Abstract
In this study we focus on the ophiuroid species associated with cold-water corals south of Iceland. The specimens were sampled with the ROV Phoca (GEOMAR) in three different areas, during the recent expedition MSM75 connected to the IceAGE_RR (Icelandic marine Animals: Genetics and Ecology_Reykjanes Ridge hydrothermal vent activity) project. In each area, several corals were sampled and the ophiuroid specimens identified to the species level. The integrative taxonomic approach, based on morphological characters and DNA barcoding with COI of the collected ophiuroids, revealed five species that live on corals: Ophiomitrella clavigera (Ljungman, 1865); Ophiomyxa serpentaria (Lyman, 1883); Ophiacantha cuspidata (Lyman, 1879); Ophiactis abyssicola (M. Sars, 1861); and Ophiolebes bacata Koehler, 1921. Some of the sampled deep-sea corals exclusively host the species O. clavigera. The collected species are therefore associated with different corals but do not demonstrate a species-specific distribution. The video data support the integrative taxonomy and confirm the ecological evidence.
Keywords:
echinoderms; brittle stars; Iceland; Reykjanes Ridge; COI; species delimitation; ROV; host preference 1. Introduction
The Nordic seas surrounding Iceland present geological and ecological interests and have been intensively studied during the last four decades [1,2,3,4]. The marine region around Iceland is indicated by a submarine mountain chain, the Greenland–Scotland-Ridge, and several water masses. The ridge forms a physical barrier that clearly separates the Arctic deep-sea basins from the North Atlantic ones, and its complex topography influences marine habitats. Cold, deep-water currents engulf Iceland from the western and eastern side with a north–south orientation. In contrast to this cold deep-water flow, warmer surface waters circulate around Iceland in a southwest to northeast direction [5,6,7,8]. The BIOFAR (1987–1992) [9] and BIOICE projects (1992–2004) [1] were the first extensive studies focusing on an inventorial effort of Icelandic benthic marine invertebrates. The follow-up project, IceAGE (Icelandic marine Animals: Genetics and Ecology, 2011–present), was then funded under a similar premise and enlarged the BIOICE sampling grit, adding a genetic focus [3]. During the expedition IceAGE_RR (Icelandic marine Animals: Genetics and Ecology_Reykjanes Ridge hydrothermal vent activity, MSM75), a special focus was placed on the geology and ecology of the Reykjanes Ridge (RR) [3,4]. The RR is an extension of the Mid-Atlantic Ridge (MAR), separated by the Bight Fracture Zone, and elongates ~900 km to the Icelandic peninsula [10]. In contrast to the rest of the MAR, the RR has an oblique orientation angle of 27°, and periodic adjustment by rift propagation formed its unique structure [10,11]. It is the longest V-shaped and volcanically influenced mid-oceanic ridge, and it’s topography is mainly dominated by faults [10]. This unique area hosts various ecosystems such as hydrothermal vents [4,5,12], seamount chains built from pillow lava, and reefs build by cold-water corals [2,13,14].
Cold-water coral reefs form a diverse ecosystem in the deep sea [15,16,17]. During their growth, cold-water corals produce a three-dimensional hard substrate and provide a habitat for many other organisms such as ophiuroids, crustaceans, and polychaete worms [16,17,18,19]. Cold-water corals are found in all seas, but high densities have been reported in the northeast Atlantic; mostly in water temperatures ranging from 4 °C to 12 °C [13,15,17,18,20]. In 1996, Copley et al. [2] performed an extensive study on the fauna of the Reykjanes Ridge and reported species of the class Ophiuroidea that are continuously found with pieces of corals. Additionally, Buhl-Mortensen et al. [15] found associations of echinoderms, in particular ophiuroids and other organisms, with some species of gorgonians.
Ophiuroids are a widespread class and have been frequently reported from coral reefs [21,22,23,24,25]. They can occur in high biomass from the shore to the hadal trenches, and from tropical waters to the Arctic and Antarctic regions [26,27]. With 2123 described species [28], ophiuroids are the most speciose class within the echinoderms. They are common dwellers in the benthic environment and inhabit many habitats with contrasting characteristics [21,29,30,31]. Brittle stars often represent the largest proportion of the megabenthic community and are found in high abundance. Because of their widespread distribution, ophiuroids are a suitable class for genetic studies focusing on phylogeny or biogeography [21,30,32,33,34,35,36,37,38]. In benthic organisms, genetic connectivity is often linked to the reproductive strategy, which can differ among species. In ophiuroids, the most widespread reproduction type is spawning, wherein each individual can release up to 880,000 eggs into the water before developing via a planktotrophic development stage to a benthic juvenile, but it is also still unknown for many species [39,40,41,42]. Ophiuroid larvae can also be lecithotrophic, with the production of fewer eggs containing a yolk sack [39,40,43]. The third reproduction type is brooding. Here, about 2–2000 eggs are carried in the adult bursa slits and develop directly to the juvenile state without any pelagic phase [39,41,44].
In recent years, molecular tools and DNA barcoding in particular have provided a useful method for fast, efficient, and reliable species identification and discovery [45,46,47,48]. It is based on the concept that intraspecific diversity for the COI gene is lower than interspecific diversity. The resulting difference is called a “barcode gap” [48]. DNA barcoding not only shortcuts the difficulties of a morphology-based identification, e.g., when diagnostic characters are damaged during collection, but also connects the different stages of animal development [49]. A 658-bp region of the cytochrome c oxidase I (COI) gene has an effective marker as a species delimitation tool in different groups of marine organisms [37,46,50,51], particularly in brittle stars [33,34,38,52]. Currently, records are available for 10,798 ophiuroid specimens, representing 604 species on the Barcode of Life Data System, BoLD.
In this work, to create a baseline for further studies, we used an integrative approach based on morphological characters and DNA barcoding to study ophiuroid species associated with cold-water corals collected with a remotely operated vehicle (ROV) from Reykjanes Ridge south of Iceland. We further analyzed image data to embed the molecular study in an ecological view.
2. Materials and Methods
2.1. Morphological Identification
All specimens used in this study were sampled during the IceAGE_RR expedition MSM75 [3] in 2018. During the expedition, three cold-water coral reefs found at depths ranging from 239 m to 1579 m along the Reykjanes Ridge were investigated using the ROV Phoca (GEOMAR). The sampling areas used in this study are divided into the following stations: Area 2 with stations 67-7, 188-4, 188-5, 188-6; Area 3 with stations 80-2, 80-5, 80-11, 111-6, 149-2, 149-3; and Area 4 with station 127-2 (Figure 1). All collected specimens were treated as described in Taylor et al. [4], fixated in 96% ethanol, and stored at −20 °C at the German Centre for Marine Biodiversity Research (DZMB), Senckenberg am Meer, Wilhelmshaven. Each specimen was then observed using a Leica M125 microscope and given an individual number (sample ID). A picture was taken from the oral and ventral sides (Leica EC3 Camera) and a tissue sample was collected from an arm segment for molecular analyses. The arm segment tissues were also fixated in 96% ethanol and stored at −20 °C. Morphological identifications were performed using Mortensen [53] and Paterson [54]. The morphology of the five species is represented in Figure 2.
