Exploring the Biodiversity of a European NATURA 2000 Mediterranean Lagoon through eDNA Metabarcoding
Abstract
1. Introduction
2. Materials and Methods
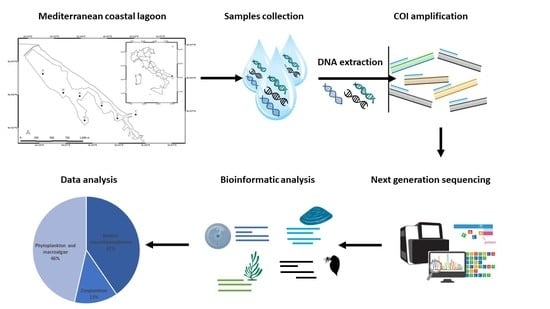
2.1. Sampling Protocol and eDNA Extraction
2.2. DNA Amplification and High-Throughput Sequencing
3. Results
3.1. Species Biodiversity Assessment by eDNA Metabarcoding
3.2. Molecular and Morphological Species’ Identification Are Congruent and Complementary
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Exposito-Alonso, M.; Booker, T.R.; Czech, L.; Gillespie, L.; Hateley, S.; Kyriazis, C.C.; Lang, P.L.M.; Leventhal, L.; Nogues-Bravo, D.; Pagowski, V.; et al. Genetic diversity loss in the Anthropocene. Science 2022, 377, 1431–1435. [Google Scholar] [CrossRef] [PubMed]
- Chapin, F.S.; Zavaleta, E.S.; Eviner, V.T.; Naylor, R.L.; Vitousek, P.M.; Reynolds, H.L.; Hooper, D.U.; Lavorel, S.; Sala, O.E.; Hobbie, S.E.; et al. Consequences of changing biodiversity. Nature 2000, 405, 234–242. [Google Scholar] [CrossRef] [PubMed]
- European Commission, Directorate-General for Environment. EU Biodiversity Strategy for 2030: Bringing Nature Back into Our Lives; Publications Office of the European Union: Luxembourg, 2021. [Google Scholar]
- Council Directive 92/43/EEC of 21 May 1992 on the Conservation of Natural Habitats and of Wild Fauna and Flora. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:31992L0043 (accessed on 11 November 2022).
- Baldwin, C.C.; Collette, B.B.; Parenti, L.R.; Smith, D.G.; Springer, V.G. Collecting fishes. Methods and Techniques of Underwater Research. In Proceedings of the 16th Annual Scientific Diving Symposium, Washington, DC, USA, 12–13 October 1996; American Academy of Underwater Sciences: Dauphin Island, AL, USA; pp. 11–33. [Google Scholar]
- Jones, J.B. Environmental impact of trawling on the seabed: A review. N. Z. J. Mar. Freshw. Res. 1992, 26, 59–67. [Google Scholar] [CrossRef]
- Leese, F.; Altermatt, F.; Bouchez, A.; Ekrem, T.; Hering, D.; Meissner, K.; Mergen, P.; Pawlowski, J.; Piggott, J.; Rimet, F.; et al. DNAqua-Net: Developing new genetic tools for bioassessment and monitoring of aquatic ecosystems in Europe. Res. Ideas Outcomes 2016, 2, e11321. [Google Scholar] [CrossRef]
