” animals feed on marine microorganisms including microalgae, and it is therefore possible that Shahe Narna-like virus 6 in fact infects ingested-algae rather than arthropod host, in a similar manner to other members of the Narnaviridae.
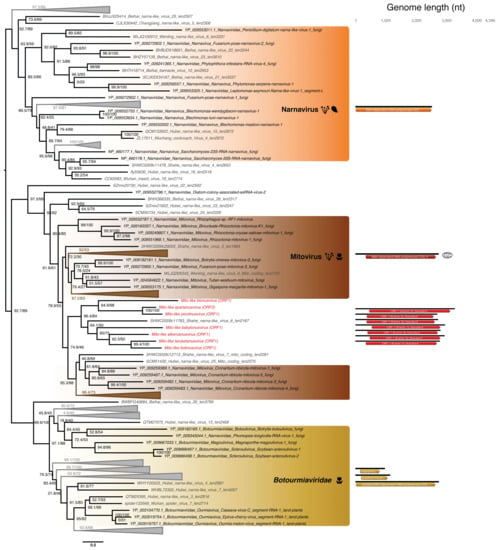
Figure 6.
Phylogeny of the Narnaviridae-Botourmiaviridae group based on the RdRp. Newly discovered viruses from Ostreobium sp. are highlighted in red. RdRp sequences from unassigned RNA virus retrieved from [65] are marked in grey. Left, phylogenetic tree estimated using IQ-Tree with bootstrap replicates and SH-aLTR set to 1000 (values in parenthesis). Right, genomic organization of both viral genomes identified in this study (red) and representative species of each major family and genus used in the phylogeny (Cassava virus C—Botourmiaviridae; Saccharomyces 23S RNA narnavirus—Narnavirus genus; Chenopodium quinoa mitovirus 1 Mitovirus genus). Annotations of Cassava virus C coding sequences: RdRp (Segment I); Putative movement protein (Segment II); Coat protein (Segment III). Branch lengths are scaled according to the number of amino acid substitutions per site.
Based on the 50% RdRp sequence identity used as a species demarcation criterion in the Narnaviridae (ICTV report 2009), we identified seven new mito-like viral species (Table S3, bottom). To place of these new species among the Narnaviridae we performed a PASC analysis using the putative full-length genomes from the seven new viral species. This revealed that the identity levels of the new sequences fell in the range expected of intra-genus diversity (Figure 4B). We therefore propose the existence of a new subgroup of mitoviruses, comprising these seven new species as well as the Shahe Narna-like virus 6 (Figure 6). Whether this clade is associated with mitochondria is currently unclear, and predicted ORFs were obtained using both standard and mitochondrial genetic codes.
3.5.3. Tombusviridae-Like ss(+)RNA Viruses
One sequence from Ostreobium sp., the Tombus-like chagrupourvirus, exhibited similarity to members of the Tombusviridae family of ssRNA(+) viruses (Figure 7), grouping with viruses previously identified as infecting plant or plant pathogenic fungi. This could again illustrate a shared evolutionary history between green algae and land plants, and that horizontal virus transfers can occur between plants and their pathogenic fungi. Of note is that the closest relative of Tombus-like chagrupourvirus documented to date, Hubei-Tombus-like virus 12, was isolated from freshwater animals (mollusca Nodularia douglasiae) [65]. According to the lack of distinguishable animal-related contigs in the Ostreobium sp. sample (File S2), and the average abundance level associated to this virus (0.7% of total non rRNA reads, Table 2), we assume this Hubei-Tombus-like virus 12 may, together with the newly tombus-like sequence identified here, constitute a new clade of green algae-infecting viruses.

Figure 7.
Phylogeny of the Tombusviridae RdRp. The tombus-like sequence identified in this study is labeled in red. Unclassified sequences from [65] are highlighted in grey. For clarity, some families and genera have been collapsed. Left, phylogenetic tree estimated using IQ-Tree with bootstrap replicates and SH-aLTR set to 1000 (values in parenthesis). Right, genomic organizations of the new viruses as well as their closest homologs and representative species from each family/genus as follows: Black beetle virus (Nodaviridae); Carrot mottle virus (Dianthovirus); Carnation ringspot virus (Dianthovirus); Beet black scorch virus (Betanecrovirus); Cucumber leaf spot virus (Aureusvirus); Maize necrotic streak virus (Tombusvirus); Carnation mottle virus (Alphacarmovirus). The tree is mid-pointed rooted and branch lengths are scaled according to the number of amino acid substitutions per site.