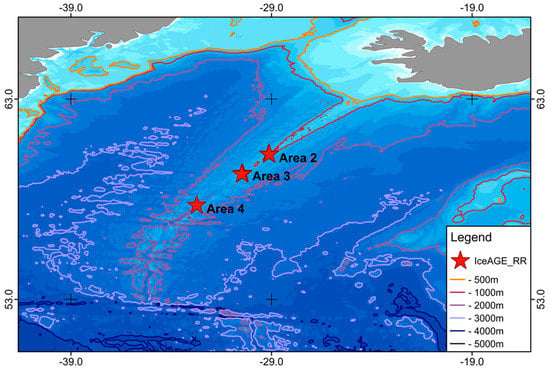
Figure 1.
Sampling areas in the North Atlantic south of Iceland. Depth contours are displayed between 500 and 5000 m.
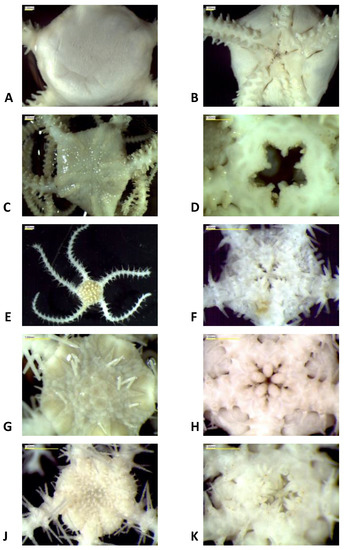
Figure 2.
Morphology of the five species in dorsal and ventral views. (A,B): Ophiomyxa serpentaria Lyman, 1883; (C,D): Ophiolebes bacata Koehler, 1921; (E,F): Ophiomitrella clavigera (Ljungman, 1865); (G,H): Ophiactis abyssicola (M. Sars, 1861); (I,J): Ophiacantha cuspidata Lyman, 1879.
2.2. Molecular Species Delimitation
DNA extractions were carried out using 40 μL Chelex (InstaGene™Matrix) according to the protocol of Estoup et al. [55]. PCR protocols followed Christodoulou et al. [52].
The sequencing and sequence editing were conducted as they were in Khodami et al. [34]. Additionally, six sequences from the same morphospecies or genus were either downloaded from Genbank or completed by the DZMB databank and added to the dataset (KX459004.1; KU895176; KJ620586.1; KF663499.1; HQ946175.1; HQ919150.1; MT152642.1; DZMB54597; DZMB42400B; DZMB42400A; DZMB37435B; Supplementary Material Table S1). Sequences were aligned using MAFFT v7.017 [56] and blasted against GenBank. The alignment was exported as a FASTA file for further analysis. For each specimen, a picture and the sequence data were uploaded on the Barcode of Life Data System (BoLD, www.barcodinglife.org, accessed on 19 April 2022) in the project IARRO (Associated ophiuroid fauna on cold water corals of the Reykjanes Ridge; DOI: dx.doi.org/10.5883/DS-IAORR) and will also be available on GenBank (ON341454–ON341716).
Three different methods for species delimitation were applied to assess the number of putative species. The first one, Barcode Index Number System [57], is a distance-based method. Within BoLD, the newly submitted sequences are compared with the sequences already available. They are clustered based on their molecular divergence using algorithms that aim to discover discontinuities between these clusters. A specific code (Barcode Index Number or BIN) is assigned to each cluster, either already existing or newly generated if submitted sequences do not match with sequences of already known BINs.
Automatic Barcode Gap Discovery (ABGD) is the second, distance-based method. It assigns the inserted specimens into species based on the pure distribution of pairwise differences (p-distance). The ABGD method initially calculates the indicative barcode gap to partition the sequences and gap detection, then continues recursively on previously obtained clusters to redefine partitions [48].
The third one is the Generalized Mixed Yule Coalescent (GMYC) method. It is a likelihood method for delimiting species by fitting within- and between-species branching models to reconstruct ultrametric gene trees. The model is described by Pons et al. and Monaghan et al. [58,59]. Species in this model are delimited by the descendent nodes of branches crossing the barcode gap threshold. This approach defines each species based on the most recent common ancestor on the phylogenetic tree and assumes that the most recent diversification event occurred before the oldest within-species coalescent event. The method should be implemented on a pre-analyzed Bayesian phylogenetic tree. Beast package (Version 1.8.4, available on: http://beast.bio.ed.ac.uk, accessed on 20 February 2020) has been used to prepare the ultrametric rooted tree from COI sequences of ophiuroids, setting the tree prior to “Speciation: Yule Process” for 506 generations with a sampling frequency of 5000 Generations. TreeAnnotator has been used to summarize the trees to a consensus tree with posterior probabilities. This tree has been used to run GMYC analysis by the SPLITS package (http://splits.r-forge.r-project.org, accessed on 20 February 2020) in the statistic program R (Version 3.3.2, available at: https://www.r-project.org, accessed on 5 January 2020). The genetically delimited species were examined for diagnostic morphological characteristics to confirm the species delimitation by morphological evidence in parallel to genetic analysis.
The distribution maps for each species are based on an extended data set, including the BIOICE data (Stöhr, previously unpublished material) and the public Ocean Biodiversity Information System (OBIS), and were completed by the stations of IceAGE_RR. The full distribution was added to QGIS (version 3.12, http://qgis.org, accessed on 30 November 2020) with which the distribution maps for each species were also created.
2.3. Image Data Analysis
Images used for ophiuroid assessment data generation were reviewed in random order to minimize time or sequence-related bias [60]. Specimens were identified to the collected morphospecies, measured using the BIIGLE 2.0 software [61], and assigned to a defined habitat. The data were then exported from BIIGLE and the statistical analyses were carried out with the following packages in R. The present-absence matrix was analyzed with the package “vegan”; a Nonmetric Multidimensional Scaling (nMDS) was carried out to determine if there were differences in the distribution of the ophiuroids among the defined habitats. The package “BiodiversityR” was applied to measure the Shannon index (H). The differences of the ophiuroid assemblages between the habitats were then calculated with a multilevel pairwise comparison using the package “pairwiseAdonis”. Additionally, an indicator species was defined for each habitat with the package “indicspecies”.