- Tiralongo, F.; Crocetta, F.; Riginella, E.; Lillo, A.O.; Tondo, E.; Macali, A.; Mancini, E.; Russo, F.; Coco, S.; Paolillo, G.; et al. Snapshot of rare, exotic and overlooked fish species in the Italian seas: A citizen science survey. J. Sea Res. 2020, 164, 101930. [Google Scholar] [CrossRef]
- Pawlowski, M.; Branstrator, D.; Hrabik, T. Major shift in the phenology of crustacean biomass in western Lake Superior associated with temperature anomaly. J. Great Lakes Res. 2018, 44, 788–797. [Google Scholar] [CrossRef]
- Andrew, R.L.; Bernatchez, L.; Bonin, A.; Buerkle, C.A.; Carstens, B.C.; Emerson, B.C.; Garant, D.; Giraud, T.; Kane, N.C.; Rogers, S.M.; et al. A road map for molecular ecology. Mol. Ecol. 2013, 22, 2605–2626. [Google Scholar] [CrossRef]
- Pawlowski, J.; Bonin, A.; Boyer, F.; Cordier, T.; Taberlet, P. Environmental DNA for biomonitoring. Mol. Ecol. 2021, 30, 29–31. [Google Scholar] [CrossRef]
- Pawlowski, J.; Kelly-Quinn, M.; Altermatt, F.; Apothéloz-Perret-Gentil, L.; Beja, P.; Boggero, A.; Borja, A.; Bouchez, A.; Cordier, T.; Domaizon, I.; et al. The future of biotic indices in the ecogenomic era: Integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems. Sci. Total Environ. 2018, 637–638, 1295–1310. [Google Scholar] [CrossRef]
- Tzafesta, E.; Zangaro, F.; Specchia, V.; Pinna, M. An Overview of DNA-Based Applications for the Assessment of Benthic Macroinvertebrates Biodiversity in Mediterranean Aquatic Ecosystems. Diversity 2021, 3, 112. [Google Scholar] [CrossRef]
- Closek, C.J.; Santora, J.A.; Starks, H.A.; Schroeder, I.D.; Andruszkiewicz, E.A.; Sakuma, K.M.; Bograd, S.J.; Hazen, E.L.; Field, J.C.; Boehm, A.B. Marine Vertebrate Biodiversity and Distribution within the Central California Current Using Environmental DNA (eDNA) Metabarcoding and Ecosystem Surveys. Front. Mar. Sci. 2019, 732. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; de Vere, N.; et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 21, 5872–5895. [Google Scholar] [CrossRef] [PubMed]
