Abstract
Ligustrum japonicum is a common ornamental tree species in China. However, leaf spot disease has emerged in Rizhao City, Shandong Province of China in recent years. Members of Pseudocercospora are usually known as plant pathogens, mainly causing leaf spots and blights. Species of this genus are distinguished mainly based on morphological differences on the host plants, as well as the molecular data. A new species named Pseudocercospora rizhaoensis on Ligustrum japonicum is introduced herein based on morphology and molecular data of combined ITS, LSU, act, tef1 and rpb2 sequences. Koch’s postulates were confirmed by a pathogenicity test, re-isolation and identification.
1. Introduction
Pseudocercospora Speg. (Mycosphaerellaceae, Mycosphaerellales) is a large cosmopolitan genus of plant pathogenic fungi that are commonly associated with leaf and fruit spots as well as blights on a wide range of plant hosts [1,2,3]. Pseudocercospora typed by P. vitis (Lév.) Speg. is distinguished from the other cercosporoid fungi by pale to dark olivaceous caespituli, pigmented conidia with unthickened and not refractive scars on the conidiogenous cells and hila at the basal ends of conidium in vivo [2,3].
Members of Pseudocercospora are distributed worldwide, but they are mostly abundant and diverse in tropical and subtropical areas and reproduce mainly by means of conidia [1,4,5,6,7,8,9]. Some species are associated with important plant diseases; for example, Pseudocercospora fijiensis (M. Morelet) Deighton is the causal agent of black Sigatoka leaf diseases of banana in Uganda and Tanzania [10]; P. actinidiae Deighton causes sooty spot disease on kiwifruit in Brazil [11]; P. griseola (Sacc.) Crous and U. Braun results in bean angular leaf spot disease in Ethiopia [12].
Species of Pseudocercospora are distinguished based on the morphology produced on the host plants and sequence data [1]. In addition, host information can also separate species of this genus, which is supported by the overall DNA phylogeny of ITS, LSU, act and tef1 [1]. Subsequently, the rpb2 locus was recommended to be added to the phylogeny for recognition of species within the genus Pseudocercospora [4]. Ligustrum japonicum Thunb. of the family Oleaceae Hoffmanns. and Link is native to central and southern Japan and Korea, and widely planted as an ornamental in parks and landscapes in China. During the surveys of plant diseases in Shandong Province, China, a colored cercosporoid fungus with fasciculate conidiophores, slightly thickened and darkened conidial scars and hilum was discovered, which causes a severe foliar disease on Ligustrum japonicum. Morphologically and phylogenetically, it was shown to be a species of Pseudocercospora. We compared its morphological features and molecular data to the known Pseudocercospora species and concluded that this species is new to science. Illustrations and detailed descriptions are provided for this new species herein.
2. Materials and Methods
2.1. Sample Survey, Collection and Fungal Isolation
Diseased leaf samples of Ligustrum japonicum were observed and collected in Rizhao City, Shandong Province of China (Figure 1), packed in paper bags and brought to the laboratory for isolation. The infected leaves were first surface-sterilized for 1 min in 75% ethanol, 3 min in 1.25% sodium hypochlorite, and 1 min in 75% ethanol, and then rinsed for 2 min in distilled water and blotted on dry sterile filter paper [13]. Then, the diseased areas of the samples were cut into 0.5 × 0.5 cm pieces using a double-edge blade, and transferred onto the surface of potato dextrose agar plates (PDA; 200 g potatoes, 20 g dextrose, 20 g agar per L) and incubated at 25 °C to obtain pure cultures. The cultures were deposited in the China Forestry Culture Collection Center (CFCC; http://cfcc.caf.ac.cn) and the specimen was deposited in the Herbarium of the Chinese Academy of Forestry (CAF; http://museum.caf.ac.cn).

Figure 1.
Symptoms of leaf spots on Ligustrum japonicum.