3. Results
3.1. Morphological and Molecular Species Delimitation
A total of 684 ophiuroids were picked by the ROV at three areas along the Reykjanes Ridge south of Iceland (Figure 1) and morphologically identified to the species level (Figure 2). DNA extractions were performed on 288 specimens. In total, 270 new sequences and 11 public sequences of 658 bp were included in the analysis based on COI. All, BIN, ABGD, and GMYC methods are congruent and delineate five species, which where morphologically identified as follows: Ophiomitrella clavigera (Ljungman, 1865); Ophiomyxa serpentaria (Lyman, 1883); Ophiacantha cuspidata (Lyman, 1879); Ophiactis abyssicola (M. Sars, 1861); and Ophiolebes bacata (Koehler, 1921). The morphology of the five species is represented in Figure 2. Figure 3 displays the maximum likelihood tree with the highlighted five species of this study. The maximum likelihood reconstruction has a high branch support with values greater than 92%. The mean p-distance within a species was 0.3% and the mean p-distance between species was 26.1%.
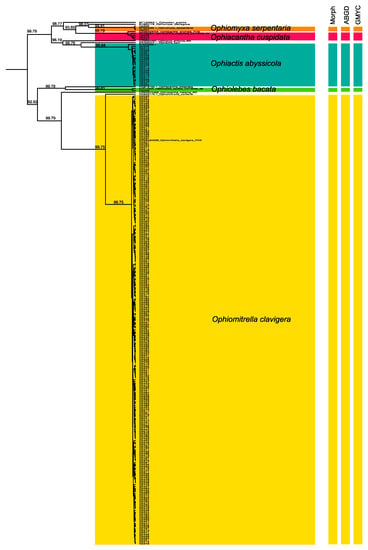
Figure 3.
Gene tree of Bayesian analysis of COI (numbers on branches represent posterior probabilities >0.92) and species delimitation tools for COI fragment. The vertical bars represent alternative taxonomies, respectively supported by Automatic Barcoding Gap Discovery (ABGD) and General Mixed Yule Coalescent (GMYC), in addition to morphology. The colors are related to the identified clades.
3.2. Ecological Analysis
The distribution map illustrated in Figure 4 presents different patterns for each species. It is striking that three species are only distributed south of Iceland (O. serpentaria, O. cuspidata and O. bacata). Ophiomitrella clavigera is mostly recorded from the south of Iceland but also from a single station in the shallow (between 100–300 m) north of Iceland. Ophiactis abyssicola has a wide distribution across the North Atlantic as well as in the Arctic region. The specimens of the IceAGE_RR project are, in most cases, the first record of these species from the RR.
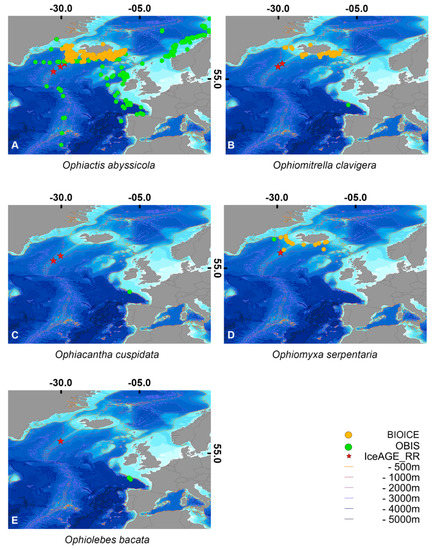
Figure 4.
The maps represent the distribution of each ophiuroid species by reference (A) Ophiactis abyssicola, (B) Ophiomitrella clavigera, (C) Ophiacantha cuspidata, (D) Ophiomyxa serpentaria, (E) Ophiolebes bacata. The red stars indicate the stations where the ophiuroids were sampled during IceAGE_RR. The green dots illustrate the stations that were reported by OBIS (accessed 24 September 2021). The yellow dots are records from BIOICE material.
A total of 5174 ophiuroid individuals were observed in the four ROV dives during the present study. The ophiuroids were classified into five morphospecies: O. abyssicola, O. clavigera, O. cuspidata, O. serpentaria, O. bacata, and “other” for the specimens that couldn’t be categorized into a morphospecies. Only O. abyssicola, O. clavigera, and O. cuspidata were found in the analyzed images. The habitats are pictured in Figure 5 and were defined as: “Pillow”, “Coral”, “Pillow + Coral”, “Pillow + Coral rubble”, and “Pillow + Sediment.” Distribution of the ophiuroid species differed with each habitat (Figure 6A–E). Ophiomitrella clavigera was mostly found on corals but rarely in the other habitats. The other species were equally distributed among the remaining habitats. The comparison of the species assemblages is displayed in an nMDS plot of the ophiuroid diversity divided into the different habitat types (Figure 6F). Generally, a separation can be seen between the habitat type “coral” and the rest of the habitat types. The Shannon index (H) between all habitats is 0.26 (p = 0.32). However, the pairwise adonis indicates a significant (p = 0.01 *) difference among all habitats, except for the comparison between the habitats “Pillow” and “Pillow + Sediment” (p = 0.52). The test for the indicator species per habitat calculated O. clavigera (0.669; p = 0.005 **) for the habitat “Coral”, O. cuspidata (0.308; p = 0.03 *) for the habitat “Pillow + Coral Rubble” and O. abyssicola (0.765; p = 0.005 **) for all the other habitat types.
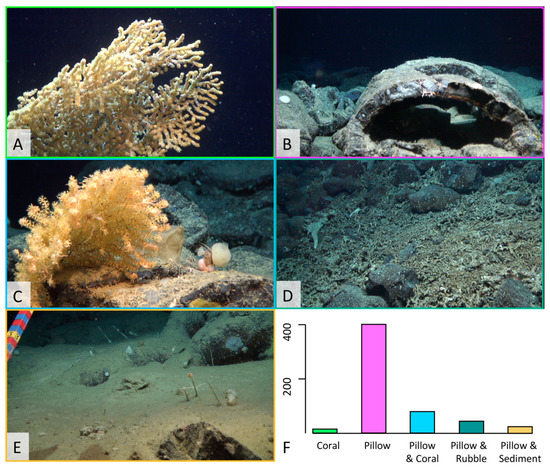
Figure 5.
(A–E): The images demonstrate a representative of each defined habitat type. (A): Coral; (B): Pillow; (C): Pillow + Coral; (D): Pillow + Coral Rubble; (E): Pillow + Sediment. (F): The bar plot displaying the proportional occurrence of each habitat.
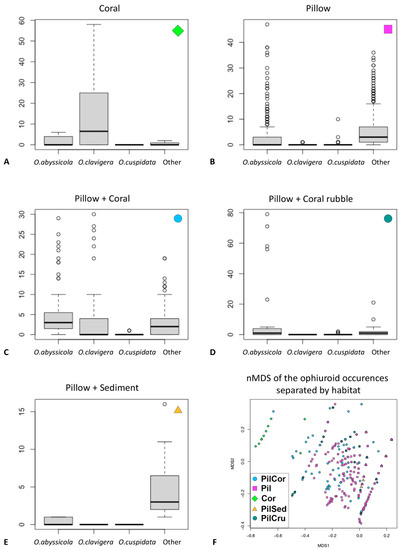
Figure 6.
