Downregulation of Gene Expression by Alpha Satellite Transcripts
Abstract
1. Introduction
2. Results
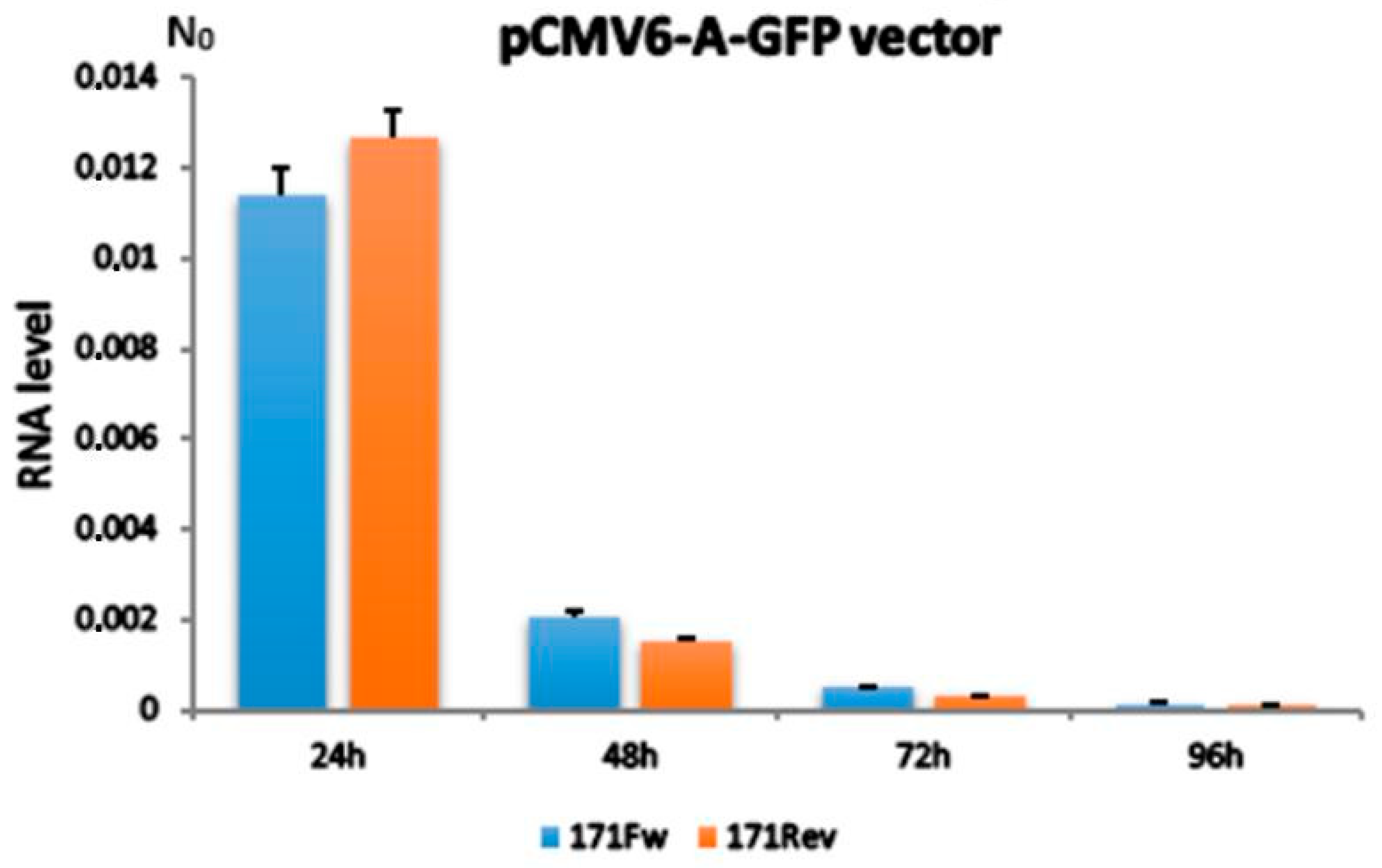
2.1. Alpha Satellite Transcription After Transfection
2.2. Expression Analysis of Alpha-Associated Genes After Transfection
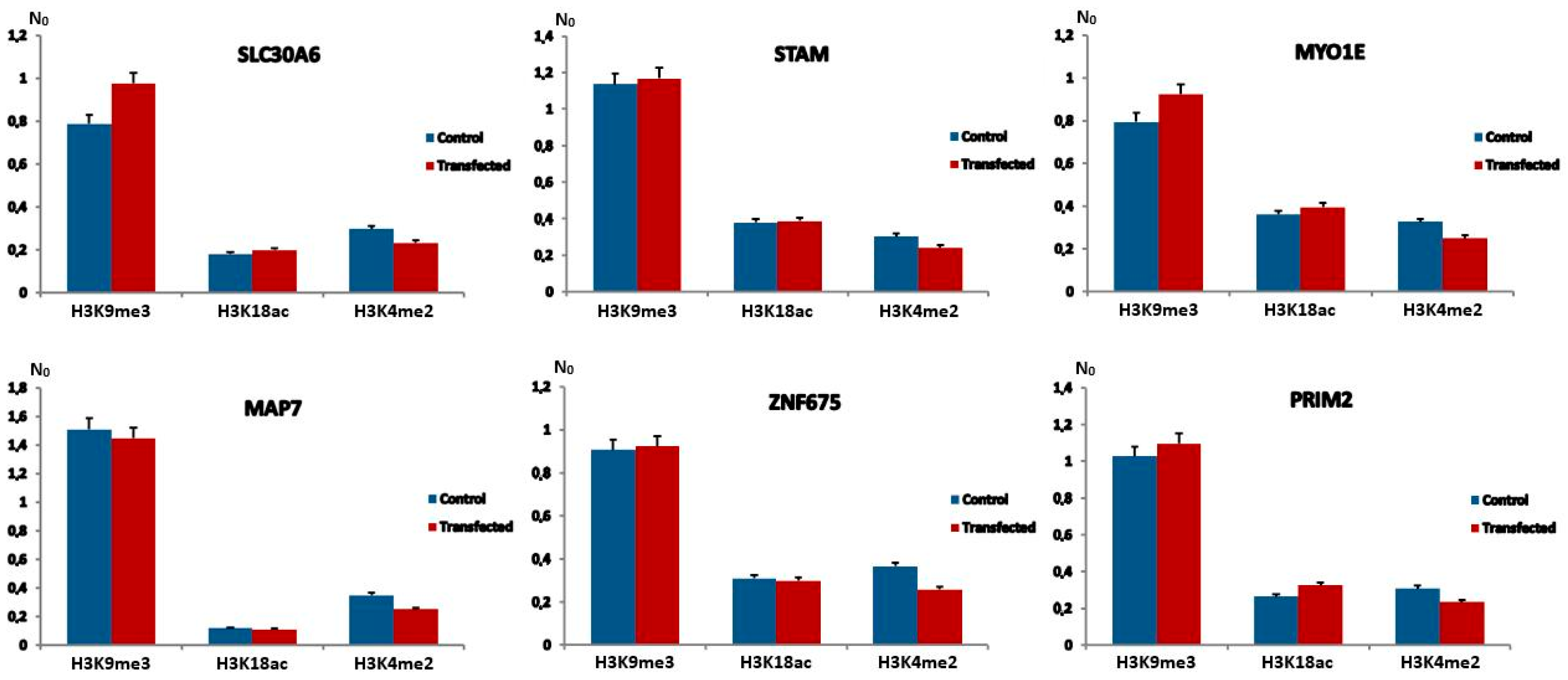
2.3. H3K9me3, H3K18ac and H3K4me2 Levels at Alpha Repeats Dispersed Within Genes After Transfection
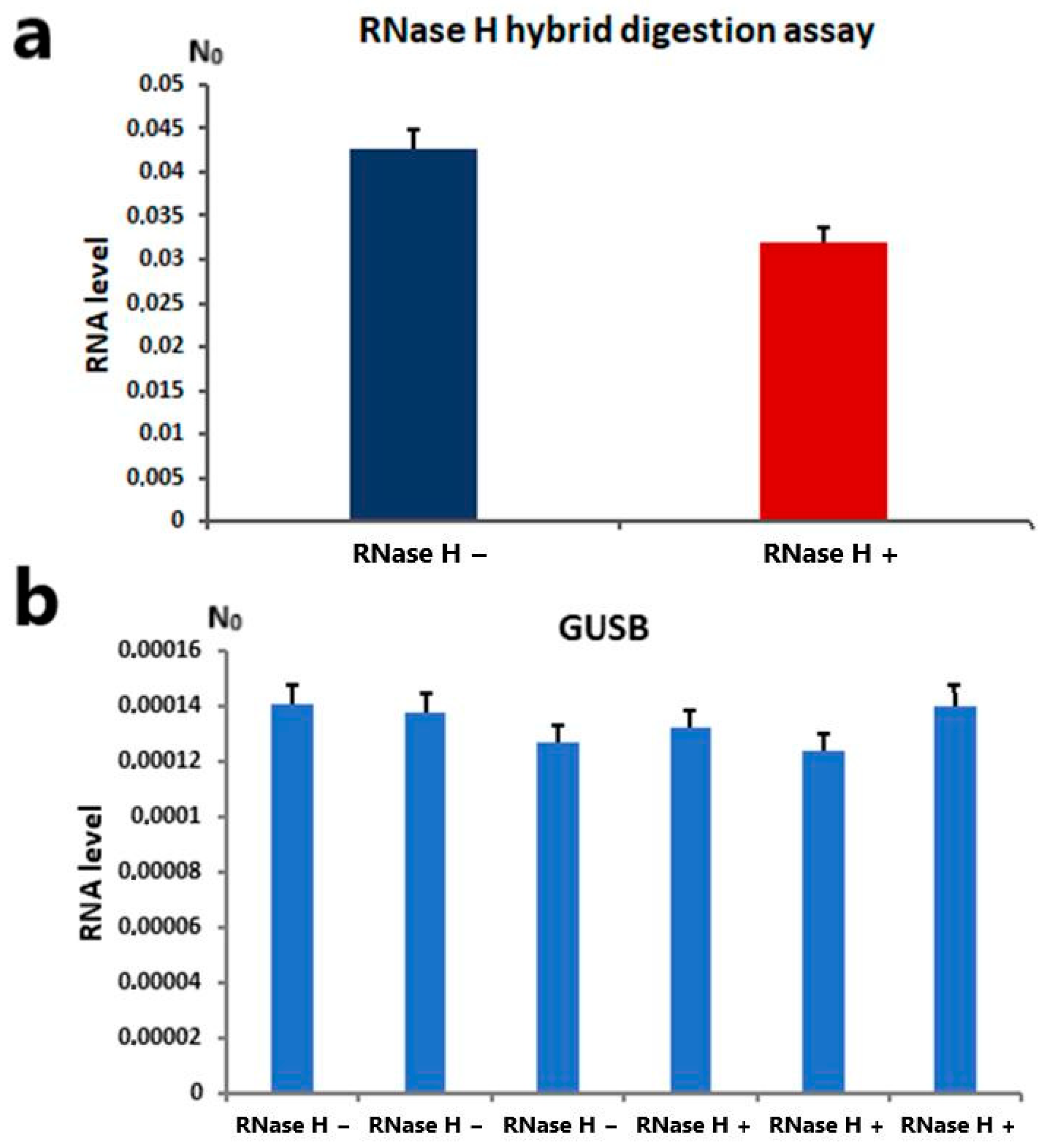
2.4. Alpha Satellite RNA Level Analysis After RNase H Digestion of RNA:DNA Hybrids
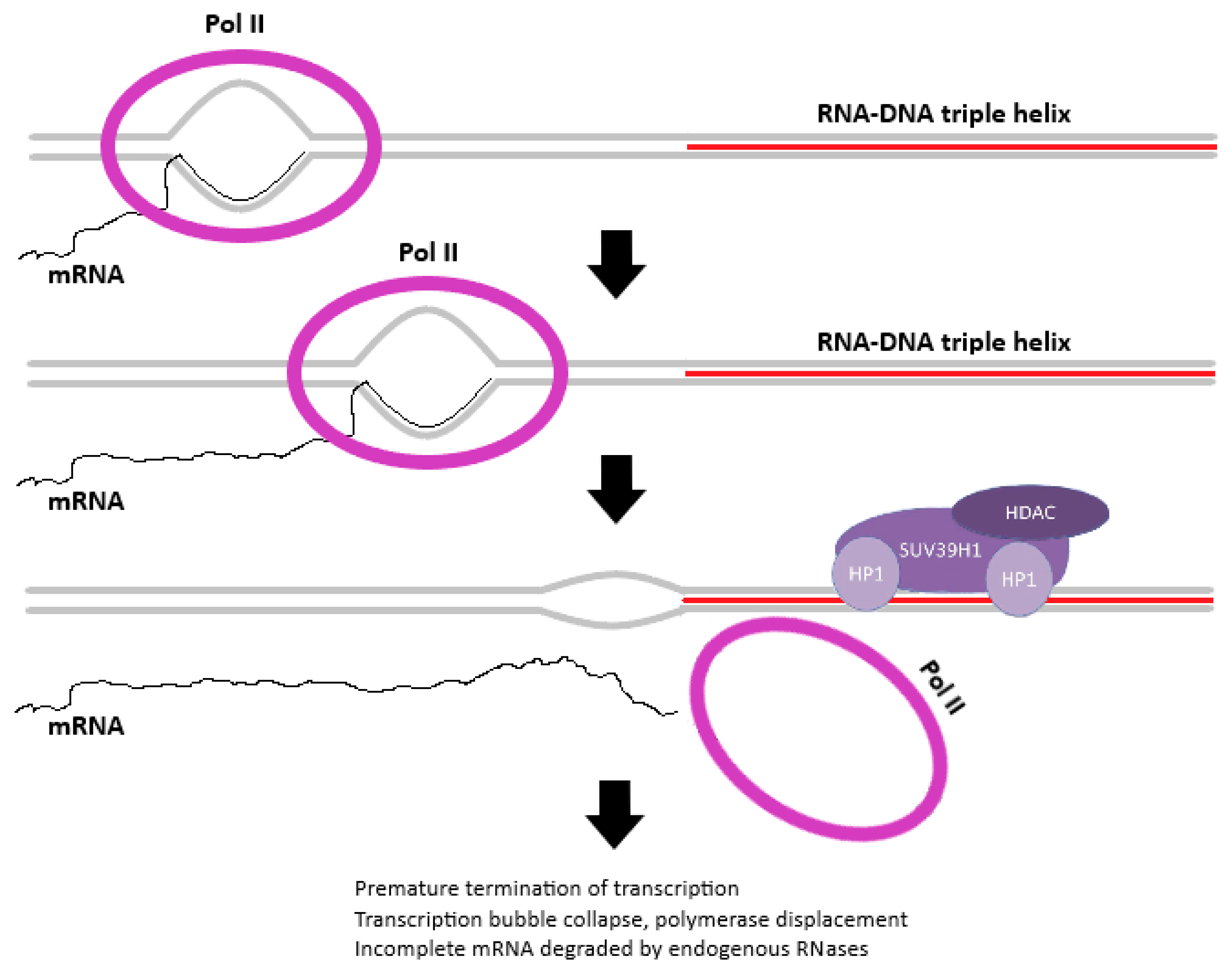
3. Discussion
4. Materials and Methods
4.1. Human Cell Line
4.2. Construction of Vectors
4.3. MJ90hTERT Cell Line Transfection
4.4. RNA Isolation and Reverse Transcription
4.5. Quantitative Real-Time PCR (qPCR) Analyses
4.6. Chromatin Immunoprecipitation
4.7. Alpha Satellite RNA:DNA Hybrid Detection Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, C.; Wevrick, R.; Fisher, R.B.; Ferguson-Smith, M.A.; Lin, C.C. Human centromeric DNAs. Hum. Genet. 1997, 100, 291–304. [Google Scholar] [CrossRef]
- McNulty, S.M.; Sullivan, B.A. Alpha satellite DNA biology: Finding function in the recesses of the genome. Chromosom. Res. 2018, 26, 115–138. [Google Scholar] [CrossRef] [PubMed]
- Feliciello, I.; Pezer, Z.; Kordiš, D.; Bruvo-Mađarić, B.; Ugarković, Đ. Evolutionary history of α satellite DNA repeats dispersed within human genome euchromatin. Genome Biol. Evol. 2020, 12, 2125–2138. [Google Scholar] [CrossRef]