2.2. DNA Extraction, Sequencing and Phylogenetic Analyses
Genomic DNA was extracted from colonies grown on cellophane-covered PDA using a cetyltrimethylammonium bromide (CTAB) method [14]. DNA was checked by electrophoresis in 1% agarose gel, and the quality and quantity were measured using a NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA). The nuclear ribosomal DNA internal transcribed spacers (ITS), large subunit rRNA (LSU), actin (act), translation elongation factor 1-alpha (tef1) and the second lar-gest RNA polymerase subunit (rpb2) regions were amplified using the primer pairs ITS4 (TCC TCC GCT TAT TGA TAT GC) and ITS5 (GGA AGT AAA AGT CGT AAC AAG G) [15], LROR (GTA CCC GCT GAA CTT AAG C) and LR5 (TCC TGA GGG AAA CTT CG) [16], ACT-512 (ATG TGC AAG GCC GGT TTC GC) and ACT-783 (TAC GAG TCC TTC TGG CCC AT) [17], EF1-668 (CAT CGA GAA GTT CGA GAA GG) and EF1-1251 (GGA RGT ACC AGT SAT CAT GTT) [17,18] and RPB2-5f2 (GGGGWGAYCAGAAGAAGGC) and fRPB2-7cR (CCCATRGCTTGYTTRCCCAT) [19], respectively. The PCR conditions were set as follows: an initial denaturation step of 5 min at 95 °C, followed by 35 cycles of denaturation at 94 °C for 1 min, 50 s at 52 °C (ITS and LSU) or 54 °C (tef1, act and rpb2) [20]. The final extension step was done at 72 °C for 7 min. The PCR products were examined by electrophoresis on 1.5% (w/v) agarose gels stained with ethidium bromide in 1 × TBE buffer. DNA sequencing was performed by the Shanghai Invitrogen Biological Technology Company Limited (Beijing, China).
DNA sequences were generated by using SeqMan v.7.1.0 from the DNASTAR Lasergene software suite (DNASTAR Inc., Madison, WI, USA). Reference sequences used in the paper were downloaded from GenBank, and the GenBank accession numbers are listed in Table 1. Trocophora simplex was used as the outgroup taxon given its proposed relationship to Pseudocercospora. Sequences were aligned using MAFFT v.6 [21] and corrected manually using MEGA 7.0.21. The phylogenetic analyses of the combined loci were performed using the Maximum Likelihood (ML) and Bayesian Inference (BI) methods. ML was implemented on the CIPRES Science Gateway portal (https://www.phylo.org, accessed on 3 October 2022) using RAxML-HPC BlackBox 8.2.10 [22] employing a GTRGAMMA substitution model with 1000 bootstrap replicates. Bayesian inference was performed using a Markov Chain Monte Carlo (MCMC) algorithm in MrBayes v.3.2.6 [23]. The six simultaneous Markov chains were run for 1 M generations; starting from random trees and sampling trees every 100 th generation and 25% of aging samples were discarded, running until the average standard deviation of the split frequencies dropped below 0.01. The phylogram was visualized in FigTree v.1.3.1 (http://tree.bio.ed.ac.uk/software, accessed on 4 October 2022) and edited in Adobe Illustrator CS5 (Adobe Systems Inc., San Jose, CA, USA). The nucleotide sequence data of the new taxon were deposited in GenBank, and the GenBank accession numbers of all accessions included in the phylogenetic analyses are listed in Table 1.

Table 1.
Strains and GenBank accession numbers used in this study.
2.3. Morphological Identification and Characterization
The morphology of the new species was studied based on the fruiting bodies formed on the diseased leaves. The fruiting bodies were observed and photographed under a dissecting microscope (M205 C, Leica, Wetzlar, Germany). The conidiogenous cells and conidia were immersed in tap water, and then the microscopic photographs were captured with an Axio Imager 2 microscope (Zeiss, Oberkochen, Germany) equipped with an Axiocam 506 color camera using Differential Interference Contrast (DIC) illumination. For measurements, more than 50 conidia were randomly selected. Culture characteristics were recorded from PDA after 20 d of incubation at 25 °C in the dark.
2.4. Pathogenicity Testing
Two isolates of Pseudocercospora rizhaoensis (ex-type strain: CFCC 57581; CFCC 52288) were used for inoculations, and agar plugs were used as the negative control. Detached healthy Ligustrum japonicum leaves were used for artificial inoculation experiments. The leaves were surface-sterilized with 75% ethanol, rinsing three times in sterile water, and then we waited for the surface moisture to dry. Discs of agar were cut from the actively growing margins of the cultures and these were placed on the non-wounded in vitro leaves. Each group was performed three times and cultured at 25 °C in the dark.
3. Results
3.1. Phylogeny
The combined sequence dataset (ITS, LSU, tef1, act and rpb2) was analyzed to infer the phylogenetic placement of our new isolates within the genus Pseudocercospora. The dataset consisted of 193 sequences, including an outgroup taxon, Trocophora simplex (CBS124744). A total of 2755 characters, including gaps (510 for ITS, 788 for LSU, 543 for tef1, 241 for act, and 673 for rpb2), were included in the phylogenetic analysis. The best ML tree (lnL = − 33,732.84) revealed by RA×ML is shown in Figure 2. The topologies resulting from ML and BI analyses of the concatenated dataset were congruent (Figure 2). The phylogenetic tree showed that isolates CFCC 57581 and CFCC 57582 from the present study formed into a distinguished clade from the other known Pseudocercospora species.