- Bienert, F.; De Danieli, S.; Miquel, C.; Coissac, E.; Poillot, C.; Brun, J.J.; Taberlet, P. Tracking earthworm communities from soil DNA. Mol. Ecol. 2021, 8, 2017–2030. [Google Scholar] [CrossRef] [PubMed]
- Anglès d’Auriac, M.B.; Strand, D.A.; Mjelde, M.; Demars, B.O.; Thaulow, J. Detection of an invasive aquatic plant in natural water bodies using environmental DNA. PLoS ONE 2019, 7, e0219700. [Google Scholar] [CrossRef] [PubMed]
- Klymus, K.E.; Marshall, N.T.; Stepien, C.A. Environmental DNA (eDNA) metabarcoding assays to detect invasive invertebrate species in the Great Lakes. PLoS ONE 2017, 5, e0177643. [Google Scholar] [CrossRef] [PubMed]
- Strand, D.A.; Johnsen, S.I.; Rusch, J.C.; Agersnap, S.; Larsen, W.B.; Knudsen, S.W.; Møller, P.R.; Vrålstad, T. Monitoring a Norwegian freshwater crayfish tragedy: eDNA snapshots of invasion, infection and extinction. J. Appl. Ecol. 2019, 7, 1661–1673. [Google Scholar] [CrossRef]
- Kirse, A.; Bourlat, S.J.; Langen, K.; Fonseca, V.G. Metabarcoding Malaise traps and soil eDNA reveals seasonal and local arthropod diversity shifts. Sci. Rep. 2021, 1, 10498. [Google Scholar] [CrossRef]
- Epp, L.S.; Boessenkool, S.; Bellemain, E.P.; Haile, J.; Esposito, A.; Riaz, T.; Erséus, C.; Gusarov, V.I.; Edwards, M.E.; Johnsen, A.; et al. New environmental metabarcodes for analysing soil DNA: Potential for studying past and present ecosystems. Mol. Ecol. 2012, 8, 1821–1833. [Google Scholar] [CrossRef]
- Sønstebø, J.H.; Gielly, L.; Brysting, A.K.; Elven, R.; Edwards, M.; Haile, J.; Willerslev, E.; Coissac, E.; Rioux, D.; Sannier, J.; et al. Using next-generation sequencing for molecular reconstruction of past Arctic vegetation and climate. Mol. Ecol. Resour. 2010, 6, 1009–1018. [Google Scholar] [CrossRef]
- Tzafesta, E.; Saccomanno, B.; Zangaro, F.; Vadrucci, M.R.; Specchia, V.; Pinna, M. DNA Barcode Gap Analysis for Multiple Marker Genes for Phytoplankton Species Biodiversity in Mediterranean Aquatic Ecosystems. Biology 2022, 11, 1277. [Google Scholar] [CrossRef]
- Pinna, M.; Saccomanno, B.; Marini, G.; Zangaro, F.; Kabayeva, A.; Khalaj, M.; Shaimardan, L.; D’Attis, S.; Tzafesta, E.; Specchia, V. Testing the Influence of Incomplete DNA Barcode Libraries on Ecological Status Assessment of Mediterranean Transitional Waters. Biology 2021, 11, 1092. [Google Scholar] [CrossRef] [PubMed]
