Discovery of New Everninomicin Analogs from a Marine-Derived Micromonospora sp. by Metabolomics and Genomics Approaches
Abstract
1. Introduction
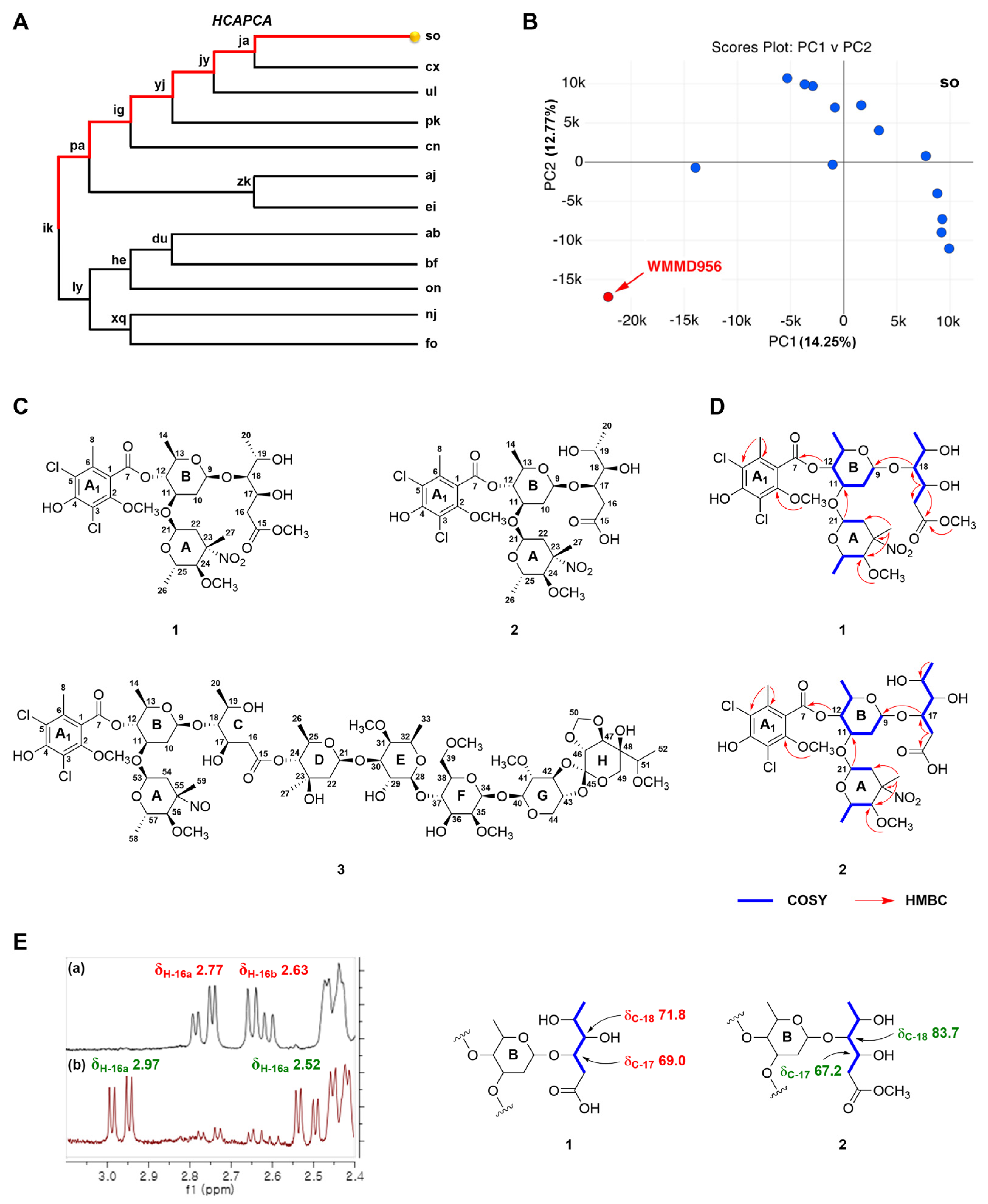
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Biological Material
3.3. Strain Genome Sequencing, Assembly, and Validation
3.4. Annotation of Biosynthetic Gene Clusters (BGCs)
3.5. Species Classification and BGC Analysis
3.6. Molecular Networking
3.7. Fermentation
3.8. Extraction and Isolation
3.9. Screening for Inhibitors of MRSA Growth
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Abdelmohsen, U.R.; Balasubramanian, S.; Oelschlaeger, T.A.; Grkovic, T.; Pham, N.B.; Quinn, R.J.; Hentschel, U. Potential of marine natural products against drug-resistant fungal, viral, and parasitic infections. Lancet Infect. Dis. 2017, 17, e30–e41. [Google Scholar] [CrossRef] [PubMed]
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Huang, J.; Muhammad, M.; Deng, Z.; Gao, J. Genome mining as a biotechnological tool for the discovery of novel marine natural products. Crit. Rev. Biotechnol. 2020, 40, 571–589. [Google Scholar] [CrossRef] [PubMed]
- Bauman, K.D.; Butler, K.S.; Moore, B.S.; Chekan, J.R. Genome mining methods to discover bioactive natural products. Nat. Prod. Rep. 2021, 38, 2100–2129. [Google Scholar] [CrossRef] [PubMed]
- Cortegiani, A.; Misseri, G.; Fasciana, T.; Giammanco, A.; Giarratano, A.; Chowdhary, A. Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J. Intensive Care 2018, 6, 69. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.; Forde, B.M.; Kidd, T.J.; Harris, P.N.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Chanana, S.; Thomas, C.S.; Zhang, F.; Rajski, S.R.; Bugni, T.S. HCAPCA: Automated hierarchical clustering and principal component analysis of large metabolomic datasets in R. Metabolites 2020, 10, 297. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- McCranie, E.K.; Bachmann, B.O. Bioactive oligosaccharide natural products. Nat. Prod. Rep. 2014, 31, 1026–1042. [Google Scholar] [CrossRef] [PubMed]