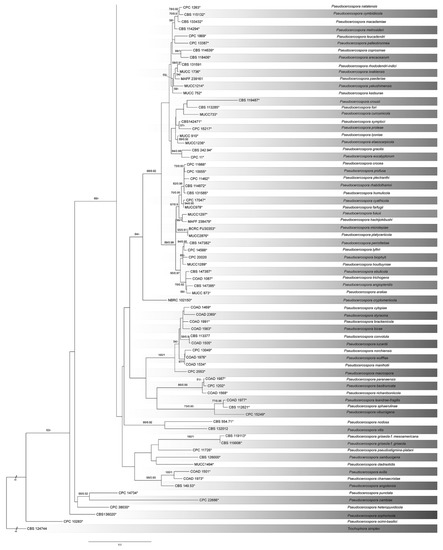
Figure 2.
Phylogram of Pseudocercospora resulting from a maximum likelihood analysis, based on a combined matrix of ITS, LSU, act, rpb2 and tef1. Numbers above the branches indicate ML bootstraps (left, ML BS ≥ 50%) and Bayesian Posterior Probabilities (right, BPP ≥ 0.90). Ex-type strains are marked with *, and the new species proposed in the present study is marked in blue.
3.2. Taxonomy
Pseudocercospora rizhaoensis Yun Liu, sp. nov. Figure 3.

Figure 3.
Morphology of Psedocercospora rizhaoensis. (A) Disease symptom on the host leaves; (B,E) stroma and conidiophores; (C) colony on PDA plates at 10 d; (D) colony on PDA plates at 20 d; (F) conidia. Scale bars: (B) = 100 μm; (E,F) = 10 μm.
Mycobank no.: 845997.
Etymology—Named after Rizhao City, where the holotype was collected.
Description—Leaf spots amphigenous, circular, scattered, pale brown to brown with reddish brown margin. Caespituli hypophyllous, synnematous with blackish brown conidiophores. Mycelium internal, hyaline to brown. Stromata hypophyllous, substomatal, epidermal, erumpent, well-developed, subglobose to globose, dark brown to blackish, 25–85 μm diam. Conidiophores dense, arising from the upper part of stromata, straight to sinuous-geniculated, cylindrical, unbranched, pale brown to brown, paler towards the apex, 4.5–25 × 2–3.5 μm, 0–2-septate, smooth. Conidiogenous cells integrated, terminal, proliferating percurrently or sympodially, with unthickened and truncated conidial loci. Conidia solitary, holoblastic, cylindrical to obclavate, 15–50.5 × 2.5–4.5 μm, 1–5-septate, hyaline- to pale-colored, smooth, acute at the apex, obconically truncated, unthickened and not darkened at the base.
Culture characteristics—Colonies on PDA flat, spreading, with flocculent aerial mycelium, edge entire, mouse grey, reaching 60 mm diam. after 20 d at 25 °C.
Material examined—CHINA, Shandong Province, Rizhao City, Beijinglu Street, on diseased leaves of Ligustrum japonicum, 7 August 2021, Yun Liu (CAF800066 holotype; ex-type living culture, CFCC 57581); Ibid. Shandong Province, Rizhao City, Shijiu Street, on diseased leaves of Ligustrum japonicum, 12 August 2021, Yun Liu (culture CFCC 57582).
Notes—Two isolates of Pseudocercospora from leaf spots of Ligustrum japonicum clustered into a well-supported clade distinguished from the other members within this genus (Figure 2), which is proposed as P. rizhaoensis herein. Phylogenetically, P. rizhaoensis is close to P. eupatoriella Crous and Den Breeÿen and P. ginkgoana R. Kirschner; however, these species can be distinguished by their hosts (P. rizhaoensis on Ligustrum japonicum vs. P. eupatoriella on Chromolaena odorata R. M. King and H. Robinson vs. P. ginkgoana on Ginkgo biloba L.) [1,24]. Morphologically, conidia of P. rizhaoensis is similar to those of P. eupatoriella, but wider than those of P. ginkgoana (2.5–4.5 μm in P. rizhaoensis vs. 2–2.5 μm in P. ginkgoana) [1,24].