- Zangaro, F.; Saccomanno, B.; Tzafesta, E.; Bozzeda, F.; Specchia, V.; Pinna, M. Current limitations and future prospects of detection and biomonitoring of NIS in the Mediterranean Sea through environmental DNA. NeoBiota 2021, 70, 151. [Google Scholar] [CrossRef]
- Specchia, V.; Tzafesta, E.; Marini, G.; Scarcella, S.; D’Attis, S.; Pinna, M. Gap analysis for DNA barcode reference libraries for aquatic macroinvertebrate species in the Apulia Region (Southeast of Italy). J. Mar. Sci. Eng. 2020, 7, 538. [Google Scholar] [CrossRef]
- Weigand, H.; Beermann, A.J.; Čiampor, F.; Costa, F.O.; Csabai, Z.; Duarte, S.; Geiger, M.F.; Grabowski, M.; Rimet, F.; Rulik, B.; et al. DNA barcode reference libraries for the monitoring of aquatic biota in Europe: Gap-analysis and recommendations for future work. Sci. Total Environ. 2019, 678, 499–524. [Google Scholar] [CrossRef] [PubMed]
- Zangaro, F.; Marini, G.; Specchia, V.; De Luca, M.; Visintin, F.; Bullo, G.; Richard, J.; Šalaja, N.; Rakar, B.; Lipej, B.; et al. Building a transnational biodiversity geo-database of the protected areas in the Adriatic-Ionian Macro-Region: Approaches and results from the IMPRECO Project. Biodivers. Data J. 2021, 9, e67169. [Google Scholar] [CrossRef]
- Marrocco, V.; Sicuro, A.; Zangaro, F.; Pinna, M. First record of the protected species Pinna nobilis (Linnaeus, 1758) in the Aquatina Lagoon (NATURA 2000 site IT9150003, South-East Italian coastline). Nat. Conserv. 2018, 28, 51. [Google Scholar] [CrossRef]
- Mazor, T.; Doropoulos, C.; Schwarzmueller, F.; Gladish, D.W.; Kumaran, N.; Merkel, K.; Gagic, V. Global mismatch of policy and research on drivers of biodiversity loss. Nat. Ecol. Evol. 2018, 7, 1071–1074. [Google Scholar] [CrossRef] [PubMed]
- Tickner, D.; Opperman, J.J.; Abell, R.; Acreman, M.; Arthington, A.H.; Bunn, S.E.; Cooke, S.J.; Dalton, J.; Darwall, W.; Edwards, G.; et al. Bending the curve of global freshwater biodiversity loss—An emergency recovery plan. Bioscience 2020, 4, 330–342. [Google Scholar] [CrossRef]