The 3.8 kb genome length of the Tombus-like chagrupourvirus is similar to those commonly observed in Tombusviridae and their relatives (Figure 7, right), suggesting that it comprises a full-length genome for this virus. This putative full-length genome sequence was compared to Tombusviridae reference genomic sequences using PASC to assess its taxonomic position (Figure 4C). Accordingly, the Ostreobium-associated tombus-like sequence could constitute a new Tombusviridae genus.
3.5.4. Virgaviridae-Like ssRNA(+) Viruses
One sequence identified in the Chlorarachnion reptans culture displayed detectable sequence similarity to the Virgaviridae-like RdRp supergroup (Figure 8) and is present at average abundance in the library (0.01% of all non-rRNA reads). The family Virgaviridae comprise ssRNA(+) viruses traditionally associated with plants and display diverse genomic organizations. The short length of the Virga-like bellevillovirus associated with chlorarachniophytes indicates that this sequence likely comprises a partial genome sequence only. Moreover, the multi-segment structure of the closest relatives suggests that the partial genome recovered in ALG_3 could also contain additional segments not yet identified. Although further host confirmation is required, this newly described RNA virus-like sequence would constitute the first algae virus from the Hepe-Virga group.
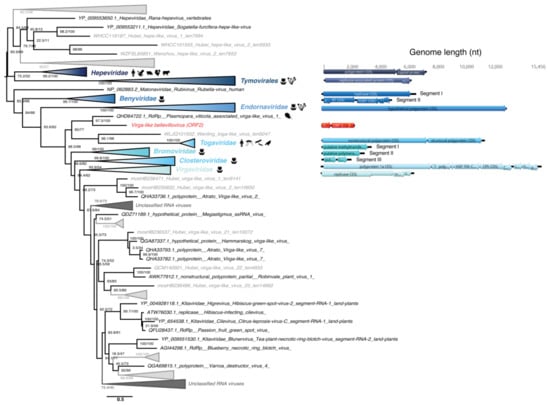
Figure 8.
Phylogeny of the Hepe-Virga group RdRp. The hepe-virga-like sequence identified in this study is labeled in red. Unclassified sequences from [65] are highlighted in grey. For clarity, some families or genera have been collapsed. Left, RdRp-based phylogenetic tree obtained using IQ-tree with bootstrap replicates and SH-aLTR set to 1000 (values in parenthesis). Right, genomic organizations of the new viruses as well as closest homologs and representative species from each family/genus as follows: Orthohepevirus A (Hepeviridae); Poinsettia mosaic virus (order Tymovirales); Wheat stripe mosaic virus (Benyviridae); Diatom colony associated dsRNA virus 15 (Endornaviridae); Cabassou virus (Togaviridae); Apple mosaic virus (Bromoviridae); Mint virus 1 (Closteroviridae); Cucumber mottle virus (Virgaviridae). The tree is mid-pointed rooted and branch lengths are scaled according to the number of amino acid substitutions per site.
3.5.5. Bunyavirales-Like ss(-)RNA Viruses
A partial viral genome, denoted Bunya-like bridouvirus, encoding a RdRp-like signal was identified in the Chlorarachnion reptans sample at an abundance of 0.01% of total non rRNA reads. However, this sequence is highly divergent and cannot be formally assigned to any previously described viral family. Despite this, it is striking that the sequence clusters with a Bunya-Arena-like virus, Shahe bunya-like virus 1 (Figure 9) previously identified in diverse Freshwater small planktonic crustaceans (Daphnia magna, Daphnia carinata and Moina macrocopa) [65] that typically feed on algae. Our phylogenetic analysis places this sequence within the diversity of the order Bunyavirales (Figure 9). In addition to the freshwater organism-associated viruses identified in [65], this contig clusters with several bunya-like unclassified negative-strand viruses isolated from the fungi Cladosporium cladosporioides and the oomycete Plasmopara viticola (Figure 9) that are both plant pathogens. The multi-segment structure of the closest classified family, the Phenuiviridae, suggests that additional segments associated with the partial Bunya-like bridouvirus genome may exist in C. reptans.

Figure 9.
Phylogeny of the Bunyavirales RdRp. The bunya-like sequence identified in this study is labeled in red. Unclassified sequences from [65] are highlighted in grey. For clarity, some families and genera have been collapsed. Left, phylogenetic tree estimated using IQ-Tree with bootstrap replicates and SH-aLTR set to 1000 (values in parenthesis). Right, genomic organizations of the new viruses as well as their closest homologs and representative species from each family/genus as follows: Melon chlorotic spot virus (Phenuiviridae); Yogue virus (Nairoviridae); Latino mammarenavirus (Arenaviridae); Seattle orthophasmavirus (Phasmaviridae); Melon yellow spot virus (Tospoviridae); Fig mosaic emaravirus (Fimoviridae); Tataguine orthobunyavirus (Peribunyaviridae). The tree is mid-pointed rooted and branch lengths are scaled according to the number of amino acid substitutions per site.