- Feliciello, I.; Picariello, O.; Chinali, G. The first characterisation of the overall variability of repetitive units in a species reveals unexpected features of satellite DNA. Gene 2005, 349, 153–164. [Google Scholar] [CrossRef]
- Feliciello, I.; Picariello, O.; Chinali, G. Intra-specific variability and unusual organization of the repetitive units in a satellite DNA from Rana dalmatina: Molecular evidence of a new mechanism of DNA repair acting on satellite DNA. Gene 2006, 383, 81–92. [Google Scholar] [CrossRef]
- Brajković, J.; Feliciello, I.; Bruvo-Mađarić, B.; Ugarković, Đ. Satellite DNA-Like Elements Associated with Genes Within Euchromatin of the Beetle Tribolium castaneum. G3 Genes/Genomes/Genet. 2012, 2, 931–941. [Google Scholar] [CrossRef]
- Feliciello, I.; Akrap, I.; Brajković, J.; Zlatar, I.; Ugarković, Đ. Satellite DNA as a driver of population divergence in the red flour beetle Tribolium castaneum. Genome Biol. Evol. 2015, 7, 228–239. [Google Scholar] [CrossRef]
- Kuhn, G.C.; Küttler, H.; Moreira-Filho, O.; Heslop-Harrison, J.S. The 1.688 Repetitive DNA of Drosophila: Concerted Evolution at Different Genomic Scales and Association with Genes. Mol. Biol. Evol. 2011, 29, 7–11. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef]
- Feliciello, I.; Akrap, I.; Ugarković, Đ. Satellite DNA Modulates Gene Expression in the Beetle Tribolium castaneum after Heat Stress. PLoS Genet. 2015, 11, e1005466, Erratum in PLoS Genet. 2015, 11, e1005547. [Google Scholar]