- Weitnauer, G.; Hauser, G.; Hofmann, C.; Linder, U.; Boll, R.; Pelz, K.; Glaser, S.J.; Bechthold, A. Novel avilamycin derivatives with improved polarity generated by targeted gene disruption. Chem. Biol. 2004, 11, 1403–1411. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Simard, M.; Bergeron, Y.; Beauchamp, D.; Bergeron, M.G. In vivo activity and pharmacokinetics of ziracin (SCH27899), a new long-acting everninomicin antibiotic, in a murine model of penicillin-susceptible or penicillin-resistant pneumococcal pneumonia. Antimicrob. Agents Chemother. 2000, 44, 1010–1018. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Limbrick, E.M.; Yñigez-Gutierrez, A.E.; Dulin, C.C.; Derewacz, D.K.; Spraggins, J.M.; McCulloch, K.M.; Iverson, T.; Bachmann, B.O. Methyltransferase contingencies in the pathway of everninomicin D antibiotics and analogues. ChemBioChem 2020, 21, 3349–3358. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Wang, L.; Zhang, H.; Zhang, L.; Tan, B.; Huang, Q.; Zhu, Y.; Zhang, C. Biosynthesis and engineered overproduction of everninomicins with promising activity against multidrug-resistant bacteria. ACS Synth. Biol. 2023, 12, 1520–1532. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H.; Borgwardt, K. GTDB-Tk v2: Memory friendly classification with the genome taxonomy database. Bioinformatics 2022, 38, 5315–5316. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.-A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Kautsar, S.A.; van der Hooft, J.J.; de Ridder, D.; Medema, M.H. BiG-SLiCE: A highly scalable tool maps the diversity of 1.2 million biosynthetic gene clusters. Gigascience 2021, 10, giaa154. [Google Scholar] [CrossRef] [PubMed]
- Kautsar, S.A.; Blin, K.; Shaw, S.; Weber, T.; Medema, M.H. BiG-FAM: The biosynthetic gene cluster families database. Nucleic Acids Res. 2021, 49, D490–D497. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
| Position | 1 | 2 | ||
|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | 114.2 | 114.2 | ||
| 2 | 153.7 | 154.0 | ||
| 3 | 116.4 | 116.1 | ||
| 4 | n.d. a | n.d. a | ||
| 5 | 119.2 | 119.0 | ||
| 6 | 134.4 | 134.3 | ||
| 7 | 166.2 | 166.4 | ||
| 8 | 17.0 | 2.38, s | 18.2 | 2.37, s |
| 2-OCH3 | 61.1 | 3.91, s | 62.2 | 3.90, s |
| 9 | 99.8 | 4.71, d (9.3) | 101.5 | 4.81, d (9.0) |
| 10a | 35.8 | 2.45, m | 37.2 | 2.45, m |
| 10b | 1.61, m | 1.59, m | ||
| 11 | 72.4 | 3.99, m | 73.6 | 3.99, m |
| 12 | 76.2 | 4.80, t (9.4) | 77.2 | 4.79, m |
| 13 | 70.6 | 3.60, m | 72.2 | 3.69, m |