4. Discussion
We aimed to better characterize the RNA virus diversity in two major algal lineages, the chlorarachniophytes and the ulvophytes, for which no RNA viruses had previously been reported. Our investigation of the RNA virus diversity in samples from six microalgae species led to the identification of 18 new and divergent RNA viruses, although with clear homology to five established viral families. While an unequivocal host assignment cannot be formally established on these metagenomic data alone, that the algae studied were the dominant host species in the metagenomic sequencing data, in one case (Ostreobium sp.) representing 99% of all assigned contigs, makes it likely that most if not all of these 18 viruses infect algae hosts. In addition, we identified a number of narnaviruses, a group previously observed in algae [69,70], and our observation of a Bunyavirales-like sequence is similarly in accord with a study that presented evidence for the presence of bunya/phlebo-like viruses in brown algae [71]. As such, the apparent domination of DNA viruses in microalgae at least partially reflects major sampling biases. The concept that there is a potentially large dark matter of algal viruses is further supported by the high proportion of unassigned contigs observed: we speculate that these likely contain a nonneglectable number of highly divergent viral reads.
4.1. RNA Virome Similarities between Green Algae and Land Plants
Among the 18 novel RNA virus species described here, seven of those detected in the green algae Ostreobium sp. and K. allantoideum were seemingly related to the Tombusviridae and Amalgaviridae families of plant RNA viruses. Such similarities in RNA virome composition between green algae and land plants are consistent with previous analyses based on the Plant Genome project transcriptomic data that identified partitivirus-like signals in Chlorophyte algae [22]. However, the very limited sequence similarity among these viral families strongly suggests an ancient divergence among them, perhaps even before the chlorophyte-streptophyte split some 850–1100 million years ago (Ma) [57]. The close link between land plant and green algae RNA virosphere is further supported by the recent observation that plant viruses are able to infect nonvascular plants such as mosses and algae [72,73].
The detectable sequence similarity observed between the Partitiviridae and Amalgaviridae also suggests that they share common ancestry [68], despite a wide range of hosts and genomic organizations. As such, it is important to determine whether amalgaviruses are restricted to plants [74,75,76] or, as the Partitiviridae, infect many divergent eukaryotic hosts such as fungi, plants and protists [77,78].
4.2. Divergent Homologs to Fungal Mitoviruses Detected in Ostreobium sp.
While we cannot formally identify host associations from our RNA-seq data alone, no fungal-associated contigs were detected in any of the libraries, strongly arguing against the mitovirus-like viruses detected in Ostreobium sp. as being fungal viruses. The presence of a potential new group of protist-associated mitoviruses is of importance as they have traditionally been viewed as restricted to fungal hosts and were only very recently identified in plants [79]. Similar to virus transfer between fungi and land plants, it is possible that the symbiosis and co-evolution between green algae and fungi [80,81] explains the close phylogenetic relationships of their viromes, perhaps including horizontal gene transfer events. Indeed, coral holobionts are the location of frequent interactions between endolithic algae, such as Ostreobium sp., and fungi [82,83,84]. Considering the high levels of sequence divergence between our viral sequences and those associated with fungi and within the clade formed by Ostreobium sp.-associated viruses, it seems likely that any such horizontal gene transfer events are not recent and may have occurred in Ulvophyceae or even Chlorophyta ancestors. It will be of considerable interest to examine this new group of mitoviruses across a larger set of green microalgae species, particularly whether their putative mitochondrial subcellular location is the result of an escape from cytoplasmic dsRNA silencing (as suggested for newly characterized plant mitoviruses [85]) or if they are relics of the eukaryotic endosymbiosis event, particularly as mitoviruses have bacterial counterparts—the Leviviridae [60]. More broadly, these newly-reported mitovirus-like sequences further illustrate the enormous diversity of hosts infected by the Narnaviridae, including such eukaryotic microorganisms as Apicomplexa, Excavates and Oomycetes hosts [86,87,88,89].