- Ljubić, S.; Matulić, M.; Đermić, D.; Feliciello, M.C.; Procino, A.; Ugarković, Đ.; Feliciello, I. Antibiotics induce overexpression of alpha satellite DNA accompanied with epigenetic changes at alpha satellite arrays as well as genome-wide. Epigenet. Chromatin 2025, 18, 62. [Google Scholar] [CrossRef]
- Sermek, A.; Feliciello, I.; Ugarković, Đ. Distinct Regulation of the Expression of Satellite DNAs in the Beetle Tribolium castaneum. Int. J. Mol. Sci. 2021, 22, 296. [Google Scholar] [CrossRef] [PubMed]
- Bury, L.; Moodie, B.; Ly, J.; McKay, L.S.; Miga, K.H.; Cheeseman, I.M. α-satellite RNA transcripts are repressed by centromere-nucleolus associations. eLife 2020, 9, e59770. [Google Scholar] [CrossRef] [PubMed]
- Teng, Z.; Yang, L.; Zhang, Q.; Chen, Y.; Wang, X.; Zheng, Y.; Tian, A.; Tian, D.; Lin, Z.; Deng, W.M.; et al. Topoisomerase I is an evolutionarily conserved key regulator for satellite DNA transcription. Nat. Commun. 2024, 15, 5151. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Shu, X.; Zhou, S.; Mi, Y.; Bian, H.; Li, T.; Li, T.; Ying, X.; Cheng, C.; Liu, D.; et al. Nuclear m6A modification regulates satellite transcription and chromosome segregation. Nat. Chem. Biol. 2025. online ahead of print. [Google Scholar] [CrossRef]
- Johnson, W.L.; Yewdell, W.T.; Bell, J.; McNulty, S.M.; Duda, Z.; O′Neill, R.J.; Sullivan, B.A.; Straight, A. RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. eLife 2017, 6, e25299. [Google Scholar] [CrossRef]
