Analysis of the Genetic Structure of Slovak Holstein Cattle Using Seven Candidate Genes Related to Milk Quality
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. SNP Genotyping
2.3. Genetic Structure
3. Results and Discussion
3.1. SNP Identification and Genotyping
3.2. Genetic Structure
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Garcia-Gomez, B.; Vazquez-Oderiz, M.L.; Munoz-Ferreiro, N.; Romero-Rodriguez, M.A.; Vazquez, M. Interaction between rennet source and transglutaminase in white fresh cheese production: Effect on physicochemical and textural properties. LWT-Food Sci. Technol. 2019, 113, 108279. [Google Scholar] [CrossRef]
- Čítek, J.; Brzáková, M.; Hanusová, L.; Hanuš, O.; Večerek, L.; Samková, E.; Křížová, Z.; Hoštičková, I.; Kávová, T.; Straková, K.; et al. Gene polymorphisms influencing yield, composition and technological properties of milk from Czech Simmental and Holstein cows. Anim. Biosci. 2021, 34, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Miglior, F.; Fleming, A.; Malchiodi, F.; Brito, L.F.; Martin, P.; Baes, C.H.F. A 100-Year Review: Identification and genetic selection of economically important traits in dairy cattle. J. Dairy Sci. 2017, 100, 10251–10271. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, V.L.; Pereira Lima, M.L.; Nogueira, J.R.; de Carneiro, R.L.R.; Sesana, R.C.; Oliveira, E.J.; El Faro, L. Economic values for milk production and quality traits in south and southeast regions of Brazil. Rev. Bras. Zootec. 2014, 43, 636–642. [Google Scholar] [CrossRef]
- Smaragdov, M.G. Genomic selection as a possible accelerator of traditional selection. Russ. J. Genet. 2009, 45, 633–636. [Google Scholar] [CrossRef]
- Yudin, N.S.; Voevoda, M.I. Molecular Genetic Markers of Economically Important Traits in Dairy Cattle. Russ. J. Genet. 2015, 51, 506–517. [Google Scholar] [CrossRef]
- Kyselová, J.; Ječmínková, K.; Matějíčková, J.; Hanuš, O.; Kott, T.; Štípková, M.; Krejčová, M. Physiochemical characteristics and fermentation ability of milk from Czech Fleckvieh cows are related to genetic polymorphisms of β-casein, κ-casein, and β-lactoglobulin. Asian-Australas. J. Anim. Sci. 2019, 32, 14–22. [Google Scholar] [CrossRef]
- Grisart, B.; Coppieters, W.; Farnir, F.; Karim, L.; Ford, C.; Berzi, P.; Cambisano, N.; Mni, M.; Reid, S.; Simon, P.; et al. Positional candidate cloning of a QTL in dairy cattle: Identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 2002, 12, 222–231. [Google Scholar] [CrossRef]
- Nakamura, M.T.; Nara, T.Y. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu. Rev. Nutr. 2004, 24, 345–376. [Google Scholar] [CrossRef]
- Schennink, A.; Bovenhuis, H.; Leon-Kloosterziel, K.M.; van Arendonk, J.A.M.; Visker, M.H.P.W. Effect of polymorphisms in the FASN, OLR1, PPARGC1A, PRL and STAT5 genes on bovine milk-fat composition. Anim. Genet. 2009, 40, 909–916. [Google Scholar] [CrossRef]
- Ibeagha-Awemu, E.M.; Akwanji, K.A.; Beaudoin, F.; Zhao, X. Associations between variants of FADS genes and omega-3 and omega-6 milk fatty acids of Canadian Holstein cows. BMC Genet. 2014, 15, 25. [Google Scholar] [CrossRef]
- Li, M.; Gao, Q.; Wang, M.; Liang, Y.; Sun, Y.; Chen, Z.; Zhang, H.; Karrow, N.A.; Yang, Z.; Mao, Y. Polymorphisms in Fatty Acid Desaturase 2 Gene Are Associated with Milk Production Traits in Chinese Holstein Cows. Animals 2020, 10, 671. [Google Scholar] [CrossRef]