4.3. Detection of Plant Viruses in the Chlorarachniophytes
The apparent similarity between a Rhizarian (C. reptans) associated viral sequence and land-plant infecting viruses was striking. The Rhizaria and Archaeplastida are assumed to have diverged before the cyanobacteria primary endosymbiosis event, ca. 1.5 billion years ago [90,91]. Thus, a detectable sequence similarity between Rhizaria-associated viruses and those infecting land plants cannot be reasonably attributed to such an ancient evolutionary event. Rather, assuming these viruses actually infect C. reptans, the presence of such land plant-like viruses in Chlorarachniophyte would reflect their more recent acquisition in chlorarachniophytes through either horizontal transfer by common vector/symbiont/parasite ancestors or secondary endosymbiosis (eukaryote-to-eukaryote) events. Indeed, a secondary endosymbiosis event of a green alga in the core Chlorophyta, possibly related to Bryopsidales, led to the origin of the plastid of chlorarachniophytes between 578–318 Ma [33]. Whether this virus (i) constitutes a relic of viruses that infected this engulfed green algal endosymbiont, (ii) is part of the chlorarachniophyte cytoplasm or still associated with the periplastidial compartment (i.e., remnant cytoplasm of the endosymbiont), or (iii) interacts with the nucleomorph (remnants of the green algal endosymbiont nucleus) are key questions in the evolution of eukaryotic RNA viruses. While our data cannot provide answers and still require a formal virus-host association, it will be of interest to extend these analyses to euglenophytes and the dinoflagellate genus Lepidodinium where distinct secondary endosymbiosis with green algae have also occurred, as well as to cryptophytes that also contain the remnant nucleus of its red algae endosymbiont [92,93].
4.4. First Report of a Negative-Sense RNA Virus in Microalgae
Our detection of a Bunyavirales-like sequence in C. reptans is the first evidence of a negative-sense RNA virus in microalgae. Considering the extensive host range of the Bunyavirales (land plants, invertebrates, vertebrates and humans), their association with chlorarachniophyte hosts is plausible. However, given its very low abundance in C. reptans, additional work is clearly needed to retrieve the complete virus genome and to confirm the association of such bunya-like viruses with the chlorarachniophytes.
One of the greatest challenges in viral genomics is the ability to detect distant homologies, especially in rapidly evolving RNA viruses. As a first attempt to retrieve such ephemeral evolutionary signals, we scanned orphan contigs using RdRp protein structural information in addition to the standard primary amino acid sequence. Notably, both the RdRp profile and 3D protein structure comparison led to the identification of highly divergent RNA virus candidates, although these remain difficult to annotate. Efforts to better describe the repertoire of sequence and structure of viral RdRps are therefore central to unveiling the RNA virosphere in overlooked eukaryotic organisms.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4915/12/10/1180/s1, Figure S1: Gel electrophoresis of RT-PCR to validate the putative new RNA viruses in each sample used for library construction and RNA-seq. Figure S2: MAFFT alignment of the HMM-detected RdRp candidates with Pfam RdRPp profiles. Figure S3: rRNA depletion and contig assembly results. Figure S4: Genomic organization of the ALG_2 Partiti-like sequences and Bryopsis mitochondria-associated dsRNA. Table S1: RNA-seq library preparation and sequencing results, Table S2: List of primers used in this study. Table S3 RdRp-based pairwise sequence identity of the viruses newly identified here: File S1: Krona graphs obtained using the KMA and CCMetagen methods for the ALG_1 library. File S2: Krona graphs obtained using the KMA and CCMetagen methods for the ALG_2 library. File S3: Krona graphs obtained using the KMA and CCMetagen methods for the ALG_3 library.
Author Contributions
Conceptualization, J.C., H.V., E.C.H.; methodology, J.C., V.R.M.; validation, J.C.; formal analysis, J.C., V.R.M.; investigation, J.C.; resources, R.W., H.V., E.C.H.; data curation, J.C.; writing—original draft preparation, J.C.; writing—review and editing, J.C., H.V., V.R.M., R.W., E.C.H.; supervision, E.C.H.; funding acquisition, E.C.H. All authors have read and agreed to the published version of the manuscript.
Funding
E.H. is funded by an ARC Australian Laureate Fellowship, grant number FL170100022.