- Velazquez Camacho, O.; Galan, C.; Swist-Rosowska, K.; Ching, R.; Gamalinda, M.; Karabiber, F.; De La Rosa-Velazquez, I.; Engist, B.; Koschorz, B.; Shukeir, N.; et al. Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA- nucleosome association and RNA:DNA hybrid formation. eLife 2017, 6, 817. [Google Scholar] [CrossRef]
- Eymery, A.; Horard, B.; el Atifi-Borel, M.; Fourel, G.; Berger, F.; Vitte, A.-L.; Broeck, A.V.D.; Brambilla, E.; Fournier, A.; Callanan, M.; et al. A transcriptomic analysis of human centromeric and pericentric sequences in normal and tumor cells. Nucleic Acids Res. 2009, 37, 6340–6354. [Google Scholar] [CrossRef]
- Feliciello, I.; Sermek, A.; Pezer, Ž.; Matulić, M.; Ugarković, Đ. Heat stress affects H3K9me3 level at human α satellite DNA repeats. Genes 2020, 11, 663. [Google Scholar] [CrossRef]
- Ugarković, Đ.; Sermek, A.; Ljubić, S.; Feliciello, I. Satellite DNAs in Health and Disease. Genes 2022, 13, 1154. [Google Scholar] [CrossRef]
- Jolly, C.; Metz, A.; Govin, J.; Vigneron, M.; Turner, B.M.; Khochbin, S.; Vourc, H.C. Stress-induced transcription of satellite III repeats. J. Cell Biol. 2003, 164, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, K.; Adachi, S.; Natsume, T.; Iwakiri, J.; Terai, G.; Asai, K.; Hirose, T. LncRNA-dependent nuclear stress bodies promote intron retention through SR protein phosphorylation. EMBO J. 2019, 39, e102729. [Google Scholar] [CrossRef]
- Ninomiya, K.; Iwakiri, J.; Aly, M.K.; Sakaguchi, Y.; Adachi, S.; Natsume, T.; Terai, G.; Asai, K.; Suzuki, T.; Hirose, T. m6A modification of HSATIII lncRNAs regulates temperature-dependent splicing. EMBO J. 2021, 40, e107976. [Google Scholar] [CrossRef]