- Matsumoto, H.; Inada, S.; Kobayashi, E.; Abe, T.; Hasebe, H.; Sasazaki, S.; Oyama, K.; Mannen, H. Identification of SNPs in the FASN gene and their effect on fatty acid milk composition in Holstein cattle. Livest. Sci. 2012, 144, 281–284. [Google Scholar] [CrossRef]
- Li, C.; Sun, D.; Zhang, S.; Alim, M.A.; Zhang, Q.; Li, Y.; Liu, L. Genetic effects of FASN, PPARGC1A, ABCG2 and IGF1 revealing the association with milk fatty acids in a Chinese Holstein cattle population based on a post genome-wide association study. BMC Genet. 2016, 17, 110. [Google Scholar] [CrossRef]
- Mauric, M.; Masek, T.; Ljoljic, D.B.; Grbavac, J.; Starcevic, K. Effects of different variants of the FASN gene on production performance and milk fatty acid composition in Holstein × Simmental dairy cows. Vet. Med. 2019, 64, 101–108. [Google Scholar] [CrossRef]
- Chung, M.; Ha, S.; Jeong, S.; Bok, J.; Cho, K.; Baik, M.; Choi, Y. Cloning and characterization of bovine stearoyl CoA desaturasel cDNA from adipose tissues. Biosci. Biotechnol. Biochem. 2000, 64, 1526–1530. [Google Scholar] [CrossRef]
- Paton, C.M.; Ntambi, J.M. Biochemical and physiological function of stearoyl-CoA desaturase. Am. J. Physiol.-Endoc. Metab. 2009, 297, E28–E37. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, H.; Cheng, L.; Zhao, J.; Hickford, J. Variation in the stearoyl-CoA desaturase gene (SCD) and its influence on milk fatty acid composition in late-lactation dairy cattle grazed on pasture. Arch. Anim. Breed. 2020, 63, 355–366. [Google Scholar] [CrossRef]
- Dudásová, S.; Miluchová, M.; Gábor, M.; Candrák, J.; Dočkalová, K. Effects of the DGAT1 K232A polymorphism on milk production traits in Holstein cattle. Acta Fytotech. Zootech. 2021, 24, 233–237. [Google Scholar] [CrossRef]
- Pathak, R.K.; Lim, B.; Park, Y.; Kim, J.M. Unraveling structural and conformational dynamics of DGAT1 missense nsSNPs in dairy cattle. Sci. Rep. 2022, 12, 4873. [Google Scholar] [CrossRef]
- Neamt, R.I.; Saplacan, S.; Acatincai, S.; Cziszter, L.T.; Gavojdian, D.; Ilie, D.E. The influence of CSN3 and LGB polymorphisms on milk production and chemical composition in Romanian Simmental cattle. Acta Biochim. Pol. 2017, 64, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, M.; Kopuzlu, S.; Topal, M.; Bilgin, O.C. Relationships between milk protein polymorphisms and production traits in cattle: A systematic review and meta-analysis. Arch. Anim. Breed. 2018, 61, 197–206. [Google Scholar] [CrossRef]
- Čítek, J.; Hanusová, L.; Lískovcová, L.; Samková, E.; Hanuš, O.; Hasoňová, L.; Křížová, Z.; Večerek, L. Polymorphisms in CSN3, CSN2 and LGB Genes and Their Relation to Milk Production in Dairy Cattle in the Czech Republic. Acta Univ. Agric. Silvic. Mendel. Brun. 2019, 67, 19–24. [Google Scholar] [CrossRef]
- Ng-Kwai-Hang, K.F. Genetic polymorphism of milk proteins: Relationships with production traits, milk composition and technological properties. Can. J. Anim. Sci. 1998, 78, 131–147. [Google Scholar]
- Miluchová, M.; Gábor, M.; Candrák, J.; Trakovická, A.; Candráková, K. Association of HindIII-polymorphism in kappa-casein gene with milk, fat and protein yield in Holstein cattle. Acta Biochim. Pol. 2018, 65, 403–407. [Google Scholar] [CrossRef]