(A–E): The boxplots illustrate the proportional abundance of Ophiomyxa serpentaria Lyman, 1883; Ophiolebes bacata Koehler, 1921; Ophiomitrella clavigera (Ljungman, 1865); Ophiactis abyssicola (M. Sars, 1861); and Ophiacantha cuspidata Lyman, 1879 relatied to each habitat: (A): “Coral” (Cor); (B): Pillow” (Pil); (C): “Pillow + Coral” (PilCor), (D): “Pillow + Coral Rubble” (PilCru); (E): “Pillow + Sediment” (PilSed). (F): The distribution of the ophiuroid fauna depending on the habitat presented by Nonmetric Multidimensional Scaling (nMDS).
4. Discussion
4.1. Ophiuroids Distribution According to Seafloor Heterogeneity
The ophiuroid species in this work were all found on cold-water corals and recorded in previous studies from the North Atlantic [62,63,64]. Ophiomyxa serpentaria and O. abyssicola were found during different expeditions in the northeast Atlantic [63,64] and particularly on the RR [2]. There is a significant (p = 0.01) separation of the faunal composition in the nMDS plot between the different habitats, which may be explained by differences in food supply, surfaces, and structures of the habitats [65,66,67,68,69,70]. Particularly interesting is the indicator species from the coral habitat, which was identified as O. clavigera (0.669; p = 0.005). This species was observed to exclusively use gorgonians as hosts. In contrast to other coral-associated ophiuroids that sit alone or in small groups on the corals (e.g., reported for O. oedipus Lyman, 1879 or some species of the genus Asteroschema Örsted and Lütken, 1856 [22,23,71]), the specimens of O. clavigera aggregate with hundreds of individuals per coral. Additionally, their brooding lifestyle leads to the suggestion that there is a higher gene flow within these aggregations than between them. As with O. clavigera, O. abyssicola has been reported in association with different hosts, like corals or sponges, but it also occurs in high densities on the seafloor [2,19,63,64]. To use corals as hosts serves different advantages. One advantage is the species can expand their habitat from the seafloor to a higher level and reach new food resources [15,19,22] as the branches of the corals are often in a strong current with different food sources, like planktonic organisms as copepods or dead particles from the sea surface. Whereas many ophiuroids are known to be suspension feeders with an unspecified diet, they are observed with some arms entwined around the coral branch and the other arms within the current. Furthermore, the higher level can enhance larval dispersal when the eggs are released into the water currents [2,15,19,25,72]. Conversely, ophiuroids can use corals as a shelter from strong currents or predators [15,19]. However, dead coral, classified as coral rubble, was inhabited by various ophiuroid species, especially by O. cuspidata, which was the indicator species for this habitat (0.308; p = 0.03). The remnants of the corals still form a diverse habitat with hard substrates, holes, and hollows that provide shelter for the ophiuroids and is further inhabited by smaller invertebrates that serve as food supply [18,73]. The most abundant habitat was the lava pillows that form a smooth hard substrate surface with some cracks and folds in between. Mostly, the indicator species O. abyssicola was living inside these formations extending their long and spiny arms in the water column, which suggests a suspension feeding lifestyle. Additionally, individuals were also spotted a few times sitting on corals with two to three arms attached to the branch and the others extended in the water column. However, an association with a special host coral couldn’t be identified, as they were seen on varying coral species as well as highly abundant on the seafloor.
4.2. Deep Icelandic Ophiuroid Fauna
The deep-sea ophiuroid fauna around Iceland has been studied for decades taxonomically [42,54,62,74,75]. In 1985, Paterson published a book about “The deep-sea Ophiuroidea from the North Atlantic Ocean” containing approximately 120 species living below 1000 m [54]. In recent years, new taxonomic discoveries have been made [26,76], and more information about genetic diversity has been established [22,30,33,34,77,78]. The study by Copley et al. [2] focused on the faunal community living on the RR. It suggested a different bathymetric range for species with a split between 800 m and 1000 m, because of the transition of the two water masses occurring at this depth (SMW and UNADW). Ophiactis abyssicola, O. clavigera, and O. serpentaria were recorded along the RR across the whole depth range, so differentiations of the bathymetric depth cannot be supported in this study. The contrast between the results presented by Copley et al. [2] and our work is not surprising. Deep-sea environments are among the least studied on our planet [16,79] and bias in the sampling as well as cryptic speciation could be possible [52,80,81,82]. A comparable approach of barcoding the Icelandic ophiuroid fauna was caried out from a more northern area by Khodami et al. [34]. These authors found a completely different ophiuroid community than described in this study. Due to different abiotic factors (e.g., soft sediment instead of hard substrate, differences in depth and temperature) as well as the different sample set up (ROV vs. AGT, EBS, and TAD) the differences between the studies reflect the diversity of the deep-sea ophiuroid fauna around Iceland and that a wide sample set up with different sampling methods should be used.
4.3. Species Delimitation and DNA Barcoding
Over the last few decades, DNA barcoding has been increasingly used for species delimitations and has proven to be an effective method for many organisms, including ophiuroids [34,37,38,83]. The species from the present data set could also be clearly distinguished from each other by the established DNA barcoding method, so we focused more on the way of life than on the genetic analysis. However, five BINs could be assigned to our studied material. Three of the BINs are identified to the species level and publicly accessible, whereas the other two are unique, held in private datasets, or lack detailed identification. This indicates that genetic information of deep-sea megabenthic fauna is still quite unknown [52,84,85,86,87], and this dataset acts as a supplement to this knowledge. Working at such a small regional scale certainly does not mirror the complete variation within each species, and indicates the limits of DNA barcoding. It only provides partial information due to being limited to just one region in the mitochondrial gene [88]. However, this method has proved useful to extend background knowledge of taxonomy, population genetics, and molecular phylogenetics [89]. DNA barcoding reduces the difficulties in identifying damaged specimens and can overcome identification problems with morphologically highly variable species, and is helpful in connecting the larvae to the adults [49]. In this work, we had different developmental stages of O. clavigera, and DNA barcoding supported the result that all of them belong to this species. The mean intra- and interspecific p-distances (0.3% and 26.1%) were in the known range for echinoderms and do not present any signs of cryptic speciation [34,38]. It is important to point out that the taxonomic and molecular diversity in this study does not reflect the complete diversity of ophiuroid species from Icelandic and adjacent waters, but reflects only a small representation. For example, the study published by Khodami et al. [34] reported a completely different ophiuroid community with a higher molecular and taxonomic diversity. The authors further reported that results in morphology can differ from genetic evidence and that closer related species were hard to delimit, as presented for the genus Ophiacantha. Cryptic speciation or recently separated species are still problematic in genetic analyses [30,33,34,82]. However, the five species from this study, belonging to different genera, did not present any cryptic speciation and could be separated well from each other. It is worth highlighting that the genetic methods were congruent with the morphological identification.