3.3. Pathogenicity Tests
Similar leaf spot symptoms were reproduced on the Ligustrum japonicum leaves after inoculated 20 days, while no symptoms were observed on the control leaves. The respective inoculated fungi were re-isolated from leaves’ lesions and were identical to Ligustrum japonicum by using morphological characteristics and phylogeny.
4. Discussion
The genus Psedocercospora was previously considered as an anamorphic state of Mycosphaerella or having mycosphaerella-like teleomorphs, but it is now treated as a genus based on phylogeny and morphology [1,25,26,27,28]. Now members within this genus are distinguished from each other based on combined approaches of host association, conidia characters and gene sequences [1].
In the pathogenicity test, the leaves after inoculating those isolates showed the same symptoms as disease that occurred in the field, and those isolates could be re-isolated from the lesions. Based on those data, Psedocercospora rizhaoensis is considered as the causal agent of the Ligustrum japonicum leaf spot disease in China.
Pseudocercospora ligustri was recorded causing Ligustrum japonicum leaf spots in the USA and Ligustrum japonicum ‘Howardii’ leaf spots in China [29,30]. For this fungus species, no DNA data are available from the type material (IMI 91224 collected in the USA) [29], and three genes, namely ITS, act and tef1, were sequenced based on the samples collected in China (JS1201, JS1202 and JS1203) [30]. Morphologically, conidiophores of Psedocercospora rizhaoensis are narrower than those of P. ligustri (2–3.5 μm in P. rizhaoensis vs. 3–4.2 μm in P. ligustri) [29,30]. In addition, P. rizhaoensis differs from P. ligustri in the sequence data (3/470 in ITS, 3/222 in act and 1/309 in tef1) [30].
As shown in Figure 2, several species were not well-distinguished in the phylogram based on combined loci of ITS, LSU, act, rpb2 and tef1. This may be caused by the absence of sequence data for those species (Table 1). Chen et al. [25] demonstrated that ITS is the genus DNA barcode, and the act, rpb2 and tef1 genes are the species DNA barcode. More phylogenetic analyses to infer the species relationships are necessary in following studies by employing more genes.
Author Contributions
Conceptualization, Y.L.; methodology, Y.L. and S.G.; software, S.G.; validation, J.L., X.Y. and Y.L.; formal analysis, Y.L.; investigation, Y.L.; resources, Y.L.; data curation, Y.L.; writing—original draft preparation, Y.L.; writing—review and editing, Y.L.; visualization, Y.L.; supervision, Y.L.; project administration, X.Y.; funding acquisition, X.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Shandong Province Pasture Industry Technology System Project (SDAIT-23-03) and College Youth Science and Technology Support Program of Shandong Province (2021KJ087).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Crous, P.W.; Braun, U.; Hunter, G.C.; Wingfield, M.; Verkley, G.; Shin, H.-D.; Nakashima, C.; Groenewald, J. Phylogenetic lineages in Pseudocercospora. Stud. Mycol. 2013, 75, 37–114. [Google Scholar] [CrossRef] [PubMed]
- Braun, U.; Nakashima, C.; Bakhshi, M.; Zare, R.; Shin, H.D.; Alves, R.F.; Sposito, M.B. Taxonomy and phylogeny of cercosporoid ascomycetes on Diospyros spp. with special emphasis on Pseudocercospora spp. Fungal Syst. Evol. 2020, 6, 95–127. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Schoch, C.L.; Hyde, K.D.; Wood, A.R.; Gueidan, C.; De Hoog, G.S.; Groenewald, J.Z. Phylogenetic lineages in the Capnodiales. Stud. Mycol. 2009, 64, 17–47. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, C.; Motohashi, K.; Chen, C.Y.; Groenewald, J.Z.; Crous, P.W. Species diversity of Pseudocercospora from Far East Asia. Mycol. Prog. 2016, 15, 1093–1117. [Google Scholar] [CrossRef]