Acknowledgments
We acknowledge the SIH Bioinformatics Hub and the University of Sydney’s high-performance computing cluster Artemis for providing the computing resources used for this study. We also thank Fabien Burki for providing us source files to build Figure 1.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Krupovic, M.; Prangishvili, D.; Hendrix, R.W.; Bamford, D.H. Genomics of bacterial and archaeal viruses: Dynamics within the prokaryotic virosphere. Microbiol. Mol. Biol. Rev. 2011, 75, 610–635. [Google Scholar] [CrossRef] [PubMed]
- Lang, A.S.; Rise, M.L.; Culley, A.I.; Steward, G.F. RNA viruses in the sea. FEMS Microbiol. Rev. 2009, 33, 295–323. [Google Scholar] [CrossRef] [PubMed]
- Paez-Espino, D.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Thomas, A.D.; Huntemann, M.; Mikhailova, N.; Rubin, E.; Ivanova, N.N.; Kyrpides, N.C. Uncovering Earth’s virome. Nature 2016, 536, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Brum, J.R.; Ignacio-Espinoza, J.C.; Roux, S.; Doulcier, G.; Acinas, S.G.; Alberti, A.; Chaffron, S.; Cruaud, C.; De Vargas, C.; Gasol, J.M.; et al. Patterns and ecological drivers of ocean viral communities. Science 2015, 348, 1261498. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Brum, J.R.; Dutilh, B.E.; Sunagawa, S.; Duhaime, M.B.; Loy, A.; Poulos, B.T.; Solonenko, N.; Lara, E.; Poulain, J.; et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 2016, 537, 689–693. [Google Scholar] [CrossRef]
- Steward, G.F.; Culley, A.I.; Mueller, J.A.; Wood-Charlson, E.M.; Belcaid, M.; Poisson, G. Are we missing half of the viruses in the ocean? ISME J. 2013, 7, 672–679. [Google Scholar] [CrossRef]
- Miranda, J.A.; Culley, A.I.; Schvarcz, C.R.; Steward, G.F. RNA viruses as major contributors to Antarctic virioplankton. Environ. Microbiol. 2016, 18, 3714–3727. [Google Scholar] [CrossRef]
- Krishnamurthy, S.R.; Wang, D. Origins and challenges of viral dark matter. Virus Res. 2017, 239, 136–142. [Google Scholar] [CrossRef]
- Zhang, Y.-Z.; Shi, M.; Holmes, E.C. Using metagenomics to characterize an expanding virosphere. Cell 2018, 172, 1168–1172. [Google Scholar] [CrossRef]
- Edwards, R.A.; Rohwer, F. Viral metagenomics. Nat. Rev. Microbiol. 2005, 3, 504–510. [Google Scholar] [CrossRef]
- Suttle, C.A. Viruses in the sea. Nature 2005, 437, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.A.L.; Andrade, A.C.; Dos, S.P.; Boratto, P.V.d.M.; Trindade, G.d.S.; Kroon, E.G.; Abrahão, J.S. An anthropocentric view of the virosphere-host relationship. Front. Microbiol. 2017, 8, 1673. [Google Scholar] [CrossRef] [PubMed]
- Culley, A.I.; Lang, A.S.; Suttle, C.A. Metagenomic analysis of coastal RNA virus communities. Science 2006, 312, 1795–1798. [Google Scholar] [CrossRef] [PubMed]
- Culley, A.I.; Lang, A.S.; Suttle, C.A. The complete genomes of three viruses assembled from shotgun libraries of marine RNA virus communities. Virol. J. 2007, 4, 69. [Google Scholar] [CrossRef] [PubMed]
- Short, S.M.; Staniewski, M.A.; Chaban, Y.V.; Long, A.M.; Wang, D. Diversity of viruses infecting eukaryotic algae. Curr. Issues Mol. Biol. 2020, 39, 29–62. [Google Scholar] [CrossRef]
- Mihara, T.; Nishimura, Y.; Shimizu, Y.; Nishiyama, H.; Yoshikawa, G.; Uehara, H.; Hingamp, P.; Goto, S.; Ogata, H. Linking virus genomes with host taxonomy. Viruses 2016, 8, 666. [Google Scholar] [CrossRef]
- Mayer, J.A.; Taylor, F.J.R. A virus which lyses the marine nanoflagellate Micromonas pusilla. Nature 1979, 281, 299–301. [Google Scholar] [CrossRef]
- Brown, R.M. Algal viruses. Adv. Virus Res. 1972, 17, 243–277. [Google Scholar]
- Brussaard, C.P.D.; Martínez, J. Algal Bloom Viruses. Plant Viruses 2008, 2, 1–10. [Google Scholar]
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; De Clerck, O. Phylogeny and molecular evolution of the green algae. CRC Crit. Rev. Plant Sci. 2012, 31, 1–46. [Google Scholar] [CrossRef]