- Čítek, J.; Brzáková, M.; Hanusová, L.; Hanuš, O.; Večerek, L.; Samková, E.; Křížová, Z.; Hoštičková, I.; Kávová, T.; Straková, K.; et al. Technological properties of cow’s milk: Correlations with milk composition, effect of interactions of genes and other factors. Czech J. Anim. Sci. 2020, 65, 13–22. [Google Scholar] [CrossRef]
- Samoré, A.B.; Rizzi, R.; Rossoni, A.; Bagnato, A. Genetic parameters for functional longevity, type traits, SCS, milk flow and production in the Italian Brown Swiss. Ital. J. Anim. Sci. 2010, 9, e28. [Google Scholar] [CrossRef]
- Oltenacu, P.A.; Broom, D.M. The impact of genetic selection for increased milk yield on the welfare of dairy cows. Anim. Welf. 2010, 19, 39–49. [Google Scholar]
- Stavetska, R.; Dynko, Y. The Characteristic of Economically Important Traits of Dairy Cows Depending on Type of Body Constitution. EUREKA Life Sci. 2021, 2, 9–15. [Google Scholar] [CrossRef]
- Ollivier, L.; Foulley, J. Aggregate diversity: New approach combining within and between breed genetic diversity. Livest. Prod. Sci. 2005, 95, 247–254. [Google Scholar] [CrossRef]
- Biscarini, F.; Nicolazzi, E.L.; Stella, A.; Boettcher, P.J.; Gandini, G. Challenges and opportunities in genetic improvement of local livestock breeds. Front. Genet. 2015, 6, 33. [Google Scholar] [CrossRef]
- Notter, D.R. The importance of genetic diversity in livestock populations of the future. J. Anim. Sci. 1999, 77, 61–69. [Google Scholar] [CrossRef]
- Barker, J.S.F. Conservation and management of genetic diversity: A domestic animal perspective. Can. J. For. Res. 2001, 31, 588–595. [Google Scholar] [CrossRef]
- Toro, M.A.; Caballero, A. Characterization and conservation of genetic diversity in subdivided populations. Philos. Trans. R. Soc. B 2005, 360, 1367–1378. [Google Scholar] [CrossRef]
- Ruan, D.; Yang, J.; Zhuang, Z.; Ding, R.; Huang, J.; Quan, J.; Gu, T.; Hong, L.; Zheng, E.; Li, Z.; et al. Assessment of Heterozygosity and Genome-Wide Analysis of Heterozygosity Regions in Two Duroc Pig Populations. Front. Genet. 2022, 12, 812456. [Google Scholar] [CrossRef]
- Gautschi, B.; Müller, J.P.; Schmid, B.; Shykoff, J.A. Effective number of breeders and maintenance of genetic diversity in the captive bearded vulture population. Heredity 2003, 91, 9–16. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Populations. The Theory of Gene Frequencies, 2nd ed.; University of Chicago Press: Chicago, IL, USA, 1969; 512p. [Google Scholar]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Abe, T.; Saburi, J.; Hasebe, H.; Nakagawa, T.; Misumi, S.; Nade, T.; Nakajima, H.; Shoji, N.; Kobayashi, M.; Kobayashi, E. Novel mutations of the FASN gene and their effect on fatty acid composition in Japanese Black beef. Biochem. Genet. 2009, 47, 397–411. [Google Scholar] [CrossRef]
- Taniguchi, M.; Utsugi, T.; Oyama, K.; Mannen, H.; Kobayashi, M.; Tanabe, Y.; Ogino, A.; Tsuji, S. Genotype of stearoyl-CoA desaturase is associated with fatty acids composition in Japanese Black cattle. Mamm. Genome 2004, 14, 142–148. [Google Scholar] [CrossRef]
- Komisarek, J.; Michalak, A. A relationship between DGAT1 K232A polymorphism and selected reproductive traits in Polish Holstein-Friesian cattle. Anim. Sci. Pap. Rep. 2008, 26, 89–95. [Google Scholar]