5. Conclusions
Five ophiuroid species were successfully and consistently identified in all methods, using morphological characters combined with the COI marker. This study highlights the exclusive association of the brittle star species Ophiomitrella clavigera with non-species-specific gorgonian cold-water corals. The combination of video data with morphological and molecular information from actual sampled specimens highlights the possibility to better understand the variety in ecology and geography that is often mirrored in the genetic diversity. We need to understand the species in connection with their environment to be able to draw conclusions from our morphological and genetic results. Video surveys help to observe specimens in their actual environment and supplement the knowledge from classical sampling methods.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d14050358/s1, Table S1: Table of additional sequences used in the dataset.
Author Contributions
A.E., S.B. and P.M.A. designed the study, supervised all steps of the project, analyzed the data, and drafted the manuscript. S.B. organized the sampling and provided the biological material. A.E. performed the COI sequencing. A.E. and P.M.A. analyzed the COI data. S.S. did the morphological species identification and provided the BIOICE distribution data of ophiuroid species. A.E. and J.T. analyzed the image data. All authors have read and agreed to the published version of the manuscript.
Funding
The sampling of the specimens was funded via the German Science Foundation (DFG grant MERMET17-15).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The sequence and meta data of the specimens were deposit in GenBank and BoLD (DOI).
Acknowledgments
We would like to thank the following people for their key contributions to our manuscript: the crew of MS Merian during IceAGE_RR (MSM75) cruise; Tim O’Hara (Victoria Museum Melbourne), who helped to check the sequencing data and provided some complement sequences for not collected species; Antje Fischer and Karen Jeskulke (TAs DZMB HH) for their support in sorting the samples; and Sahar Khodami (DZMB Wilhelmshaven) for the help in the lab.
Conflicts of Interest
The authors declare no conflict of interest. The research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Brix, S.; Meißner, K.; Kaiser, S.; Svavarsson, J.; Yasuhara, M.; Martinez Arbizu, P. The IceAGE Project—A Follow up of BIOICE. Pol. Polar Res. 2014, 35, 141–150. [Google Scholar] [CrossRef]
- Copley, J.T.P.; Tyler, P.A.; Sheader, M.; Murton, B.J.; German, C.R. Megafauna from Sublittoral to Abyssal Depths along the Mid-Atlantic Ridge South of Iceland. Oceanol. Acta 1996, 19, 549–559. [Google Scholar]
- Devey, C.W.; Brix, S.; Barua, A.; Bodendorfer, M.; Cuno, P.; Frutos, I.; Huusmann, H.; Kurbjuhn, T.; Le Saout, M.; Linse, K.; et al. Detailed Mapping and Sampling of the Reykjanes Ridge, Cruise No. MSM75, 29 June 2018–8 August 2018, Reykjavik-Reykjavik; GEOMAR: Bonn, Germany, 2018. [Google Scholar] [CrossRef]
- Taylor, J.; Devey, C.; Le Saout, M.; Petersen, S.M.; Frutos, I.I.; Linse, K.; Loerz, A.-N.; Pałgan, D.; Tandberg, A.H.; Svavarsson, J.; et al. The Geological and Faunal Composition of Steinahóll Vent Sites, Reykjanes Ridge, Iceland. Front. Mar. Sci. 2021, 8, 520713. [Google Scholar] [CrossRef]
- German, C.R.; Briem, J.; Chin, C.; Danielsen, M.; Holland, S.; James, R.; Jónsdóttir, A.; Ludford, E.; Moser, C.; Ólafsson, J.; et al. Hydrothermal Activity on the Reykjanes Ridge: The Steinahóll Vent-Field at 63°06′ N. Earth Planet. Sci. Lett. 1994, 121, 647–654. [Google Scholar] [CrossRef]
- Malmberg, S.A.; Jónsson, S. Timing of Deep Convection in the Greenland and Iceland Seas. ICES J. Mar. Sci. 1997, 54, 300–309. [Google Scholar] [CrossRef]
- Meißner, K.; Fiorentino, D.; Schnurr, S.; Martinez Arbizu, P.; Huettmann, F.; Holst, S.; Brix, S.; Svavarsson, J. Distribution of Benthic Marine Invertebrates at Northern Latitudes—An Evaluation Applying Multi-Algorithm Species Distribution Models. J. Sea Res. 2014, 85, 241–254. [Google Scholar] [CrossRef]
- Ostmann, A.; Schnurr, S.; Martinez Arbizu, P. Marine Environment around Iceland: Hydrography, Sediments and First Predictive Models of Icelandic Deep—Sea Sediment Characteristics. Pol. Polar Res. 2014, 35, 151–176. [Google Scholar] [CrossRef]
- Nørrevang, A.; Brattegard, T.; Josefson, A.B.; Sneil, J.-A.; Tendal, O.S. List of BIOFAR Stations. Sarsia 1994, 79, 165–180. [Google Scholar] [CrossRef]
- Keeton, J.A.; Searle, R.C.; Parsons, B.; White, R.S.; Murton, B.J.; Parson, L.M.; Peirce, C.; Sinha, M.C. Bathymetry of the Reykjanes Ridge. Mar. Geophys. Res. 1997, 19, 55–64. [Google Scholar] [CrossRef]
- Hey, R.; Martinez, F.; Höskuldsson, Á.; Benediktsdóttir, Á. Propagating Rift Model for the V-Shaped Ridges South of Iceland. Geochem. Geophys. Geosyst. 2010, 11, 1–24. [Google Scholar] [CrossRef]
- Pałgan, D.; Devey, C.W.; Yeo, I.A. Volcanism and Hydrothermalism on a Hotspot–Influenced Ridge: Comparing Reykjanes Peninsula and Reykjanes Ridge, Iceland. J. Volcanol. Geotherm. Res. 2017, 348, 62–81. [Google Scholar] [CrossRef]
- Arnaud-Haond, S.; Van den Beld, I.M.J.; Becheler, R.; Orejas, C.; Menot, L.; Frank, N.; Grehan, A.; Bourillet, J.F. Two “Pillars” of Cold-Water Coral Reefs along Atlantic European Margins: Prevalent Association of Madrepora oculata with Lophelia pertusa, from Reef to Colony Scale. Deep Res. Part II Top. Stud. Oceanogr. 2017, 145, 110–119. [Google Scholar] [CrossRef]