- Urayama, S.I.; Takaki, Y.; Nunoura, T. FLDS: A comprehensive DSRNA sequencing method for intracellular RNA virus surveillance. Microbes Environ. 2016, 31, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Mushegian, A.; Shipunov, A.; Elena, S.F. Changes in the composition of the RNA virome mark evolutionary transitions in green plants. BMC Biol. 2016, 14, 68. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Illergard, K.; Ardell, D.H.; Elofsson, A. Structure is three to ten times more conserved than sequence—A study of structural response in protein cores. Proteins Struct. Funct. Bioinform. 2009, 77, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Bamford, D.H.; Grimes, J.M.; Stuart, D.I. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 2005, 15, 655–663. [Google Scholar] [CrossRef]
- Chen, J.; Guo, M.; Wang, X.; Liu, B. A comprehensive review and comparison of different computational methods for protein remote homology detection. Brief. Bioinform. 2018, 19, 231–244. [Google Scholar] [CrossRef]
- Singh, J.; Saxena, R.C. An introduction to microalgae: Diversity and significance. In Handbook of Marine Microalgae; Academic Press: Cambridge, MA, USA, 2015; pp. 11–24. ISBN 9780128007761. [Google Scholar]
- Archibald, J.M. The evolution of algae by secondary and tertiary endosymbiosis. Adv. Bot. Res. 2012, 64, 87–118. [Google Scholar]
- Burki, F.; Roger, A.J.; Brown, M.W.; Simpson, A.G.B. The New Tree of Eukaryotes. Trends Ecol. Evol. 2020, 35, 43–55. [Google Scholar] [CrossRef]
- Richmond, A.; Hu, Q. Handbook of Microalgal Culture: Applied Phycology and Biotechnology, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2013; pp. 1–719. ISBN 9781118567166. [Google Scholar]
- Guiry, M.D. How many species of algae are there? J. Phycol. 2012, 48, 1057–1063. [Google Scholar] [CrossRef]
- Chapman, R.L. Algae: The world’s most important “plants”—an introduction. Mitig. Adapt. Strateg. Glob. Chang. 2013, 18, 5–12. [Google Scholar] [CrossRef]
- Jackson, C.; Knoll, A.H.; Chan, C.X.; Verbruggen, H. Plastid phylogenomics with broad taxon sampling further elucidates the distinct evolutionary origins and timing of secondary green plastids. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Fuhrman, J.A. Marine viruses and their biogeochemical and ecological effects. Nature 1999, 399, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Forterre, P.; Prangishvili, D. The major role of viruses in cellular evolution: Facts and hypotheses. Curr. Opin. Virol. 2013, 3, 558–565. [Google Scholar] [CrossRef]
- Wetherbee, R.; Marcelino, V.R.; Costa, J.F.; Grant, B.; Crawford, S.; Waller, R.F.; Andersen, R.A.; Berry, D.; McFadden, G.I.; Verbruggen, H. A new marine prasinophyte genus alternates between a flagellate and a dominant benthic stage with microrhizoids for adhesion. J. Phycol. 2019, 55, 1210–1225. [Google Scholar] [CrossRef]
- Wetherbee, R.; Verbruggen, H. Kraftionema allantoideum, a new genus and family of Ulotrichales (Chlorophyta) adapted for survival in high intertidal pools. J. Phycol. 2016, 52, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Remmert, M.; Biegert, A.; Hauser, A.; Söding, J. HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 2012, 9, 173–175. [Google Scholar] [CrossRef]
- Soding, J. Protein homology detection by HMM-HMM comparison. Bioinformatics 2005, 21, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Marcelino, V.R.; Clausen, P.T.L.C.; Buchmann, J.P.; Wille, M.; Iredell, J.R.; Meyer, W.; Lund, O.; Sorrell, T.C.; Holmes, E.C. CCMetagen: Comprehensive and accurate identification of eukaryotes and prokaryotes in metagenomic data. Genome Biol. 2020, 21, 103. [Google Scholar] [CrossRef]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Vieira, H.H.; Bagatini, I.L.; Guinart, C.M.; Vieira, A.A.H.; Vieira, H.H.; Bagatini, I.L.; Guinart, C.M.; Vieira, A.A.H. tufA gene as molecular marker for freshwater Chlorophyceae. ALGAE 2016, 31, 155–165. [Google Scholar] [CrossRef]