- McLachlan, C.N. Breeding and Milking Cows for Milk Free of β-Casein A1. U.S. Patent 7094949, 22 August 2006. [Google Scholar]
- Schlieben, S.; Erhard, G.; Senft, B. Genotyping of bovine kappa-casein following DNA sequence amplification and direct sequencing of kappa-CNE PCR product. Anim. Genet. 1991, 22, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Brody, R.J.; Kern, S.E. Sodium boric acid: A Tris-free, cooler conductive medium for DNA electrophoresis. Biotechniques 2004, 36, 214–216. [Google Scholar] [CrossRef] [PubMed]
- Komisarek, J.; Dorynek, Z. Effect of ABCG2, PPARGC1A, OLR1 and SCD1 gene polymorphism on estimated breeding values for functional and production traits in Polish Holstein-Friesian bulls. J. Appl. Genet. 2009, 50, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [PubMed]
- Boltstein, D.; White, R.L.; Skolnik, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Crow, J.F.; Kimura, M. An Introduction to Population Genetics Theory; Harper and Row: New York, NY, USA, 1970; 591p. [Google Scholar]
- Liu, W.T.; Marsh, T.L.; Cheng, H.; Forney, L.J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 1997, 63, 4516–4522. [Google Scholar] [CrossRef]
- Dunbar, J.; Ticknor, L.O.; Kuske, C.R. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 2001, 67, 190–197. [Google Scholar] [CrossRef]
- Vincze, T.; Posfai, J.; Roberts, R.J. NEBcutter: A program to cleave DNA with restriction enzymes. Nucleic Acids. Res. 2003, 31, 3688–3691. [Google Scholar] [CrossRef]
- Harris, W.S. The omega-3 index: From biomarker to risk marker to risk factor. Curr. Atheroscler. Rep. 2009, 11, 411–417. [Google Scholar] [CrossRef]
- Beak, S.H.; Lee, Y.; Lee, E.B.; Kim, K.H.; Kim, J.G.; Bok, J.D.; Kang, S.K. Study on the fatty acid profile of phospholipid and neutral lipid in Hanwoo beef and their relationship to genetic variation. J. Anim. Sci. Technol. 2019, 61, 69–76. [Google Scholar] [CrossRef]
- Grofová, Z. Fatty acids. Med. Pro Praxi 2010, 7, 388–390. [Google Scholar]
- Roy, R.; Ordovas, L.; Zaragoza, P.; Romero, A.; Moreno, C.; Altarriba, J.; Rodellar, C. Association of polymorphisms in the bovine FASN gene with milk-fat content. Anim. Genet. 2006, 37, 215–218. [Google Scholar] [CrossRef]
- Barton, L.; Bures, D.; Kott, T.; Kottova, B. Effects of DGAT1, FABP4, FASN, PPARGC1A, SCD1, SREBP-1 and STAT5A Gene Polymorphisms on the Fatty Acid Composition in Fleckvieh Bulls. In Proceedings of the ICoMST 2011, 57th International Congress of Meat Science and Technology, Ghent, Belgium, 7–12 August 2011. [Google Scholar]
- Kawaguchi, F.; Kakiuchi, F.; Oyama, K.; Mannen, H.; Sasazaki, S. Effect of Five Polymorphisms on Percentage of Oleic Acid in Beef and Investigation of Linkage Disequilibrium to Confirm the Locations of Quantitative Trait Loci on BTA19 in Japanese Black Cattle. Life 2021, 11, 597. [Google Scholar] [CrossRef]
- Kgwatalala, P.M.; Ibeagha-Awemu, E.M.; Mustafa, A.F.; Zhao, X. Influence of stearoyl-coenzyme A desaturase 1 genotype and stage of lactation on fatty acid composition of Canadian Jersey cows. J. Dairy Sci. 2009, 92, 1220–1228. [Google Scholar] [CrossRef]
- Matsuhashi, T.; Maruyama, S.; Uemoto, Y.; Kobayashi, N.; Mannen, H.; Abe, T.; Sakaguchi, S.; Kobayashi, E. Effects of bovine fatty acid synthase, stearoyl-coenzyme A desaturase, sterol regulatory element-binding protein 1, and growth hormone gene polymorphisms on fatty acid composition and carcass traits in Japanese Black cattle. J. Anim. Sci. 2011, 89, 12–22. [Google Scholar] [CrossRef]