- Frank, N.; Freiwald, A.; López Correa, M.; Wienberg, C.; Eisele, M.; Hebbeln, D.; Van Rooij, D.; Henriet, J.P.; Colin, C.; van Weering, T.; et al. Northeastern Atlantic Cold-Water Coral Reefs and Climate. Geology 2011, 39, 743–746. [Google Scholar] [CrossRef]
- Buhl-Mortensen, L.; Vanreusel, A.; Gooday, A.J.; Levin, L.A.; Priede, I.G.; Buhl-Mortensen, P.; Gheerardyn, H.; King, N.J.; Raes, M. Biological Structures as a Source of Habitat Heterogeneity and Biodiversity on the Deep Ocean Margins. Mar. Ecol. 2010, 31, 21–50. [Google Scholar] [CrossRef]
- Ramirez-Llodra, E.; Brandt, A.; Danovaro, R.; De Mol, B.; Escobar, E.; German, C.R.; Levin, L.A.; Martinez Arbizu, P.; Menot, L.; Buhl-Mortensen, P.; et al. Deep, Diverse and Definitely Different: Unique Attributes of the World’s Largest Ecosystem. Biogeosciences 2010, 7, 2851–2899. [Google Scholar] [CrossRef]
- Roberts, J.M.; Wheeler, A.J.; Freiwald, A. Reefs of the Deep: The Biology and Geology of Cold-Water Coral Ecosystems. Science 2006, 312, 543–547. [Google Scholar] [CrossRef]
- Mortensen, P.B.; Fosså, J.H. Species Diversity and Spatial Distribution of Invertebrates on Deep–Water Lophelia Reefs in Norway. Proc. 10th Int. Coral Reef Symp. 2006, 1868, 1849–1868. [Google Scholar]
- Buhl-Mortensen, L.; Mortensen, P. Symbiosis in Deep-Water Corals. Symbiosis 2004, 37, 33–61. [Google Scholar]
- Davies, A.J.; Guinotte, J.M. Global Habitat Suitability for Framework-Forming Cold-Water Corals. PLoS ONE 2011, 6, e18483. [Google Scholar] [CrossRef]
- Bribiesca-Contreras, G.; O’Hara, T.D.; Verbruggen, H.; Hugall, A.F. Global Biogeographic Structuring of Tropical Shallow—Water Brittle Stars. J. Biogeogr. 2019, 46, 1287–1299. [Google Scholar] [CrossRef]
- Cho, W.; Shank, T.M. Incongruent Patterns of Genetic Connectivity among Four Ophiuroid Species with Differing Coral Host Specificity on North Atlantic Seamounts. Mar. Ecol. 2010, 31, 121–143. [Google Scholar] [CrossRef]
- Mosher, C.V.; Watling, L. Partners for Life: A Brittle Star and Its Octocoral Host. Mar. Ecol. Prog. Ser. 2009, 397, 81–88. [Google Scholar] [CrossRef]
- Starmer, J.A. An Annotated Checklist of Ophiuroids (Echinodermata) from Guam. Micronesica 2003, 35, 547–562. [Google Scholar]
- Bos, A.R.; Hoeksema, B.W. Mushroom Corals (Fungiidae) in the Davao Gulf, Philippines, with Records of Associated Fish and Other Cryptofauna. Raffles Bull. Zool. 2017, 65, 198–206. [Google Scholar]
- O’Hara, T.D.; Stöhr, S.; Hugall, A.F.; Thuy, B.; Martynov, A. Morphological Diagnoses of Higher Taxa in Ophiuroidea (Echinodermata) in Support of a New Classification. Eur. J. Taxon. 2018, 416, 1–35. [Google Scholar] [CrossRef]
- Stöhr, S.; O’Hara, T.D.; Thuy, B. Global Diversity of Brittle Stars (Echinodermata: Ophiuroidea). PloS ONE 2012, 7, e31940. [Google Scholar] [CrossRef]
- Stöhr, S.; O’Hara, T.; Thuy, B. (Eds.) World Ophiuroidea Database. 2022. Available online: https://www.marinespecies.org/ophiuroidea (accessed on 19 April 2022).
- Christodoulou, M.; O’Hara, T.D.; Hugall, A.F.; Arbizu, P.M. Dark Ophiuroid Biodiversity in a Prospective Abyssal Mine Field. Curr. Biol. 2019, 29, 3909–3912. [Google Scholar] [CrossRef]
- Galaska, M.P.; Sands, C.J.; Santos, S.R.; Mahon, A.R.; Halanych, K.M. Geographic Structure in the Southern Ocean Circumpolar Brittle Star Ophionotus Victoriae (Ophiuridae) Revealed from MtDNA and Single-Nucleotide Polymorphism Data. Ecol. Evol. 2017, 7, 475–485. [Google Scholar] [CrossRef]
- O’Hara, T.D.; Hugall, A.F.; Woolley, S.N.C.; Bribiesca-contreras, G.; Bax, N.J. Contrasting processes drive ophiuroid phylodiversity across shallow and deep seafloors. Nature 2019, 565, 636–639. [Google Scholar] [CrossRef]
- Hugall, A.F.; O’Hara, T.D.; Hunjan, S.; Nilsen, R.; Moussalli, A. An Exon-Capture System for the Entire Class Ophiuroidea. Mol. Biol. Evol. 2015, 33, 281–294. [Google Scholar] [CrossRef]
- Jossart, Q.; Sands, C.J.; Sewell, M.A. Dwarf Brooder versus Giant Broadcaster: Combining Genetic and Reproductive Data to Unravel Cryptic Diversity in an Antarctic Brittle Star. Heredity 2019, 123, 622–633. [Google Scholar] [CrossRef]
- Khodami, S.; Martinez Arbizu, P.; Stöhr, S. Molecular Species Delimitation of Icelandic Brittle Stars (Echinodermata: Ophiuroidea). Pol. Polar Res. 2014, 35, 243–260. [Google Scholar] [CrossRef]
- O’Hara, T.D.; Hugall, A.F.; Thuy, B.; Moussalli, A. Phylogenomic Resolution of the Class Ophiuroidea Unlocks a Global Microfossil Record. Curr. Biol. 2014, 24, 1874–1879. [Google Scholar] [CrossRef]
- Woolley, S.N.C.; Foster, S.D.; O’Hara, T.D.; Wintle, B.A.; Dunstan, P.K. Characterising Uncertainty in Generalised Dissimilarity Models. Methods Ecol. Evol. 2017, 8, 985–995. [Google Scholar] [CrossRef]
- Ward, R.D.; Holmes, B.H.; O’Hara, T.D. DNA Barcoding Discriminates Echinoderm Species. Mol. Ecol. 2008, 8, 1202–1211. [Google Scholar] [CrossRef] [PubMed]
- Boissin, E.; Hoareau, T.B.; Paulay, G.; Bruggemann, J.H. DNA Barcoding of Reef Brittle Stars (Ophiuroidea, Echinodermata) from the Southwestern Indian Ocean Evolutionary Hot Spot of Biodiversity. Ecol. Evol. 2017, 7, 11197–11203. [Google Scholar] [CrossRef]
- McEdward, L.R.; Miner, B.G. Larval and Life-Cycle Patterns in Echinoderms. Can. J. Zool. 2001, 79, 1125–1170. [Google Scholar] [CrossRef]
