Truncated Equinine B Variants Reveal the Sequence Determinants of Antimicrobial Selectivity
Abstract
1. Introduction
2. Results and Discussion
2.1. Structure-Function Guided Design of the Peptide Library
2.2. Evaluation of Peptide Specificity
2.3. In Silico Structural Profiling
2.4. Evaluation of Membrane Permeability and Activity
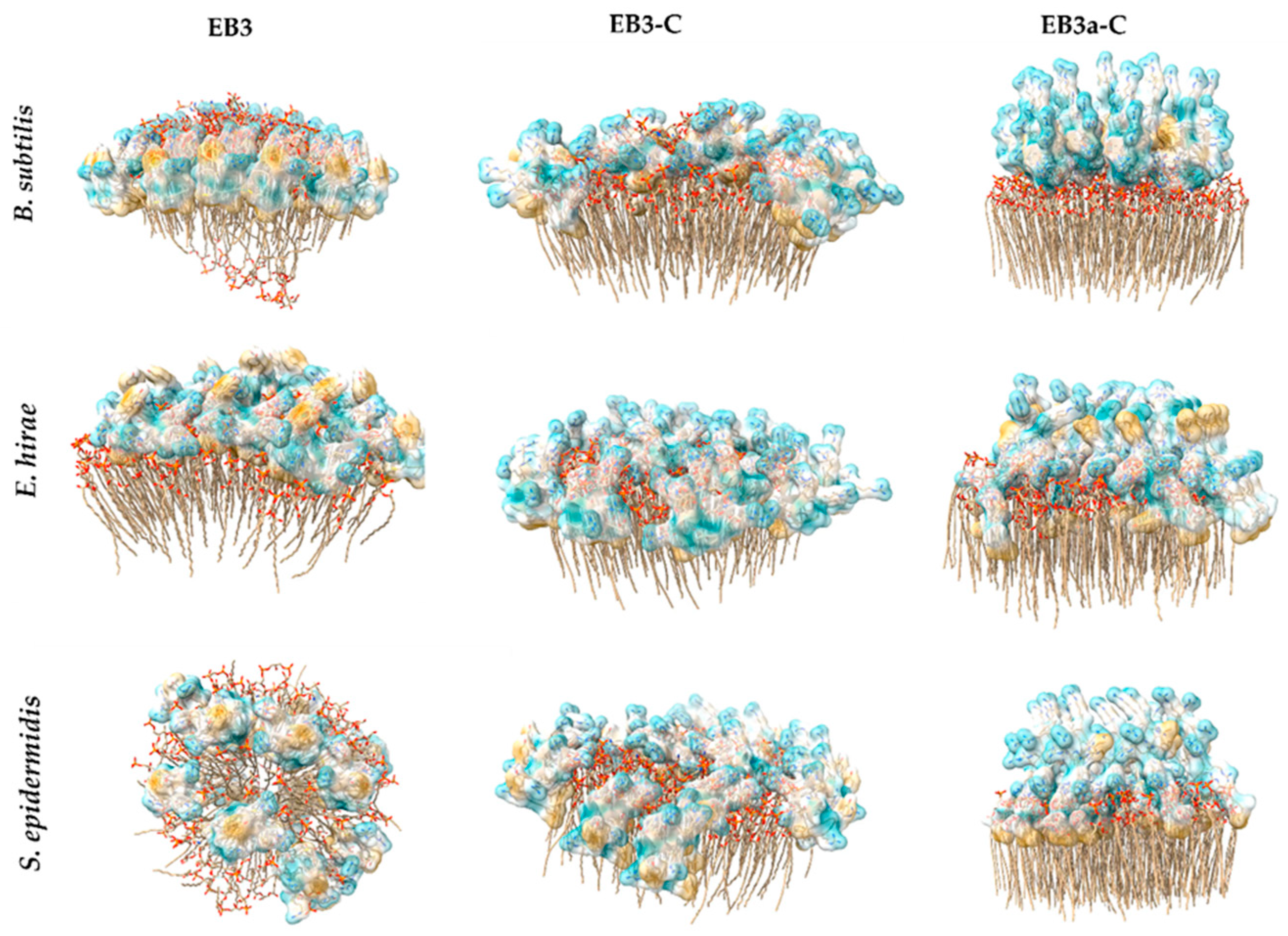
2.5. In Silico Evaluation of Peptide–Membrane Interactions
3. Materials and Methods
3.1. General
3.2. Peptide Synthesis
3.3. Biological Activity
3.3.1. Minimum Inhibitory Concentration (MIC) Assay
3.3.2. Hemolysis Assay
3.3.3. In Silico Analysis of Peptides Structural Properties
3.3.4. Membrane Permeability Assay
3.3.5. Membrane Depolarization Assay
3.3.6. In Silico Peptide–Membrane Interactions
3.4. Nano-Ultra-Performance Liquid Chromatography-Electrospray Ionization-Quadrupole Mass Spectrometry Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AR | Antibiotic resistance |
| AMPs | Antimicrobial peptides |
| HBTU | N,N,N′,N′-Tetramethyl-O-(1H-benzotriazol-1-yl)uronium hexafluorophosphate |
| NMM | N-methylmorpholine |
| TFA | Trifluoroacetic acid |
| TIS | Triisopropylsylane |
| SPE | Solid Phase Extraction |
| SPPS | Solid Phase Peptide Synthesis |
| MIC | Minimum Inhibitory Concentration |
| PI | Propidium iodide |
| MHB | Mueller–Hinton Broth |
| DMSO | Dimethyl sulfoxide |
| DMF | Dimethylformamide |
Appendix A
| Peptide ID | Peptide Sequence | Precursor m/z | Rt/min | |
|---|---|---|---|---|
| 1 | EB1 | GQCQRKCLGHCS | [660.2925 + 2H]2+ | 11.53 |
| 2 | EB-2 | KKCPKHPQCRK | [676.8743 + 2H]2+ | 09.17 |
| 3 | EB3 | RCIRRCFGYCL | [695.3392 + 2H]2+ | 18.06 |
| 4 | EB1a-K | GQQRRCLGHCS | [622.7909 + 2H]2+ | 10.17 |
| 5 | EB1-K | GQCQRRCLGHCS | [674.2955 + 2H]2+ | 10.77 |
| 6 | EB2-K | RRCPRHPQCRR | [732.8866 + 2H]2+ | 09.17 |
| 7 | EB1-C | GQRQRKRLGHRS | [739.9304 + 2H]2+ | 09.76 |
| 8 | EB2-K | KKRPKHPQRRK | [729.9662 + 2H]2+ | 08.76 |
| 9 | EB3-C | RRIRRRFGYRL | [774.9771 + 2H]2+ | 09.07 |
| 10 | EB3a-C | RRIRRRGYRL | [701.4429 + 2H]2+ | 09.23 |
References
- Boman, H.G. Antibacterial Peptides: Basic Facts and Emerging Concepts. J. Intern. Med. 2003, 254, 197–215. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Steiner, H.; Hultmark, D.; Engström, Å.; Bennich, H.; Boman, H.G. Sequence and Specificity of Two Antibacterial Proteins Involved in Insect Immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Haug, B.E.; Strom, M.B.; Svendsen, J.S.M. The Medicinal Chemistry of Short Lactoferricin-Based Antibacterial Peptides. Curr. Med. Chem. 2007, 14, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, D.; Veiga, A.S.; Castanho, M.A.R.B. From Antimicrobial to Anticancer Peptides. A Review. Front. Microbiol. 2013, 4, 294. [Google Scholar] [CrossRef] [PubMed]