- Strassert, J.F.H.; Jamy, M.; Mylnikov, A.P.; Tikhonenkov, D.V.; Burki, F. New phylogenomic analysis of the enigmatic phylum telonemia further resolves the eukaryote tree of life. Mol. Biol. Evol. 2019, 36, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Del Cortona, A.; Jackson, C.J.; Bucchini, F.; Van Bel, M.; D’hondt, S.; Skaloud, P.; Delwiche, C.F.; Knoll, A.H.; Raven, J.A.; Verbruggen, H.; et al. Neoproterozoic origin and multiple transitions to macroscopic growth in green seaweeds. Proc. Natl. Acad. Sci. USA 2020, 117, 2551–2559. [Google Scholar] [CrossRef] [PubMed]
- Cocquyt, E.; Verbruggen, H.; Leliaert, F.; Clerck, O. De Evolution and cytological diversification of the green seaweeds (Ulvophyceae). Mol. Biol. Evol. 2010, 27, 2052–2061. [Google Scholar] [CrossRef]
- Venkataraman, S.; Prasad, B.V.L.S.; Selvarajan, R. RNA Dependent RNA polymerases: Insights from structure, function and evolution. Viruses 2018, 10, 76. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Kazlauskas, D.; Iranzo, J.; Lucía-Sanz, A.; Kuhn, J.H.; Krupovic, M.; Dolja, V.V.; Koonin, E. V Origins and evolution of the global RNA virome. mBio 2018, 9, e02318–e02329. [Google Scholar] [CrossRef]
- Koga, R.; Horiuchi, H.; Fukuhara, T. Double-stranded RNA replicons associated with chloroplasts of a green alga, Bryopsis cinicola. Plant Mol. Biol. 2003, 51, 991–999. [Google Scholar] [CrossRef]
- Le Alsumard, T.C.; Golubic, S.; Priess, K. Fungi in corals: Symbiosis or disease? Interaction between polyps and fungi causes pearl-like skeleton biomineralization. Mar. Ecol. Prog. Ser. 1995, 117, 137–148. [Google Scholar] [CrossRef]
- Sabanadzovic, S.; Valverde, R.A.; Brown, J.K.; Martin, R.R.; Tzanetakis, I.E. Southern tomato virus: The link between the families Totiviridae and Partitiviridae. Virus Res. 2009, 140, 130–137. [Google Scholar] [CrossRef]
- Koga, R.; Fukuhara, T.; Nitta, T. Molecular characterization of a single mitochondria-associated double-stranded RNA in the green alga Bryopsis. Plant Mol. Biol. 1998, 36, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Bao, Y.; Chetvernin, V.; Tatusova, T. Improvements to pairwise sequence comparison (PASC): A genome-based web tool for virus classification. Arch. Virol. 2014, 159, 3293–3304. [Google Scholar] [CrossRef] [PubMed]
- Atkins, J.F.; Loughran, G.; Bhatt, P.R.; Firth, A.E.; Baranov, P.V. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016, 44, 7007–7078. [Google Scholar] [CrossRef]
- Krupovic, M.; Dolja, V.V.; Koonin, E.V. Plant viruses of the Amalgaviridae family evolved via recombination between viruses with double-stranded and negative-strand RNA genomes. Biol. Direct 2015, 10, 12. [Google Scholar] [CrossRef]
- Chiba, Y.; Tomaru, Y.; Shimabukuro, H.; Kimura, K.; Hirai, M.; Takaki, Y.; Hagiwara, D.; Takuro, N.; Urayma, S.-I. Viral RNA genomes identified from marine macroalgae and a diatom. Microbes Environ. 2020, 35. [Google Scholar] [CrossRef]
- Dinan, A.M.; Lukhovitskaya, N.I.; Olendraite, I.; Firth, A.E. A case for a negative-strand coding sequence in a group of positive-sense RNA viruses. Virus Evol. 2020, 6, veaa007. [Google Scholar]
- Waldron, F.M.; Stone, G.N.; Obbard, D.J. Metagenomic sequencing suggests a diversity of RNA interference-like responses to viruses across multicellular eukaryotes. PLoS Genet. 2018, 14, e1007533. [Google Scholar] [CrossRef]
- Polischuk, V.; Budzanivska, I.; Shevchenko, T.; Oliynik, S. Evidence for plant viruses in the region of Argentina Islands, Antarctica. FEMS Microbiol. Ecol. 2007, 59, 409–417. [Google Scholar] [CrossRef]
- Petrzik, K.; Vondrák, J.; Kvíderová, J.; Lukavský, J. Platinum anniversary: Virus and lichen alga together more than 70 years. PLoS ONE 2015, 10, e01207688. [Google Scholar] [CrossRef] [PubMed]