- Osorio, J.A.; Wingfield, M.J.; de Beer, Z.W.; Roux, J. Pseudocercospora mapelanensis sp. nov., associated with a fruit and leaf disease of Barringtonia racemosa in South Africa. Australas. Plant Pathol. 2015, 44, 349–359. [Google Scholar] [CrossRef]
- Yadav, S.; Verma, S.K.; Singh, V.K.; Singh, R.; Singh, A.; Kumar, S. Morphology and phylogeny of a new species, Pseudocercospora haldinae (Mycosphaerellaceae) on Haldina cordifolia from India. Phytotaxa 2021, 501, 281–292. [Google Scholar] [CrossRef]
- Silva, M.; Barreto, R.W.; Pereira, O.L.; Freitas, N.; Groenewald, J.; Crous, P. Exploring fungal megadiversity: Pseudocercospora from Brazil. Persoonia 2016, 37, 142–172. [Google Scholar] [CrossRef]
- Shivas, R.G.; Marney, T.S.; Tan, Y.P.; McTaggart, A.R. Novel species of Cercospora and Pseudocercospora (Capnodiales, Mycosphaerellaceae) from Australia. Fungal Biol. 2015, 119, 362–369. [Google Scholar] [CrossRef][Green Version]
- Kirschner, R.; Wang, H. New species and records of mycosphaerellaceous fungi from living fern leaves in East Asia. Mycol. Prog. 2015, 14, 65. [Google Scholar] [CrossRef]
- Kimunye, J.N.; Were, E.; Mussa, F.; Tazuba, A.; Jomanga, K.; Viljoen, A.; Swennen, R.; Muthoni, F.K.; Mahuku, G. Distribution of Pseudocercospora species causing Sigatoka leaf diseases of banana in Uganda and Tanzania. Plant Pathol. 2020, 69, 50–59. [Google Scholar] [CrossRef]
- Araujo, L.; Pinto, F.A.M.F.; de Andrade, C.C.L.; Gomes, L.B.; Mituti, T.; Duarte, V. Pseudocercospora actinidiae causes sooty spot disease on kiwifruit in Santa Catarina, Brazil. Australas. Plant Dis. Notes 2021, 16, 22. [Google Scholar] [CrossRef]
- Rezene, Y.; Tesfaye, K.; Clare, M.; Gepts, P. Pathotypes characterization and virulence diversity of Pseudocercospora griseola the causal agent of angular leaf spot disease collected from major common bean (Phaseolus vulgaris L.) growing areas of Ethiopia. J. Plant Pathol. Microbiol. 2018, 9, 1000445. [Google Scholar] [CrossRef]
- Jiang, N.; Voglmayr, H.; Ma, C.Y.; Xue, H.; Piao, C.G.; Li, Y. A new Arthrinium-like genus of Amphisphaeriales in China. MycoKeys 2022, 92, 27–43. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- O’Donnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Jiang, N.; Voglmayr, H.; Bian, D.R.; Piao, C.G.; Wang, S.K.; Li, Y. Morphology and phylogeny of Gnomoniopsis (Gnomoniaceae, Diaporthales) from fagaceae leaves in China. J. Fungi 2021, 7, 792. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 2010, 26, 1899–1900. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 312–1313. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, R.; Okuda, T. A new species of Pseudocercospora and new record of Bartheletia paradoxa on leaves of Ginkgo biloba. Mycol. Prog. 2013, 12, 421–426. [Google Scholar] [CrossRef]
- Chen, Q.; Bakhshi, M.; Balci, Y.; Broders, K.; Cheewangkoon, R.; Chen, S.; Fan, X.; Gramaje, D.; Halleen, F.; Jung, M.H.; et al. Genera of phytopathogenic fungi: GOPHY 4. Stud. Mycol. 2022, 101, 417–564. [Google Scholar] [CrossRef] [PubMed]
- Videira, S.I.R.; Groenewald, J.Z.; Nakashima, C.; Braun, U.; Barreto, R.W.; de Wit, P.J.; Crous, P.W. Mycosphaerellaceae—Chaos or clarity? Stud. Mycol. 2017, 87, 257–421. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, Z.C.; He, W.; Zhang, Y. Pseudocercospora spp. from leaf spots of Euonymus japonicus in China. Mycosystema 2019, 38, 159–170. [Google Scholar]
- Bakhshi, M.; Arzanlou, M.; Babai-Ahari, A.; Groenewald, J.Z.; Crous, P.W. Multi-gene analysis of Pseudocercospora spp. from Iran. Phytotaxa 2014, 184, 245–264. [Google Scholar] [CrossRef]
- Deighton, F.C. Studies on Cercospora and allied genera. VI. Pseudocercospora Speg., Pantospora Cif., and Cercoseptoria Petr. Mycol. Pap. 1976, 140, 1–168. [Google Scholar]
- Wang, S.H.; Liu, S.M. First Report of leaf spot caused by Pseudocercospora ligustri on Ligustrum japonicum ‘Howardii’ in China. Plant Dis. 2019, 103, 153. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).