- Mao, Y.J.; Chen, R.J.; Chang, L.L.; Chen, Y.; Ji, D.J.; Wu, X.X.; Shi, X.K.; Wu, H.T.; Zhang, M.R.; Yang, Z.P.; et al. Short communication: Effects of SCD1- and DGAT1-genes on production traits of Chinese Holstein cows located in the Delta Region of Yangtze River. Livest. Sci. 2012, 145, 280–286. [Google Scholar] [CrossRef]
- Kulig, H.; Kowalewska-Łuczak, I.; Źukowski, K.; Kunicka, M. SCD1 SNP in relation to breeding value of milk production traits in polish Holstein-Friesian cows. Acta Sci. Pol. Zoot. 2013, 12, 41–48. [Google Scholar]
- Conte, G.; Mele, M.; Chessa, S.; Castiglioni, B.; Serra, A.; Pagnacco, G.; Secchiari, P. Diacylglycerol acyltransferase 1, stearoyl-CoA desaturase 1, and sterol regulatory element binding protein 1 gene polymorphisms and milk fatty acid composition in Italian Brown cattle. J. Dairy Sci. 2010, 93, 753–763. [Google Scholar] [CrossRef]
- Safina, N.Y.; Shakirov, S.K.; Ravilov, R.K.; Sharafutdinov, G.S. Associations of the SCD1 gene SNP with fatty acids composition of Holstein cows. BIO Web Conf. 2020, 27, 00060. [Google Scholar] [CrossRef]
- Mele, M.; Conte, G.; Castiglioni, B.; Chessa, S.; Macciotta, N.P.P.; Serra, A.; Buccioni, A.; Pagnacco, G.; Secchiari, P. Stearoyl-CoA desaturase gene polymorphism and milk fatty acid composition in Italian Friesian cows. J. Dairy Sci. 2007, 90, 4458–4465. [Google Scholar] [CrossRef]
- Schennink, A.; Heck, J.M.; Bovenhuis, H.; Visker, M.H.P.W.; Van Valenberg, H.J.F.; Van Arendonk, J.A.M. Milk fatty acid unsaturation: Genetic parameters and effects of stearoyl-CoA desaturase (SCD1) and acyl CoA: Diacylglycerol acyltransferase 1 (DGAT1). J. Dairy Sci. 2008, 91, 2135–2143. [Google Scholar] [CrossRef] [PubMed]
- Grummer, R.R. Effect of feed on composition of milk fat. J. Dairy Sci. 1991, 74, 3244–3257. [Google Scholar] [CrossRef]
- Kadlecová, V.; Němečková, D.; Ječmínková, K.; Stádník, L. Association of bovine DGAT1 and leptin genes polymorphism with milk production traits and energy balance indicators in primiparous Holstein cows. Mljekarstvo 2014, 64, 19–26. [Google Scholar]
- Schennink, A.; Stoop, W.M.; Visker, M.H.P.; Wheck, J.M.L.; Bovenhuis, H.; Van Der Poel, J.J.; Van Alenberg, H.J.F.; Van Arendonk, J.A.M. DGAT1 underlies large genetic variation in milk-fat composition of dairy cows. Anim. Genet. 2007, 38, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, A.C.; Bovenhuis, H.; Visker, M.H.; Van Arendonk, J.A. Genome wide association of milk fatty acids in Dutch dairy cattle. BMC Genet. 2011, 12, 43. [Google Scholar] [CrossRef]
- Oleński, K.; Cieślińska, A.; Suchocki, T.; Szyda, J.; Kamiński, S. Polymorphism in coding and regulatory sequences of beta-casein gene is associated with milk production traits in Holstein-Friesian cattle. Anim. Sci. Pap. Rep. 2012, 30, 5–12. [Google Scholar]
- Ganguly, I.; Kumar, S.; Gaur, G.K.; Singh, U.; Kumar, A.; Kumar, S.; Mann, S.; Sharma, A. Status of β-casein (CSN2) Polymorphism in Frieswal (HF X Sahiwal Crossbred) Cattle. Int. J. Biotechnol. Bioeng. Res. 2013, 4, 249–256. [Google Scholar]