- Takasaki, K.; Aihara, H.; Imanaka, T.; Matsudaira, T.; Tsukahara, K.; Usui, A.; Osaki, S.; Doi, H. Water pre-filtration methods to improve environmental DNA detection by real-time PCR and metabarcoding. PLoS ONE 2021, 16, e0250162. [Google Scholar]
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Bohem, J.T.; Machida, R.J. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: Application for characterizing coral reef fish gut contents. Front. Zool. 2013, 10, 34. [Google Scholar] [CrossRef]
- Geller, J.; Meyer, C.; Parker, M.; Hawk, H. Redesign of PCR primers for mitochondrial cytochrome c oxidase subunit I for marine invertebrates and application in all-taxa biotic surveys. Mol. Ecol. Resour. 2013, 13, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.; Ratnasingham, S.; De Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. 2003, 270, S96–S99. [Google Scholar] [CrossRef] [PubMed]
- Pentinsaari, M.; Salmela, H.; Mutanen, M.; Roslin, T. Molecular evolution of a widely-adopted taxonomic marker (COI) across the animal tree of life. Sci. Rep. 2016, 6, 35275. [Google Scholar] [CrossRef]
- Chariton, A.A.; Stephenson, S.; Morgan, M.J.; Steven, A.; Colloff, M.J.; Court, L.N.; Hardy, C.M. Metabarcoding of benthic eukaryote communities predicts the ecological condition of estuaries. Environ. Pollut. 2015, 203, 165–174. [Google Scholar] [CrossRef]
- Caroppo, C. Le comunità fitoplanctoniche del Lago di Acquatina (Mar Adriatico meridionale). Thalass. Salentina 2009, 31, 29–36. [Google Scholar]
- Zingone, A.; Escalera, L.; Aligizaki, K.; Fernández-Tejedor, M.; Ismael, A.; Montresor, M.; Mozetič, P.; Taş, S.; Totti, C. Toxic marine microalgae and noxious blooms in the Mediterranean Sea: A contribution to the Global HAB Status Report. Harmful Algae 2021, 102, 101843. [Google Scholar] [CrossRef]
- Núñez-Vázquez, E.; Gárate-Lizárraga, I.; Band-Schmidt, C.J.; Cordero-Tapia, A.; López-Cortés, D.J.; Hernández-Sandoval, F.H.; Heredia-Tapia, A.; Bustillos-Guzmán, J.J. Impact of harmful algal blooms on wild and cultured animals in the Gulf of California. J. Environ. Biol. 2011, 32, 413–423. [Google Scholar]