- Ku, W.L.; Nakamura, K.; Gao, W.; Cui, K.; Hu, G.; Tang, Q.; Ni, B.; Zhao, K. Single-cell chromatin immunocleavage sequencing (scChIC-seq) to profile histone modification. Nat. Methods 2019, 16, 323–325. [Google Scholar] [CrossRef]
- Ku, W.L.; Pan, L.; Cao, Y.; Gao, W.; Zhao, K. Profiling single-cell histone modifications using indexing chromatin immunocleavage sequencing. Genome Res. 2021, 31, 1831–1842. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118, Erratum in Nat. Rev. Mol. Cell Biol. 2021, 22, 159. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Syed, J.; Sugiyama, H. RNA-DNA Triplex Formation by Long Noncoding RNAs. Cell Chem. Biol. 2016, 23, 1325–1333. [Google Scholar] [CrossRef]
- Warwick, T.; Brandes, R.P.; Leisegang, M.S. Computational Methods to Study DNA:DNA:RNA Triplex Formation by lncRNAs. Non-Coding RNA 2023, 9, 10. [Google Scholar] [CrossRef]
- Ariel, F.; Lucero, L.; Christ, A.; Mammarella, M.F.; Jegu, T.; Veluchamy, A.; Mariappan, K.; Latrasse, D.; Blein, T.; Liu, C.; et al. R-Loop Mediated trans Action of the APOLO Long Noncoding RNA. Mol. Cell 2020, 77, 1055–1065.e4. [Google Scholar] [CrossRef]
- Mondal, T.; Subhash, S.; Vaid, R.; Enroth, S.; Uday, S.; Reinius, B.; Mitra, S.; Mohammed, A.; James, A.R.; Hoberg, E.; et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015, 6, 7743, Erratum in Nat. Commun. 2019, 10, 5290. [Google Scholar] [CrossRef]