- Manga, I.; Říha, J.; Dvořák, J. Comparison of influence markers CSN3 and CSN2 on milk performance traits in czech spotted and holstein cattle tested at first, fifth and higher lactation. Acta Fytotech. Zootech. 2006, 9, 13–15. [Google Scholar]
- Beja-Pereira, A.; Luikart, G.; England, P.R.; Bradley, D.G.; Jann, O.C.; Bertorelle, G.; Chamberlain, A.T.; Nunes, T.P.; Metodiev, S.; Ferrand, N.; et al. Gene-culture coevolution between cattle milk protein genes and human lactase genes. Nat. Genet. 2003, 35, 311–313. [Google Scholar] [CrossRef]
- Caroli, A.; Chessa, S.; Chiatti, F.; Rignanese, D.; Meléndez, B.; Rizzi, R.; Ceriotti, G. Short communication: Carora cattle show high variability in alpha(s1)-casein. J. Dairy Sci. 2008, 91, 354–359. [Google Scholar] [CrossRef]
- Hanusová, E.; Huba, J.; Oravcová, M.; Polák, P.; Vrtková, I. Genetic variants of beta-casein in Holstein Dairy Cattle in Slovakia. Slovak J. Anim. Sci. 2010, 43, 63–66. [Google Scholar]
- Elliott, R.B.; Harris, D.P.; Hill, J.P.; Bibby, N.J.; Wasmuth, H.E. Type I (insulin-dependent) diabetes mellitus and cow milk: Casein variant consumption. Diabetologia 1999, 42, 292–296. [Google Scholar] [CrossRef]
- Morris, C.A.; Tate, M.L. Method for Altering Fatty Acid Composition of Milk. New Zealand Patent PCT/NZ2003/000140, 15 January 2004. [Google Scholar]
- McLachlan, C.N. Beta-casein A1, ischaemic heart disease mortality, and other illnesses. Med. Hypotheses 2001, 56, 262–272. [Google Scholar] [CrossRef]
- Sun, Z.; Zhang, Z.; Wang, X.; Cade, R.; Elmer, Z.; Fregly, M. Relation of beta-casomorphin to apnea in sudden infant death syndrome. Peptides 2003, 24, 937–943. [Google Scholar] [CrossRef]
- Sitkowska, B.; Neja, W.; Wiśniewska, E. Relations between kappa-casein polymorphism (csn3) and milk performance traits in heifer cows. J. Cent. Eur. Agric. 2008, 9, 641–644. [Google Scholar]
- Botaro, B.G.; De Lima, Y.V.R.; Cortinhas, C.S.; Silva, E.L.F.P.; Rennó, F.P.; Dos Santos, M.V. Effect of the kappa-casein gene polymorphism, breed and seasonality on physicochemical characteristics, composition and stability of bovine milk. Rev. Bras. Zootec. 2009, 38, 2447–2454. [Google Scholar] [CrossRef]
- Doosti, A.; Arshi, A.; Vatankhah, M.; Amjadi, P. Kappa-casein gene polymorphism in Holstein and Iranian native cattle by polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP). Afr. J. Biotechnol. 2011, 10, 4957–4960. [Google Scholar] [CrossRef]
- Gouda, E.M.; Galal, M.K.; Abdelaziz, S.A. Genetic Variants and Allele Frequencies of Kappa Casein in Egyptian Cattle and Buffalo Using PCR-RFLP. J. Agric. Sci. 2013, 5, 9752–9760. [Google Scholar] [CrossRef][Green Version]
- Alipanah, M.; Klashnikova, L.; Rodionov, G. K-casein genotypic frequencies in Russian breed Black and Red Pied cattle. Iran. J. Biotechnol. 2007, 3, 191–194. [Google Scholar]
- Hamza, A.E.; Wang, X.L.; Yang, Z.P. Kappa Casein Gene Polymorphism in Holstein Chinese Cattle. Pak. Vet. J. 2010, 30, 203–206. [Google Scholar]