- Tiffany, M.A.; Barlow, S.B.; Matey, V.E.; Hulbert, S.H. Chattonella marina (Raphidophyceae) a potentially toxic alga in the Salton Sea, California. Hydrobiologia 2017, 466, 187–194. [Google Scholar] [CrossRef]
- Zingone, A.; Enevoldsen, H.O. The diversity of harmful algal blooms: A challenge for science and management. Ocean Coast. Manag. 2000, 43, 725–748. [Google Scholar] [CrossRef]
- Bates, S.S.; Hubbard, K.A.; Lundholm, N.; Montresor, M.; Leaw, C.P. Pseudo-nitzschia, Nitzschia, and domoic acid: New research since 2011. Harmful Algae 2018, 79, 3–43. [Google Scholar] [CrossRef] [PubMed]
- Cilleros, K.; Valentini, A.; Allard, L.; Dejean, T.; Etienne, R.; Grenouillet, G.; Iribar, A.; Taberlet, P.; Vigouroux, R.; Brosse, S. Unlocking biodiversity and conservation studies in high-diversity environments using environmental DNA (eDNA): A test with Guianese freshwater fishes. Mol. Ecol. Resour. 2019, 19, 27–46. [Google Scholar] [CrossRef] [PubMed]
- Klöpper, S.; John, U.; Zingone, A.; Mangoni, O.; Kooistra, W.H.C.F.; Cembella, A. Phylogeny and morphology of a Chattonella (Raphidophyceae) species from the Mediterranean Sea: What is C. subsalsa? Eur. J. Phycol. 2013, 48, 79–92. [Google Scholar] [CrossRef]
- Litaker, R.; Montresor, M.; Brosnahan, M.; Hoppenrath, M.; Murray, S.; Wolny, J.; John, U.; Sampedro, N.; Larsen, J.; Calado, A. A practical guide to new nomenclature for species within the “Alexandrium tamarense species complex”. Harmful Algae News 2018, 61, 13–15. [Google Scholar]
- Puncher, G.N.; Alemany, F.; Arrizabalaga, H.; Cariani, A.; Tinti, F. Misidentification of bluefin tuna larvae: A call for caution and taxonomic reform. Rev. Fish. Biol. Fisheries 2015, 25, 485–502. [Google Scholar] [CrossRef]
- Collins, R.A.; Bakker, J.W.; Soto, A.Z.; Corrigan, L.; Sims, D.W.; Genner, M.J.; Mariani, S. Non-specific amplification compromises environmental DNA metabarcoding with COI. Methods Ecol. Evol. 2019, 10, 1985–2001. [Google Scholar] [CrossRef]
- Moushomi, R.; Wilgar, G.; Carvalho, G.; Creer, S.; Seymour, M. Environmental DNA size sorting and degradation experiment indicates the state of Daphnia magna mitochondrial and nuclear eDNA is subcellular. Sci. Rep. 2019, 1, 12500. [Google Scholar] [CrossRef] [PubMed]