- Duda, K.J.; Ching, R.W.; Jerabek, L.; Shukeir, N.; Erikson, G.; Engist, B.; Onishi-Seebacher, M.; Perrera, V.; Richter, F.; Mittler, G.; et al. m6A RNA methylation of major satellite repeat transcripts facilitates chromatin association and RNA:DNA hybrid formation in mouse heterochromatin. Nucleic Acids Res. 2021, 49, 5568–5587. [Google Scholar] [CrossRef]
- Cukusic Kalajzic, A.; Vidacek, N.S.; Huzak, M.; Ivankovic, M.; Rubelj, I. Telomere Q-PNA-FISH--reliable results from stochastic signals. PLoS ONE 2014, 9, e92559. [Google Scholar] [CrossRef] [PubMed]
- Gorbunova, V.; Seluanov, A.; Pereira-Smith, O.M. Expression of human telomerase (hTERT) does not prevent stress-induced senescence in normal human fibroblasts but protects the cells from stress-induced apoptosis and necrosis. J. Biol. Chem. 2002, 277, 38540–38549. [Google Scholar] [CrossRef]
- Young, J.I.; Smith, J.R. DNA Methyltransferase Inhibition in Normal Human Fibroblasts Induces a p21-dependent Cell Cycle Withdrawal. J. Biol. Chem. 2001, 276, 19610–19616. [Google Scholar] [CrossRef]
- Choo, K.H.; Vissel, B.; Nagy, A.; Earle, E.; Kalitsis, P. A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 1991, 19, 1179–1182. [Google Scholar] [CrossRef] [PubMed]
- Đermić, D.; Ljubić, S.; Matulić, M.; Procino, A.; Feliciello, M.C.; Ugarković, Đ.; Feliciello, I. Reverse transcription-quantitative PCR (RT-qPCR) without the need for prior removal of DNA. Sci. Rep. 2023, 13, 11470. [Google Scholar] [CrossRef] [PubMed]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; Hoff, M.J.B.V.D.