- Azevedo, A.L.S.; Nascimento, C.S.; Steinberg, R.S.; Carvalho, M.R.S.; Peixoto, M.G.C.D.; Teodoro, R.L.; Verneque, R.S.; Guimarães, S.E.F.; Machado, M.A. Genetic polymorphism of the kappa-casein gene in Brazilian cattle. Genet. Mol. Res. 2008, 7, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.A.; Sgrò, C.M.; Kristensen, T.N. Revisiting Adaptive Potential, Population Size, and Conservation. Trends Ecol. Evol. 2017, 32, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Ørsted, M.; Hoffmann, A.A.; Sverrisdóttir, E.; Nielsen, K.L.; Kristensen, T.N. Genomic variation predicts adaptive evolutionary responses better than population bottleneck history. PLoS Genet. 2019, 15, e1008205. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.L.; Jasper, M.E.; Weeks, A.R.; Hoffmann, A.A. Unbiased population heterozygosity estimates from genome-wide sequence data. Methods Ecol. Evol. 2021, 12, 1888–1898. [Google Scholar] [CrossRef]
- Cosenza, M.; La Rosa, V.; Rosati, R.; Chiofalo, V. Genetic diversity of the Italian thoroughbred horse population. Ital. J. Anim. Sci. 2019, 1, 538–545. [Google Scholar] [CrossRef]
- Kalashnikov, V.; Khrabrova, L.; Blohina, N.; Zaitcev, A.; Kalashnikova, T. Dynamics of the Inbreeding Coefficient and Homozygosity in Thoroughbred Horses in Russia. Animals 2020, 10, 1217. [Google Scholar] [CrossRef]
- Zhang, M.; Peng, W.F.; Hu, X.J.; Zhao, Y.X.; Lv, F.H.; Yang, J. Global genomic diversity and conservation priorities for domestic animals are associated with the economies of their regions of origin. Sci. Rep. 2018, 8, 11677. [Google Scholar] [CrossRef]
- Groeneveld, L.F.; Lenstra, J.A.; Eding, H.; Toro, M.A.; Scherf, B.; Pilling, D.; Negrini, R.; Finlay, E.K.; Jianlin, H.; Groeneveld, E.; et al. GLOBALDIV Consortium. Genetic diversity in farm animals—A review. Anim. Genet. 2010, 41, 6–31. [Google Scholar] [CrossRef]
- Zhang, P.; Qiu, X.; Wang, L.; Zhao, F. Progress in Genomic Mating in Domestic Animals. Animals 2022, 12, 2306. [Google Scholar] [CrossRef]
- Kasarda, R.; Jamborová, Ľ.; Moravčíková, N. Genetic diversity and production potential of animal food resources. Acta Fytotech. Zootech. 2020, 23, 102–108. [Google Scholar] [CrossRef]
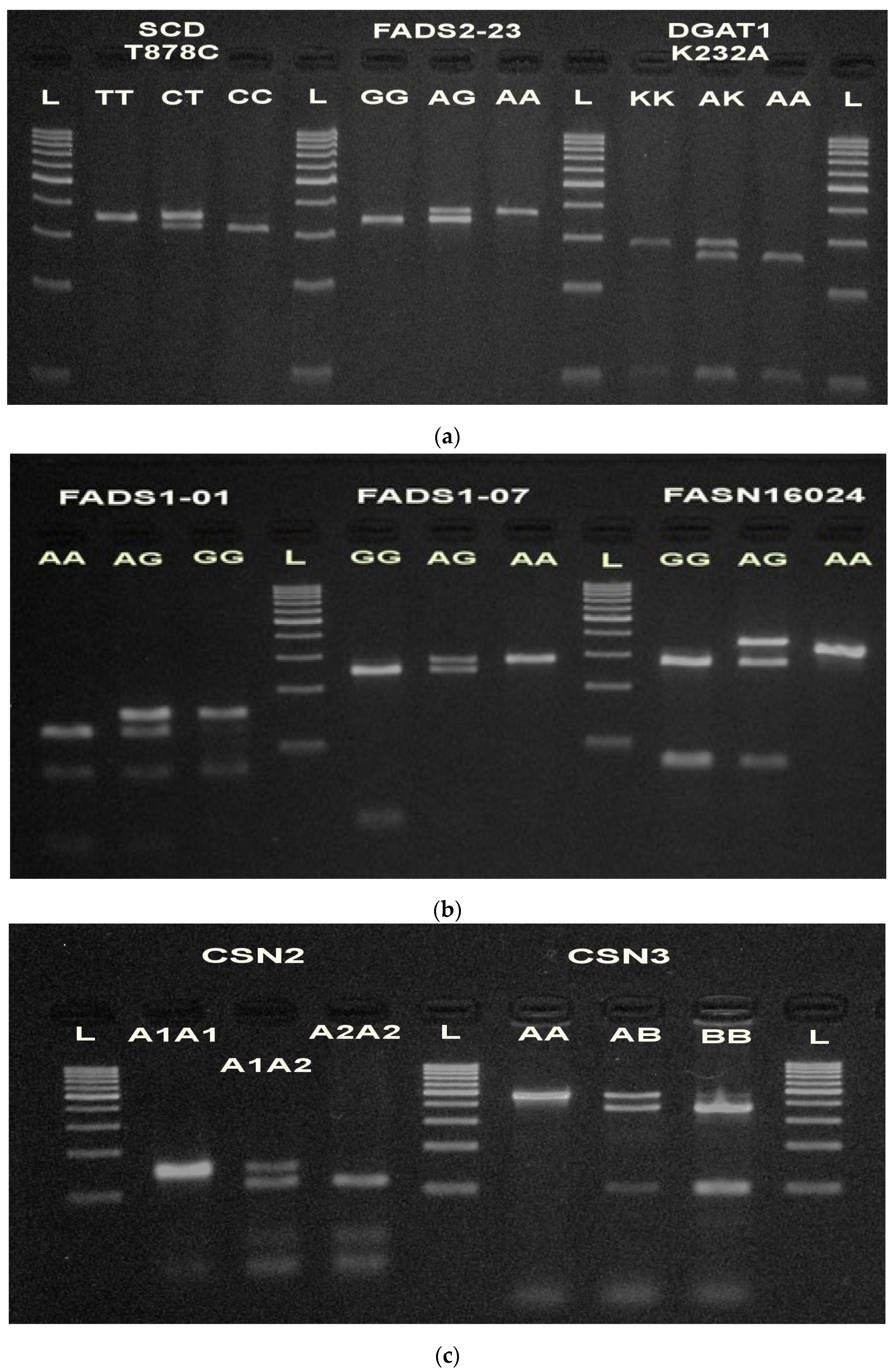

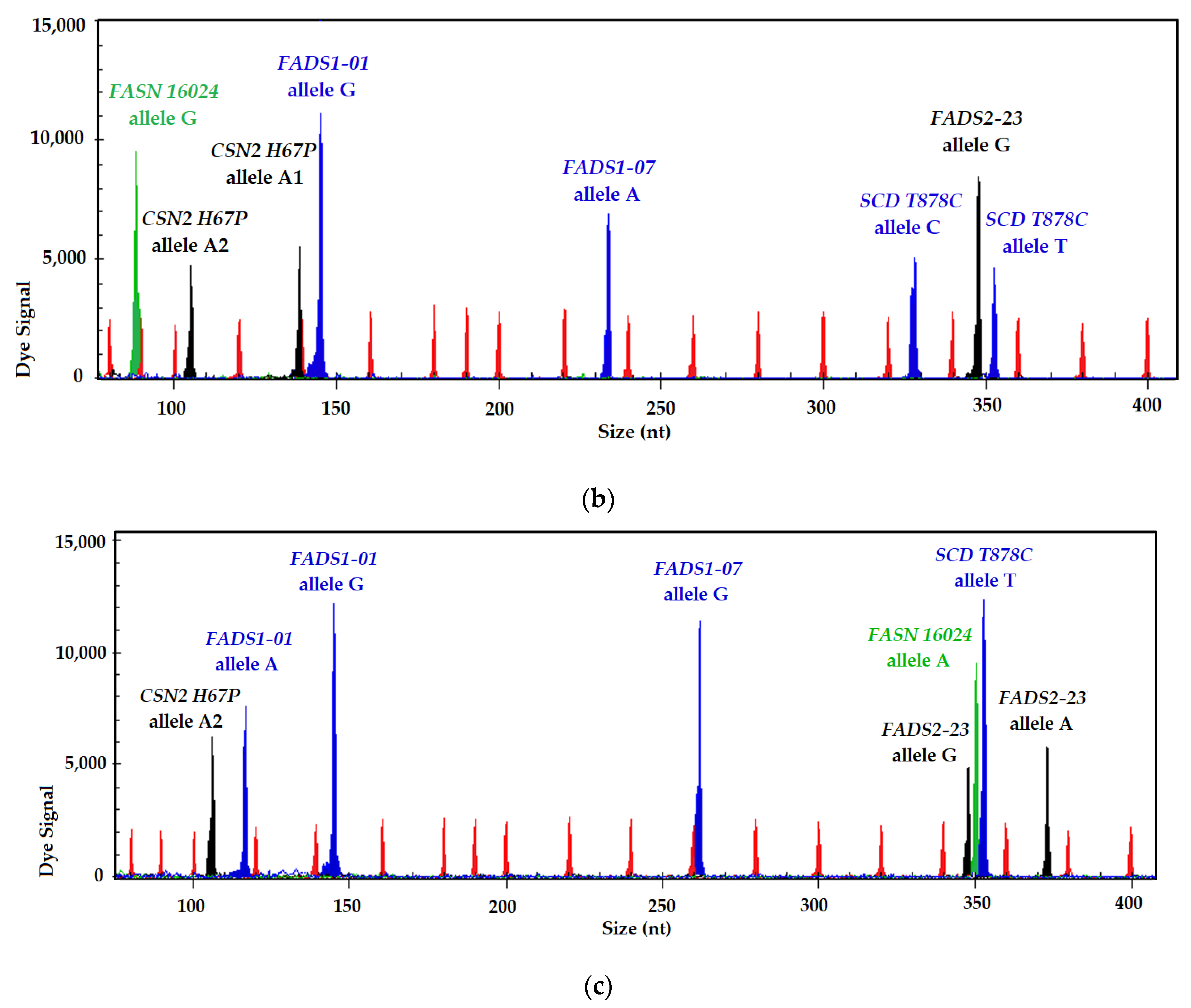
| Marker | Mutation | Location | SNP | References |
|---|---|---|---|---|
| FADS1-01 | A/G | intron | rs136261927 | [11] |
| FADS1-07 | A/G | exon | rs42187261 | [11] |
| FADS2-23 | C/G | 3′ UTR | rs109772589 | [11] |
| FASN-16024 | A/G | exon | rs480320793 | [39] |
| SCD-T878C | C/T | exon | rs41255693 | [40] |
| DGAT1-K232A | AA/GC | exon | rs109326954 | [41] |
| CSN2-H67P | A/C | exon | rs43703011 | [42] |
| CSN3-D148A | A/C | exon | rs43703016 | [43] |
| Marker | Method | Primer Sequences 5′-3′ | Primer Labeling | Ta | MgCl2 | RE | Allele Identification # | References |
|---|---|---|---|---|---|---|---|---|
| FADS1-01 | ACRS-PCR | * For: 5′-GGCAGCGGGAGAAATGGAAGG-3′ Rev: 5′-ACCCCTTAGGAGGCCACTGACCACACAG-3′ | WellRed D4 | 60 °C | 2 mM | PflMI | Allele G: 143 bp Allele A: 116 bp, 27 bp | Present study |
| FADS1-07 | ACRS-PCR | * For: 5′-TGCACCCAGATCAAATCAGTACAAGCA-3′ Rev: 5′-CACTTTCTACGTCCGTATCTTCCTCACATA-3′ | WellRed D4 | 60 °C | 2 mM | NdeI | Allele A: 261 bp Allele G: 231 bp, 30 bp | Present study |
| FADS2-23 | ACRS-PCR | For: 5′-ACCCGTAGATAGCTCCAGGAGAGGCC-3′ * Rev: 5′-GTGCTCCCATCGCAAAGCAG-3′ | WellRed D2 | 60 °C | 2 mM | MspI | Allele A: 372 bp Allele G: 347 bp, 25 bp | Present study |
| FASN 16024 | PCR-RFLP | * For: 5′-CTACCAAGCCAGGCAGGTC-3′ Rev: 5′-GCCATTGTACTTGGGCTTGT-3′ | WellRed D3 | 60 °C | 2 mM | HhaI | Allele A: 353 bp Allele G: 262 bp, 91 bp | [39] |