- Milan, D.T.; Mendes, I.S.; Damasceno, J.S.; Teixeira, D.F.; Sales, N.G.; Carvalho, D.C. New 12S metabarcoding primers for enhanced Neotropical freshwater fish biodiversity assessment. Sci. Rep. 2020, 10, 17966. [Google Scholar] [CrossRef]
- Shaw, J.L.A.; Clarke, L.J.; Wedderburn, S.D.; Barnes, T.C.; Weyrich, L.S.; Cooper, A. Comparison of environmental DNA metabarcoding and conventional fish survey methods in a river system. Biol. Conserv. 2016, 197, 131–138. [Google Scholar] [CrossRef]
- Yamamoto, S.; Masuda, R.; Sato, Y.; Sado, T.; Araki, H.; Kondoh, M.; Miya, M. Environmental DNA metabarcoding reveals local fish communities in a species-rich coastal sea. Sci. Rep. 2017, 7, 40368. [Google Scholar] [CrossRef]
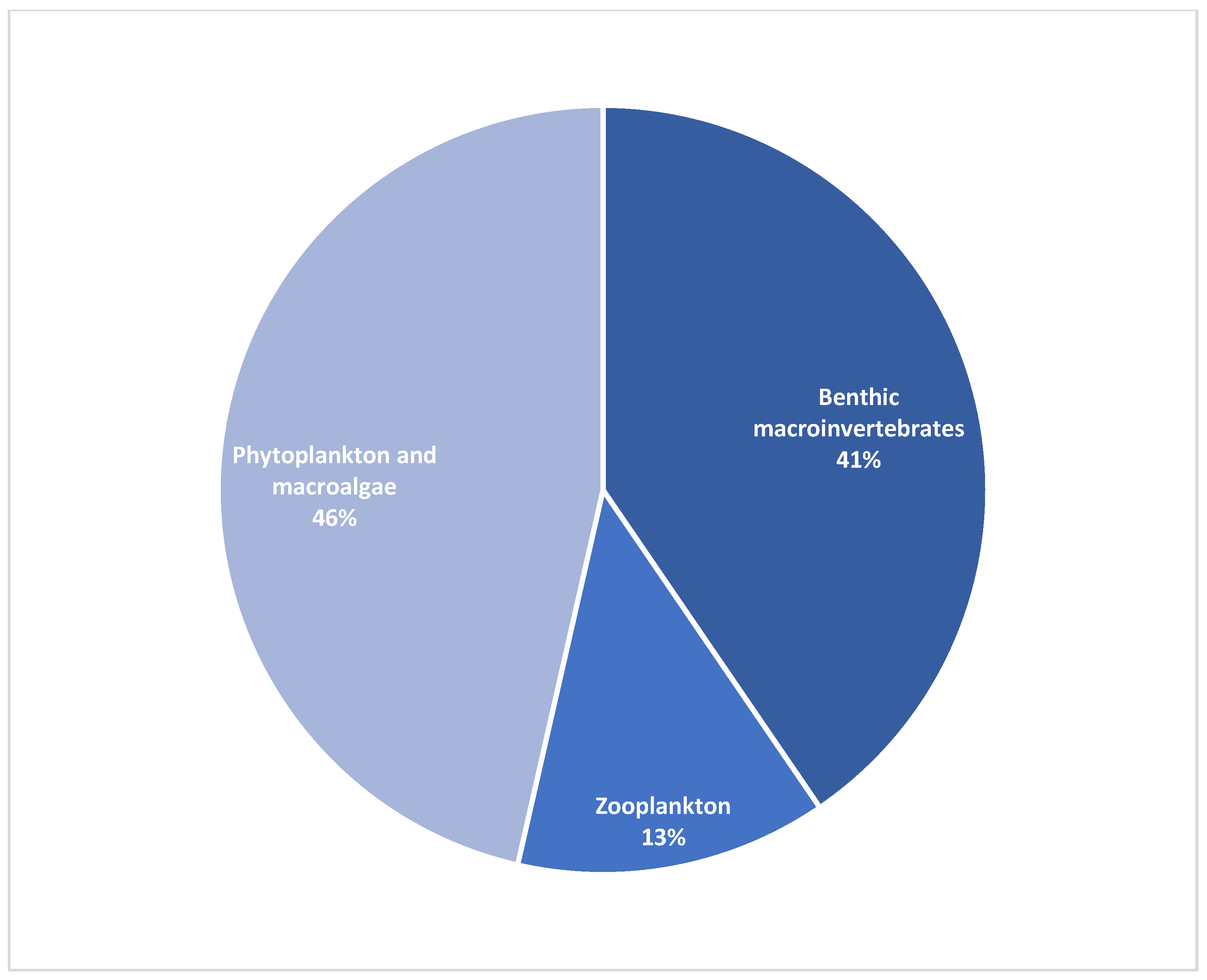
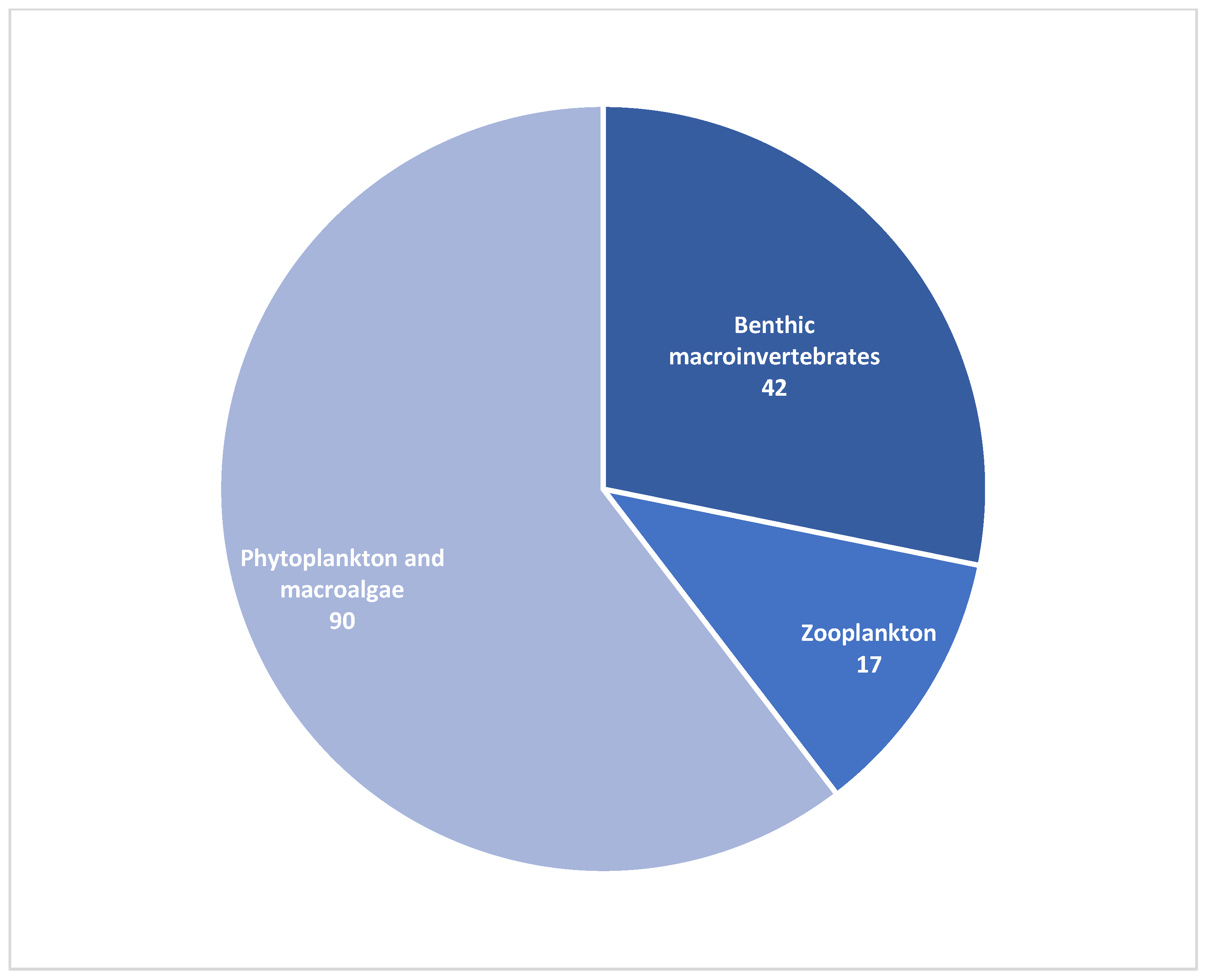
| Phylum | Percentage of Reads |
|---|---|
| Arthropoda | 40.38% |
| Rhodophyta | 22.38% |
| Ochrophyta | 13.27% |
| Cnidaria | 10.42% |
| Annelida | 10.39% |
| Mollusca | 1.77% |
| Chordata | 0.44% |
| Porifera | 0.34% |
| Echinodermata | 0.20% |
| Nemertea | 0.19% |
| Chlorophyta | 0.13% |
| Charophyta | 0.06% |
| Chrisophyta | 0.01% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Specchia, V.; Saccomanno, B.; Zangaro, F.; Tzafesta, E.; Pinna, M. Exploring the Biodiversity of a European NATURA 2000 Mediterranean Lagoon through eDNA Metabarcoding. Diversity 2022, 14, 991. https://doi.org/10.3390/d14110991
Specchia V, Saccomanno B, Zangaro F, Tzafesta E, Pinna M. Exploring the Biodiversity of a European NATURA 2000 Mediterranean Lagoon through eDNA Metabarcoding. Diversity. 2022; 14(11):991. https://doi.org/10.3390/d14110991
Chicago/Turabian StyleSpecchia, Valeria, Benedetta Saccomanno, Francesco Zangaro, Eftychia Tzafesta, and Maurizio Pinna. 2022. "Exploring the Biodiversity of a European NATURA 2000 Mediterranean Lagoon through eDNA Metabarcoding" Diversity 14, no. 11: 991. https://doi.org/10.3390/d14110991
APA StyleSpecchia, V., Saccomanno, B., Zangaro, F., Tzafesta, E., & Pinna, M. (2022). Exploring the Biodiversity of a European NATURA 2000 Mediterranean Lagoon through eDNA Metabarcoding. Diversity, 14(11), 991. https://doi.org/10.3390/d14110991