| 14 | 17.3 | 1.35, d (6.0) | 18.5 | 1.38, d (6.2) |
| 15 | 172.7 | |||
| 16a | 38.1 | 2.77, dd (16.3, 4.9) | 36.8 | 2.97, dd (16.6, 4.9) |
| 16b | 2.63, dd (16.2, 8.0) | 2.52, dd (16.6, 4.7) | ||
| 17 | 67.2 | 4.33, m | 69.0 | 4.28, m |
| 18 | 83.7 | 3.52, m | 71.8 | 3.58, m |
| 19 | 67.0 | 3.95, m | 68.4 | 4.31, m |
| 20 | 18.3 | 1.26, d (6.2) | 18.8 | 1.45, d (6.4) |
| 15-OCH3 | 50.5 | 3.70, s | ||
| 21 | 92.8 | 5.03, d (4.4) | 94.1 | 5.02, d (4.3) |
| 22a | 40.1 | 2.43, d (13.7) | 41.4 | 2.44, d (13.8) |
| 22b | 2.08, d (13.6) | 2.06, d (13.6) | ||
| 23 | 89.8 | 89.8 | ||
| 24 | 84.4 | 3.62, m | 84.3 | 3.59, m |
| 25 | 65.9 | 3.50, m | 67.1 | 3.49, m |
| 26 | 16.4 | 0.77, d (6.0) | 17.7 | 0.76, d (6.1) |
| 27 | 18.4 | 1.69, s | 19.6 | 1.67, s |
| 24-OCH3 | 54.7 | 3.34, s | 56.0 | 3.35, s |
| Compound | MRSA |
|---|---|
| IC50 (µM) | |
| 1 | 47.7 |
| 2 | 31.3 |
| 3 | >100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, T.H.; Brittin, N.J.; Alas, I.; Roberts, C.D.; Chanana, S.; Braun, D.R.; Ericksen, S.S.; Guo, S.; Rajski, S.R.; Bugni, T.S. Discovery of New Everninomicin Analogs from a Marine-Derived Micromonospora sp. by Metabolomics and Genomics Approaches. Mar. Drugs 2025, 23, 316. https://doi.org/10.3390/md23080316
Lee TH, Brittin NJ, Alas I, Roberts CD, Chanana S, Braun DR, Ericksen SS, Guo S, Rajski SR, Bugni TS. Discovery of New Everninomicin Analogs from a Marine-Derived Micromonospora sp. by Metabolomics and Genomics Approaches. Marine Drugs. 2025; 23(8):316. https://doi.org/10.3390/md23080316
Chicago/Turabian StyleLee, Tae Hyun, Nathan J. Brittin, Imraan Alas, Christopher D. Roberts, Shaurya Chanana, Doug R. Braun, Spencer S. Ericksen, Song Guo, Scott R. Rajski, and Tim S. Bugni. 2025. "Discovery of New Everninomicin Analogs from a Marine-Derived Micromonospora sp. by Metabolomics and Genomics Approaches" Marine Drugs 23, no. 8: 316. https://doi.org/10.3390/md23080316
APA StyleLee, T. H., Brittin, N. J., Alas, I., Roberts, C. D., Chanana, S., Braun, D. R., Ericksen, S. S., Guo, S., Rajski, S. R., & Bugni, T. S. (2025). Discovery of New Everninomicin Analogs from a Marine-Derived Micromonospora sp. by Metabolomics and Genomics Approaches. Marine Drugs, 23(8), 316. https://doi.org/10.3390/md23080316