- Brogden, N.K.; Brogden, K.A. Will New Generations of Modified Antimicrobial Peptides Improve Their Potential as Pharmaceuticals? Int. J. Antimicrob. Agents 2011, 38, 217–225. [Google Scholar] [CrossRef]
- Bocchinfuso, G.; Palleschi, A.; Orioni, B.; Grande, G.; Formaggio, F.; Toniolo, C.; Park, Y.; Hahm, K.; Stella, L. Different Mechanisms of Action of Antimicrobial Peptides: Insights from Fluorescence Spectroscopy Experiments and Molecular Dynamics Simulations. J. Pept. Sci. 2009, 15, 550–558. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Moore, B.S. Lessons from the Past and Charting the Future of Marine Natural Products Drug Discovery and Chemical Biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef]
- Wang, Y.-N.; Meng, L.-H.; Wang, B.-G. Progress in Research on Bioactive Secondary Metabolites from Deep-Sea Derived Microorganisms. Mar. Drugs 2020, 18, 614. [Google Scholar] [CrossRef]
- Satpute, S.K.; Banat, I.M.; Dhakephalkar, P.K.; Banpurkar, A.G.; Chopade, B.A. Biosurfactants, Bioemulsifiers and Exopolysaccharides from Marine Microorganisms. Biotechnol. Adv. 2010, 28, 436–450. [Google Scholar] [CrossRef]
- Laport, M.; Santos, O.; Muricy, G. Marine Sponges: Potential Sources of New Antimicrobial Drugs. Curr. Pharm. Biotechnol. 2009, 10, 86–105. [Google Scholar] [CrossRef]
- Macedo, M.W.F.S.; Cunha, N.B.D.; Carneiro, J.A.; Costa, R.A.D.; Alencar, S.A.D.; Cardoso, M.H.; Franco, O.L.; Dias, S.C. Marine Organisms as a Rich Source of Biologically Active Peptides. Front. Mar. Sci. 2021, 8, 667764. [Google Scholar] [CrossRef]
- La Corte, C.; Catania, V.; Dara, M.; Parrinello, D.; Staropoli, M.; Trapani, M.R.; Cammarata, M.; Parisi, M.G. Equinins as Novel Broad-Spectrum Antimicrobial Peptides Isolated from the Cnidarian Actinia Equina (Linnaeus, 1758). Mar. Drugs 2024, 22, 172. [Google Scholar] [CrossRef]
- Ovchinnikova, T.V.; Balandin, S.V.; Aleshina, G.M.; Tagaev, A.A.; Leonova, Y.F.; Krasnodembsky, E.D.; Men’shenin, A.V.; Kokryakov, V.N. Aurelin, a Novel Antimicrobial Peptide from Jellyfish Aurelia Aurita with Structural Features of Defensins and Channel-Blocking Toxins. Biochem. Biophys. Res. Commun. 2006, 348, 514–523. [Google Scholar] [CrossRef]