- Schoener, A. Fecundity and Possible Mode of Development of Some Deep-Sea Ophiuroids. Limnol. Ocenaography 1972, 17, 193–199. [Google Scholar] [CrossRef]
- Tyler, P.A. Reproductive Patterns in Deep Sea Ophiuroids from the North East Atlantic. Proc. Th Eur. Colloq. Echinoderms 1979, 417–420. [Google Scholar]
- Stöhr, S. Who’s Who among Baby Brittle Stars (Echinodermata: Ophiuroidea): Postmetamorphic Development of Some North Atlantic Forms. Zool. J. Linn. Soc. 2005, 143, 543–576. [Google Scholar] [CrossRef]
- McClain, C.R.; Hardy, S.M. The Dynamics of Biogeographic Ranges in the Deep Sea. Proc. R. Soc. B Biol. Sci. 2010, 277, 3533–3546. [Google Scholar] [CrossRef]
- Schoener, A. Post-Larval Development of Five Deep-Sea Ophiuroids. Deep Res. 1967, 14, 645–660. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. B-Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Layton, K.K.S.; Corstorphine, E.A.; Hebert, P.D.N. Exploring Canadian Echinoderm Diversity through DNA Barcodes. PLoS ONE 2016, 11, e0166118. [Google Scholar] [CrossRef]
- Puillandre, N.; Modica, M.V.; Gustave, O.; Place, L.L.; West, C.P. Large-Scale Species Delimitation Method for Hyperdiverse Groups. Mol. Ecol. 2012, 21, 2671–2691. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Lambert, A.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for Primary Species Delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Meiklejohn, K.A.; Wallman, J.F.; Dowton, M. DNA Barcoding Identifies All Immature Life Stages of a Forensically Important Flesh Fly (Diptera: Sarcophagidae). J. Forensic Sci. 2013, 58, 184–187. [Google Scholar] [CrossRef]
- Bribiesca-Contreras, G.; Solis-Martin, F.A.; Laguarda-Figueras, A.; Zaldivar-Riveron, A. Identification of Echinoderms (Echinodermata) from an Anchialine Cave in Cozumel Island, Mexico, Using DNA Barcodes. Mol. Ecol. Resour. 2013, 13, 1137–1145. [Google Scholar] [CrossRef]
- Moreau, C.; Danis, B.; Jossart, Q.; Eléaume, M.; Sands, C.; Achaz, G.; Agüera, A.; Saucède, T. Is Reproductive Strategy a Key Factor in Understanding the Evolutionary History of Southern Ocean Asteroidea (Echinodermata)? Ecol. Evol. 2019, 9, 8465–8478. [Google Scholar] [CrossRef]
- Christodoulou, M.; O’Hara, T.D.; Hugall, A.; Khodami, S.; Rodrigues, C.F.; Hilario, A.; Vink, A.; Martinez Arbizu, P. Unexpected High Abyssal Ophiuroid Diversity in Polymetallic Nodule Fields of the Northeast Pacific Ocean, and Implications for Conservation. Biogeosci. Discuss. 2020, 17, 1845–1876. [Google Scholar] [CrossRef]
- Mortensen, T. Handbook of the Echinoderms of the British Isles; Backhuys: Rotterdam, The Netherlands, 1927; ISBN 9788578110796. [Google Scholar]
- Paterson, G.L.J.J. The Deep-Sea Ophiuroidea of the North Atlantic Ocean. Bull. Br. Mus. (Nat. Hist. Zool.) 1985, 49, 1–162. [Google Scholar]
- Estoup, A.; Largiader, C.R.; Perrot, E.; Chourrout, D. Rapid One-Tube DNA Extraction for Reliable PCR Detection of Fish Polymorphic Markers and Transgenes. Mol. Mar. Biol. Biotechnol. 1996, 5, 295–298. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. The Barcode of Life Data System. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Pons, J.; Barraclough, T.G.; Gomez-zurita, J.; Cardoso, A. Sequence-Based Species Delimitation for the DNA Taxonomy of Undescribed Insects. Syst. Biol. 2006, 55, 595–609. [Google Scholar] [CrossRef]
- Monaghan, M.T.; Wild, R.; Elliot, M.; Fujisawa, T.; Balke, M.; Inward, D.J.G.; Lees, D.C.; Ranaivosolo, R.; Eggleton, P.; Barraclough, T.G.; et al. Accelerated Species Inventory on Madagascar Using Coalescent-Based Models of Species Delineation. Syst. Biol. 2009, 58, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Durden, J.M.; Bett, B.J.; Schoening, T.; Morris, K.J.; Nattkemper, T.W.; Ruhl, H.A. Comparison of Image Annotation Data Generated by Multiple Investigators for Benthic Ecology. Mar. Ecol. Prog. Ser. 2016, 552, 61–70. [Google Scholar] [CrossRef]
- Langenkämper, D.; Zurowietz, M.; Schoening, T.; Nattkemper, T.W. BIIGLE 2.0—Browsing and Annotating Large Marine Image Collections. Front. Mar. Sci. 2017, 4, 83. [Google Scholar] [CrossRef]
- Smirnov, I.S.; Piepenburg, D.; Ahearn, C.; Juterzenka, K.V. Deep-Sea Fauna of European Seas: An Annotated Species Check-List of Benthic Invertebrates Living Deeper than 2000 m in the Seas Bordering Europe. Ophiuroidea. Invertebr. Zool. 2014, 11, 192–209. [Google Scholar] [CrossRef]
- Pearson, M.; Gage, J.D. Diets of Some Deep-Sea Brittle Stars in the Rockall Trough. Mar. Biol. 1984, 82, 247–258. [Google Scholar] [CrossRef]
- Tyler, P.A.; Emson, R.H.; Sumida, P.; Howell, K. Ophiuroid Distribution at Sublittoral and Bathyal Depths Round the Faroe Islands, NE Atlantic Ocean. Ann. Soc. Sci. Faroensis. Suppl. 2005, 41, 175–194. [Google Scholar]
- Amon, D.J.; Ziegler, A.F.; Dahlgren, T.G.; Glover, A.G.; Goineau, A.; Gooday, A.J.; Wiklund, H.; Smith, C.R. Insights into the Abundance and Diversity of Abyssal Megafauna in a Polymetallic-Nodule Region in the Eastern Clarion-Clipperton Zone. Sci. Rep. 2016, 6, 30492. [Google Scholar] [CrossRef]
- De Leo, F.C.; Smith, C.R.; Rowden, A.A.; Bowden, D.A.; Clark, M.R. Submarine Canyons: Hotspots of Benthic Biomass and Productivity in the Deep Sea. Proc. R. Soc. B Biol. Sci. 2010, 277, 2783–2792. [Google Scholar] [CrossRef]