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Pfaffl, M.W.; Zhao, S.; Spiess, A.N.; Boggy, G.; Blom, J.; Rutledge, R.G.; Sisti, D.; Lievens, A.; de Preter, K.; et al. Evaluation of qPCR curve analysis methods for reliable biomarker discovery: Bias, resolution, precision, and implications. Methods 2013, 59, 32–46. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ljubić, S.; Matulić, M.; Đermić, D.; Feliciello, M.C.; Procino, A.; Passaro, F.; Ugarković, Đ.; Feliciello, I. Downregulation of Gene Expression by Alpha Satellite Transcripts. Int. J. Mol. Sci. 2025, 26, 11204. https://doi.org/10.3390/ijms262211204
Ljubić S, Matulić M, Đermić D, Feliciello MC, Procino A, Passaro F, Ugarković Đ, Feliciello I. Downregulation of Gene Expression by Alpha Satellite Transcripts. International Journal of Molecular Sciences. 2025; 26(22):11204. https://doi.org/10.3390/ijms262211204
Chicago/Turabian StyleLjubić, Sven, Maja Matulić, Damir Đermić, Maria Chiara Feliciello, Alfredo Procino, Francesco Passaro, Đurđica Ugarković, and Isidoro Feliciello. 2025. "Downregulation of Gene Expression by Alpha Satellite Transcripts" International Journal of Molecular Sciences 26, no. 22: 11204. https://doi.org/10.3390/ijms262211204
APA StyleLjubić, S., Matulić, M., Đermić, D., Feliciello, M. C., Procino, A., Passaro, F., Ugarković, Đ., & Feliciello, I. (2025). Downregulation of Gene Expression by Alpha Satellite Transcripts. International Journal of Molecular Sciences, 26(22), 11204. https://doi.org/10.3390/ijms262211204