- Malanovic, N.; Leber, R.; Schmuck, M.; Kriechbaum, M.; Cordfunke, R.A.; Drijfhout, J.W.; De Breij, A.; Nibbering, P.H.; Kolb, D.; Lohner, K. Phospholipid-Driven Differences Determine the Action of the Synthetic Antimicrobial Peptide OP-145 on Gram-Positive Bacterial and Mammalian Membrane Model Systems. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 2437–2447. [Google Scholar] [CrossRef]
- Shenkarev, Z.O.; Panteleev, P.V.; Balandin, S.V.; Gizatullina, A.K.; Altukhov, D.A.; Finkina, E.I.; Kokryakov, V.N.; Arseniev, A.S.; Ovchinnikova, T.V. Recombinant Expression and Solution Structure of Antimicrobial Peptide Aurelin from Jellyfish Aurelia Aurita. Biochem. Biophys. Res. Commun. 2012, 429, 63–69. [Google Scholar] [CrossRef]
- Huertas, N.; Monroy, Z.; Medina, R.; Castañeda, J. Antimicrobial Activity of Truncated and Polyvalent Peptides Derived from the FKCRRQWQWRMKKGLA Sequence against Escherichia Coli ATCC 25922 and Staphylococcus Aureus ATCC 25923. Molecules 2017, 22, 987. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. The Antimicrobial Peptide Database Is 20 Years Old: Recent Developments and Future Directions. Protein Sci. 2023, 32, e4778. [Google Scholar] [CrossRef]
- Merrifield, R.B. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar] [CrossRef]
- Guzmán, F.; Aróstica, M.; Román, T.; Beltrán, D.; Gauna, A.; Albericio, F.; Cárdenas, C. Peptides, Solid-Phase Synthesis and Characterization: Tailor-Made Methodologies. Electron. J. Biotechnol. 2023, 64, 27–33. [Google Scholar] [CrossRef]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Faster de Novo Structure Prediction for Linear Peptides in Solution and in Complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef]
- Pašalić, L.; Pem, B.; Jakas, A.; Čikoš, A.; Groznica, N.; Mlinarić, T.; Accorsi, M.; Mangiarotti, A.; Dimova, R.; Bakarić, D. Peptide Interaction with Mixed Lipid Bilayers Alters Packing and Hydrocarbon Chain Conformations. J. Liposome Res. 2025, 35, 548–565. [Google Scholar] [CrossRef]
- Malanovic, N.; Lohner, K. Antimicrobial Peptides Targeting Gram-Positive Bacteria. Pharmaceuticals 2016, 9, 59. [Google Scholar] [CrossRef]