- Hardy, S.M.; Smith, C.R.; Thurnherr, A.M. Can the Source-Sink Hypothesis Explain Macrofaunal Abundance Patterns in the Abyss? A Modelling Test. Proc. R. Soc. B Biol. Sci. 2015, 282, 20150193. [Google Scholar] [CrossRef]
- Stöhr, S.; Segonzac, M. Deep-Sea Ophiuroids (Echinodermata) from Reducing and Non-Reducing Environments in the North Atlantic Ocean. J. Mar. Biol. Assoc. UK 2005, 85, 383–402. [Google Scholar] [CrossRef]
- Woolley, S.N.C.; Derek, P.; Dunstan, P.K.; Guillera-Arroita, G.; Lahoz_Monfort, J.J.; O’Hara, T.D. Deep-Sea Diversity Patterns Are Shaped by Energy Availability. Nature 2016, 533, 393–396. [Google Scholar] [CrossRef]
- Zeppilli, D.; Pusceddu, A.; Trincardi, F.; Danovaro, R. Seafloor Heterogeneity Influences the Biodiversity—Ecosystem Functioning Relationships in the Deep Sea. Nat. Publ. Gr. 2016, 6, 26352. [Google Scholar] [CrossRef]
- Okanishi, M.; Olbers, J.M.; Fujita, T. A Taxonomic Review of the Genus Asteromorpha Lütken (Echinodermata: Ophiuroidea: Euryalidae). Raffles Bull. Zool. 2013, 61, 461–480. [Google Scholar] [CrossRef]
- Schönberg, C.H.L.; Hosie, A.M.; Fromont, J.; Marsh, L.; O’Hara, T.D. Apartment-Style Living on a Kebab Sponge. Mar. Biodivers. 2016, 46, 331–332. [Google Scholar] [CrossRef]
- O’Hara, T.D.; Rowden, A.A.; Williams, A. Cold-Water Coral Habitats on Seamounts: Do They Have a Specialist Fauna? Divers. Distrib. 2008, 14, 925–934. [Google Scholar] [CrossRef]
- Martynov, A.V.; Litvinova, N.M. Deep-Water Ophiuroidea of the Northern Atlantic with Descriptions of Three New Species and Taxonomic Remarks on Certain Genera and Species. Mar. Biol. Res. 2008, 4, 76–111. [Google Scholar] [CrossRef]
- Rodrigues, C.F.; Paterson, G.L.J.; Cabrinovic, A.; Cunha, M.R. Deep-Sea Ophiuroids (Echinodermata: Ophiuroidea: Ophiurida) from the Gulf of Cadiz (NE Atlantic). Zootaxa 2011, 26, 1–26. [Google Scholar] [CrossRef]
- Stöhr, S.; O’Hara, T.D. Deep-Sea Ophiuroidea (Echinodermata) from the Danish Galathea II Expedition, 1950–1952, with Taxonomic Revisions. Zootaxa 2021, 4963, 505–529. [Google Scholar] [CrossRef]
- Hunter, R.L.; Halanych, K.M. Evaluating Connectivity in the Brooding Brittle Star Astrotoma agassizii across the Drake Passage in the Southern Ocean. J. Hered. 2008, 99, 137–148. [Google Scholar] [CrossRef]
- Galaska, M.P.; Sands, C.J.; Santos, S.R.; Mahon, A.R.; Halanych, K.M. Crossing the Divide: Admixture across the Antarctic Polar Front Revealed by the Brittle Star Astrotoma agassizii. Biol. Bull. 2017, 232, 198–211. [Google Scholar] [CrossRef]
- Thurber, A.R.; Sweetman, A.K.; Narayanaswamy, B.E.; Jones, D.O.B.; Ingels, J.; Hansman, R.L. Ecosystem Function and Services Provided by the Deep Sea. Biogeosciences 2014, 11, 3941–3963. [Google Scholar] [CrossRef]
- Pérez-Portela, R.; Arranz, V.; Rius, M.; Turon, X. Cryptic Speciation or Global Spread? The Case of a Cosmopolitan Marine Invertebrate with Limited Dispersal Capabilities. Sci. Rep. 2013, 3, 3197. [Google Scholar] [CrossRef]
- Schwentner, M.; Lörz, A.N. Population Genetics of Cold-Water Coral Associated Pleustidae (Crustacea, Amphipoda) Reveals Cryptic Diversity and Recent Expansion off Iceland. Mar. Ecol. 2021, 42, e12625. [Google Scholar] [CrossRef]
- Sponer, R.; Deheyn, D.; Roy, M.S. Large Genetic Distances within a Population of Amphipholis squamata (Echinodermata; Ophiuroidea) Do Not Support Colour Varieties as Sibling Species. Mar. Ecol. Prog. Ser. 2001, 219, 169–175. [Google Scholar] [CrossRef][Green Version]
- Laming, S.R.; Christodoulou, M.; Martinez Arbizu, P.; Hilário, A. Comparative Reproductive Biology of Deep-Sea Ophiuroids Inhabiting Polymetallic-Nodule Fields in the Clarion-Clipperton Fracture Zone. Front. Mar. Sci. 2021, 8, 663798. [Google Scholar] [CrossRef]
- Gebruk, A.V.; Kremenetskaia, A.; Rouse, G.W. A Group of Species “Psychropotes longicauda” (Psychropotidae, Elasipodida, Holothuroidea) from the Kuril-Kamchatka Trench Area (North-West Pacific). Prog. Oceanogr. 2020, 180, 102222. [Google Scholar] [CrossRef]
- Gooday, A.J.; Holzmann, M.; Goineau, A.; Pearce, R.B.; Voltski, I.; Weber, A.A.T.; Pawlowski, J. Five New Species and Two New Genera of Xenophyophores (Foraminifera: Rhizaria) from Part of the Abyssal Equatorial Pacific Licensed for Polymetallic Nodule Exploration. Zool. J. Linn. Soc. 2018, 183, 723–748. [Google Scholar] [CrossRef]
- Lim, S.C.; Wiklund, H.; Glover, A.G.; Dahlgren, T.G.; Tan, K.S. A New Genus and Species of Abyssal Sponge Commonly Encrusting Polymetallic Nodules in the Clarion-Clipperton Zone, East Pacific Ocean. Syst. Biodivers. 2017, 15, 507–519. [Google Scholar] [CrossRef]
- Taboada, S.; Riesgo, A.; Wiklund, H.; Paterson, G.L.J.; Koutsouveli, V.; Santodomingo, N.; Dale, A.C.; Smith, C.R.; Jones, D.O.B.; Dahlgren, T.G.; et al. Implications of Population Connectivity Studies for the Design of Marine Protected Areas in the Deep Sea: An Example of a Demosponge from the Clarion-Clipperton Zone. Mol. Ecol. 2018, 27, 4657–4679. [Google Scholar] [CrossRef]
- Halanych, K.M.; Mahon, A.R. Challenging Dogma Concerning Biogeographic Patterns of Antarctica and the Southern Ocean. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 355–378. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Singer, G.A.C.; Hebert, P.D.N.; Hickey, D.A. DNA Barcoding: How It Complements Taxonomy, Molecular Phylogenetics and Population Genetics. Trends Genet. 2007, 23, 167–172. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).