- Goh, C.J.; Park, D.; Lee, J.S.; Sebastiani, F.; Hahn, Y. Identification of a novel plant amalgavirus (Amalgavirus, Amalgaviridae) genome sequence in Cistus incanus. Acta Virol. 2018, 62, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Zhan, B.; Cao, M.; Wang, K.; Wang, X.; Zhou, X. Detection and characterization of cucumis melo cryptic virus, cucumis melo amalgavirus 1, and melon necrotic spot virus in Cucumis melo. Viruses 2019, 11, 81. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Goh, C.J.; Park, D.; Hahn, Y. Identification of a novel plant RNA virus species of the genus Amalgavirus in the family Amalgaviridae from chia (Salvia hispanica). Genes Genom. 2019, 41, 507–514. [Google Scholar] [CrossRef]
- Roossinck, M.J. Lifestyles of plant viruses. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 1899–1905. [Google Scholar] [CrossRef]
- Nibert, M.L.; Ghabrial, S.A.; Maiss, E.; Lesker, T.; Vainio, E.J.; Jiang, D.; Suzuki, N. Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res. 2014, 188, 128–141. [Google Scholar] [CrossRef]
- Nibert, M.L.; Vong, M.; Fugate, K.K.; Debat, H.J. Evidence for contemporary plant mitoviruses. Virology 2018, 518, 14–24. [Google Scholar] [CrossRef]
- Roossinck, M.J. Evolutionary and ecological links between plant and fungal viruses. New Phytol. 2019, 221, 86–92. [Google Scholar] [CrossRef]
- Bonfante, P. Algae and fungi move from the past to the future. eLife 2019, 8, e49448. [Google Scholar] [CrossRef]
- Ricci, F.; Marcelino, V.R.; Blackall, L.L.; Kühl, M.; Medina, M.; Verbruggen, H. Beneath the surface: Community assembly and functions of the coral skeleton microbiome. Microbiome 2019, 7, 1–10. [Google Scholar] [CrossRef]
- Raghukumar, C.; Ravindran, J. Fungi and Their Role in Corals and Coral Reef Ecosystems. In Biology of Marine Fungi; Raghukumar, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 89–113. [Google Scholar]
- Marcelino, V.R.; Verbruggen, H. Multi-marker metabarcoding of coral skeletons reveals a rich microbiome and diverse evolutionary origins of endolithic algae. Sci. Rep. 2016, 6, 31508. [Google Scholar] [CrossRef] [PubMed]
- Nerva, L.; Vigani, G.; Di Silvestre, D.; Ciuffo, M.; Forgia, M.; Chitarra, W.; Turina, M. Biological and molecular characterization of chenopodium quinoa mitovirus 1 reveals a distinct small RNA response compared to those of cytoplasmic RNA viruses. J. Virol. 2019, 93, e01918–e01998. [Google Scholar] [CrossRef] [PubMed]
- Charon, J.; Grigg, M.J.; Eden, J.-S.; Piera, K.A.; Rana, H.; William, T.; Rose, K.; Davenport, M.P.; Anstey, N.M.; Holmes, E.C. Novel RNA viruses associated with Plasmodium vivax in human malaria and Leucocytozoon parasites in avian disease. PLoS Pathog. 2019, 15, e1008216. [Google Scholar] [CrossRef]
- Zangger, H.; Ronet, C.; Desponds, C.; Kuhlmann, F.M.; Robinson, J.; Hartley, M.-A.; Prevel, F.; Castiglioni, P.; Pratlong, F.; Bastien, P.; et al. Detection of Leishmania RNA virus in Leishmania parasites. PLoS Negl. Trop. Dis. 2013, 7, e2006. [Google Scholar] [CrossRef] [PubMed]
- Grybchuk, D.; Akopyants, N.S.; Kostygov, A.Y.; Konovalovas, A.; Lye, L.-F.; Dobson, D.E.; Zangger, H.; Fasel, N.; Butenko, A.; Frolov, A.O.; et al. Viral discovery and diversity in trypanosomatid protozoa with a focus on relatives of the human parasite Leishmania. Proc. Natl. Acad. Sci. USA 2018, 115, E506–E515. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Myers, K.; Fry, W.E.; Hillman, B.I. A member of the virus family Narnaviridae from the plant pathogenic oomycete Phytophthora infestans. Arch. Virol. 2012, 157, 165–169. [Google Scholar] [CrossRef]
- Yoon, H.S.; Hackett, J.D.; Ciniglia, C.; Pinto, G.; Bhattacharya, D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 2004, 21, 809–818. [Google Scholar] [CrossRef]
- Parfrey, L.W.; Lahr, D.J.G.; Knoll, A.H.; Katz, L.A. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc. Natl. Acad. Sci. USA 2011, 108, 13624–13629. [Google Scholar] [CrossRef]
- Burki, F.; Keeling, P.J. Rhizaria. Curr. Biol. 2014, 24, R103–R107. [Google Scholar] [CrossRef]
- Sven, B. Gould Algae’s complex origins. Nature 2012, 492, 46–48. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).