- Malanovic, N.; Lohner, K. Gram-Positive Bacterial Cell Envelopes: The Impact on the Activity of Antimicrobial Peptides. Biochim. Biophys. Acta (BBA)-Biomembr. 2016, 1858, 936–946. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Piller, P.; Wolinski, H.; Cordfunke, R.A.; Drijfhout, J.W.; Keller, S.; Lohner, K.; Malanovic, N. Membrane Activity of LL-37 Derived Antimicrobial Peptides against Enterococcus Hirae: Superiority of SAAP-148 over OP-145. Biomolecules 2022, 12, 523. [Google Scholar] [CrossRef]
- Barns, K.J.; Weisshaar, J.C. Real-Time Attack of LL-37 on Single Bacillus Subtilis Cells. Biochim. Biophys. Acta (BBA)-Biomembr. 2013, 1828, 1511–1520. [Google Scholar] [CrossRef]
- Sochacki, K.A.; Barns, K.J.; Bucki, R.; Weisshaar, J.C. Real-Time Attack on Single Escherichia Coli Cells by the Human Antimicrobial Peptide LL-37. Proc. Natl. Acad. Sci. USA 2011, 108, E77–E81. [Google Scholar] [CrossRef] [PubMed]
- Nagant, C.; Pitts, B.; Nazmi, K.; Vandenbranden, M.; Bolscher, J.G.; Stewart, P.S.; Dehaye, J.-P. Identification of Peptides Derived from the Human Antimicrobial Peptide LL-37 Active against Biofilms Formed by Pseudomonas Aeruginosa Using a Library of Truncated Fragments. Antimicrob. Agents Chemother. 2012, 56, 5698–5708. [Google Scholar] [CrossRef]
- Sevcsik, E.; Pabst, G.; Richter, W.; Danner, S.; Amenitsch, H.; Lohner, K. Interaction of LL-37 with Model Membrane Systems of Different Complexity: Influence of the Lipid Matrix. Biophys. J. 2008, 94, 4688–4699. [Google Scholar] [CrossRef]
- Hodzic, A.; Vejzovic, D.; Topciu, A.; Kuhlmann, K.; Kumar, R.; Mroginski, M.A.; De Miguel, A.; Hofmann, P.; Zangger, K.; Weingarth, M.; et al. SAAP-148 Oligomerizes into a Hexamer Forming a Hydrophobic Inner Core. ChemBioChem 2025, 26, e202500112. [Google Scholar] [CrossRef] [PubMed]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for Structure Building and Analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Thevenet, P.; Shen, Y.; Maupetit, J.; Guyon, F.; Derreumaux, P.; Tuffery, P. PEP-FOLD: An Updated de Novo Structure Prediction Server for Both Linear and Disulfide Bonded Cyclic Peptides. Nucleic Acids Res. 2012, 40, W288–W293. [Google Scholar] [CrossRef]
- Beaufays, J.; Lins, L.; Thomas, A.; Brasseur, R. In Silico Predictions of 3D Structures of Linear and Cyclic Peptides with Natural and Non-proteinogenic Residues. J. Pept. Sci. 2012, 18, 17–24. [Google Scholar] [CrossRef]
- Snider, C.; Jayasinghe, S.; Hristova, K.; White, S.H. MPEx: A Tool for Exploring Membrane Proteins. Protein Sci. 2009, 18, 2624–2628. [Google Scholar] [CrossRef]
- White, S.H.; Wimley, W.C. MEMBRANE PROTEIN FOLDING AND STABILITY: Physical Principles. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 319–365. [Google Scholar] [CrossRef] [PubMed]