| SCD T878C | ACRS-PCR | * For: 5′-GCCCTGTGAGAGTGGAAAATCAGGT-3′ Rev: 5′-TCTTGCTGTGGACTGCTGACTTACG-3′ | WellRed D4 | 60 °C | 2 mM | Hin6I | Allele T: 350 bp Allele C: 323 bp, 27 bp | [45] |
| DGAT1 K232A | ACRS-PCR | For: 5′-TGCCGCTTGCTCGTAGCTTTGGCC-3′ Rev: 5′-ACCTGGAGCTGGGTGAGGAACAGC-3′ | ------- | 66 °C | 1.5 mM | BglI | Allele A: 254 bp, 96 bp, 28 bp Allele K: 282 bp, 96 bp | [41] |
| CSN2 H67P | ACRS-PCR | * F: 5′-CCTTCTTTCCAGGATGAACTCCAGG-3′ R: 5′-GAGTAAGAGGAGGGATGTTTTGTGGGAGGCTCT-3′ | WellRed D2 | 60 °C | 2 mM | DdeI | Allele A1: 138 bp Allele A2: 103 bp, 35 bp | [42] |
| CSN3 D148A | PCR-RFLP | F: 5′-GCTGAGCAGGTATCCTAGTTAT-3′ R: 5′-CTTCTTTGATGTCTCCTTAGAG-3′ | --------- | 60 °C | 2 mM | HindIII | Allele A: 443 bp Allele B: 348 bp, 95 bp | [43] |
| Marker | Genotype Frequencies | Allelic Frequencies | χ2 | P | |||
|---|---|---|---|---|---|---|---|
| AA | AG | GG | A | G | |||
| FADS1-01 | 0.0381 | 0.3714 | 0.5905 | 0.2238 | 0.7762 | 1.033 | 0.6055 |
| FADS1-07 | 0.3191 | 0.5333 | 0.1476 | 0.5857 | 0.4143 | 2.056 | 0.3577 |
| FADS2-23 | 0.0810 | 0.4192 | 0.50 | 0.2905 | 0.7095 | 0.058 | 0.9713 |
| FASN-16024 | 0.0238 | 0.3000 | 0.6762 | 0.1738 | 0.8262 | 0.416 | 0.8120 |
| CC | CT | TT | C | T | |||
| SCD-T878C | 0.5286 | 0.3952 | 0.0762 | 0.7262 | 0.2738 | 0.008 | 0.9958 |
| AA | AK | KK | A | K | |||
| DGAT1-K232A | 0.6762 | 0.2810 | 0.0428 | 0.8167 | 0.1833 | 0.801 | 0.6700 |
| A1A1 | A1A2 | A2A2 | A1 | A2 | |||
| CSN2-H67P | 0.1381 | 0.4619 | 0.40 | 0.3690 | 0.6310 | 0.013 | 0.9933 |
| AA | AB | BB | A | B | |||
| CSN3-D148A | 0.6952 | 0.2762 | 0.0286 | 0.8333 | 0.1667 | 0.007 | 0.9967 |
| Marker | Alleles | He(obs) | He(exp) | PIC | E | ENA | V% |
|---|---|---|---|---|---|---|---|
| FADS1-01 | A; G | 0.3714 | 0.3474 | 0.2870 | 0.6526 | 1.5323 | 34.91 |
| FADS1-07 | A; G | 0.533 | 0.4853 | 0.367 | 0.5147 | 1.9429 | 48.76 |
| FADS2-23 | A; G | 0.4192 | 0.4122 | 0.3272 | 0.5878 | 1.7013 | 41.42 |
| FASN-16024 | A; G | 0.3000 | 0.2872 | 0.2460 | 0.7127 | 1.4029 | 28.86 |
| SCD-T878C | C; T | 0.3952 | 0.3977 | 0.3187 | 0.6023 | 1.6603 | 39.96 |
| DGAT1-K232A | A; K | 0.2810 | 0.2994 | 0.2546 | 0.7006 | 1.4273 | 30.08 |
| CSN2-H67P | A1; A2 | 0.4619 | 0.4659 | 0.3571 | 0.5344 | 1.8713 | 46.78 |
| CSN3-D148A | A; B | 0.2762 | 0.2778 | 0.2392 | 0.7222 | 1.3847 | 27.91 |
| Population | ENA | He(obs) | He(exp) | FIS |
|---|---|---|---|---|
| Holstein cows | 1.6154 | 0.3797 | 0.3716 | −0.0218 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miluchová, M.; Gábor, M.; Gašper, J. Analysis of the Genetic Structure of Slovak Holstein Cattle Using Seven Candidate Genes Related to Milk Quality. Diversity 2022, 14, 989. https://doi.org/10.3390/d14110989
Miluchová M, Gábor M, Gašper J. Analysis of the Genetic Structure of Slovak Holstein Cattle Using Seven Candidate Genes Related to Milk Quality. Diversity. 2022; 14(11):989. https://doi.org/10.3390/d14110989
Chicago/Turabian StyleMiluchová, Martina, Michal Gábor, and Juraj Gašper. 2022. "Analysis of the Genetic Structure of Slovak Holstein Cattle Using Seven Candidate Genes Related to Milk Quality" Diversity 14, no. 11: 989. https://doi.org/10.3390/d14110989
APA StyleMiluchová, M., Gábor, M., & Gašper, J. (2022). Analysis of the Genetic Structure of Slovak Holstein Cattle Using Seven Candidate Genes Related to Milk Quality. Diversity, 14(11), 989. https://doi.org/10.3390/d14110989