- Malanovic, N.; Marx, L.; Blondelle, S.E.; Pabst, G.; Semeraro, E.F. Experimental Concepts for Linking the Biological Activities of Antimicrobial Peptides to Their Molecular Modes of Action. Biochim. Biophys. Acta (BBA)-Biomembr. 2020, 1862, 183275. [Google Scholar] [CrossRef] [PubMed]




| Compound | Sequence | Escherichia coli | Staphylococcus epidermidis |
|---|---|---|---|
| MIC90/µgmL−1 | MIC90/µgmL−1 | ||
| EB1 | GQCQRKCLGHCS | >100 | >100 |
| EB2 | KKCPKHPQCRK | >100 | >100 |
| EB3 | RCIRRCFGYCL | 100 | 25 |
| EB1a-K | GQQRRCLGHCS | >100 | >100 |
| EB-1K | GQCQRRCLGHCS | >100 | >100 |
| EB-2K | RRCPRHPQCRR | >100 | >100 |
| EB1-C | GQRQRKRLGHRS | >100 | >100 |
| EB2-C | KKRPKHPQRRK | >100 | >100 |
| EB3-C | RRIRRRFGYRL | >100 | 25–50 |
| EB3a-C | RRIRRRGYRL | >100 | >100 |
| Compound | ΔG | Total Hydrydrophobic Moment |
|---|---|---|
| EB1 | 9.93 | 5.3 |
| EB2 | 16.35 | 4.48 |
| EB3 | 1.73 | 6.97 |
| EB1a-K | 16.86 | 5.47 |
| EB1-K | 16.84 | 4.42 |
| EB2-K | 20.29 | 2.17 |
| EB1-C | 15.42 | 4.59 |
| EB2-C | 20.01 | 4.01 |
| EB3-C | 7.22 | 5.58 |
| EB3a-C | 8.93 | 5.47 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Staropoli, M.; Schwaiger, T.; Tuzlak, J.; Biba, R.; Petrowitsch, L.; Fessler, J.; Roje, M.; Cammarata, M.; Malanović, N.; Jakas, A. Truncated Equinine B Variants Reveal the Sequence Determinants of Antimicrobial Selectivity. Mar. Drugs 2026, 24, 46. https://doi.org/10.3390/md24010046
Staropoli M, Schwaiger T, Tuzlak J, Biba R, Petrowitsch L, Fessler J, Roje M, Cammarata M, Malanović N, Jakas A. Truncated Equinine B Variants Reveal the Sequence Determinants of Antimicrobial Selectivity. Marine Drugs. 2026; 24(1):46. https://doi.org/10.3390/md24010046
Chicago/Turabian StyleStaropoli, Mariele, Theresa Schwaiger, Jasmina Tuzlak, Renata Biba, Lukas Petrowitsch, Johannes Fessler, Marin Roje, Matteo Cammarata, Nermina Malanović, and Andreja Jakas. 2026. "Truncated Equinine B Variants Reveal the Sequence Determinants of Antimicrobial Selectivity" Marine Drugs 24, no. 1: 46. https://doi.org/10.3390/md24010046
APA StyleStaropoli, M., Schwaiger, T., Tuzlak, J., Biba, R., Petrowitsch, L., Fessler, J., Roje, M., Cammarata, M., Malanović, N., & Jakas, A. (2026). Truncated Equinine B Variants Reveal the Sequence Determinants of Antimicrobial Selectivity. Marine Drugs, 24(1), 46. https://doi.org/10.3390/md24010046








