Preinitiation Complex Loading onto mRNAs with Long versus Short 5′ TLs
Abstract
1. Introduction
2. Results and Discussion
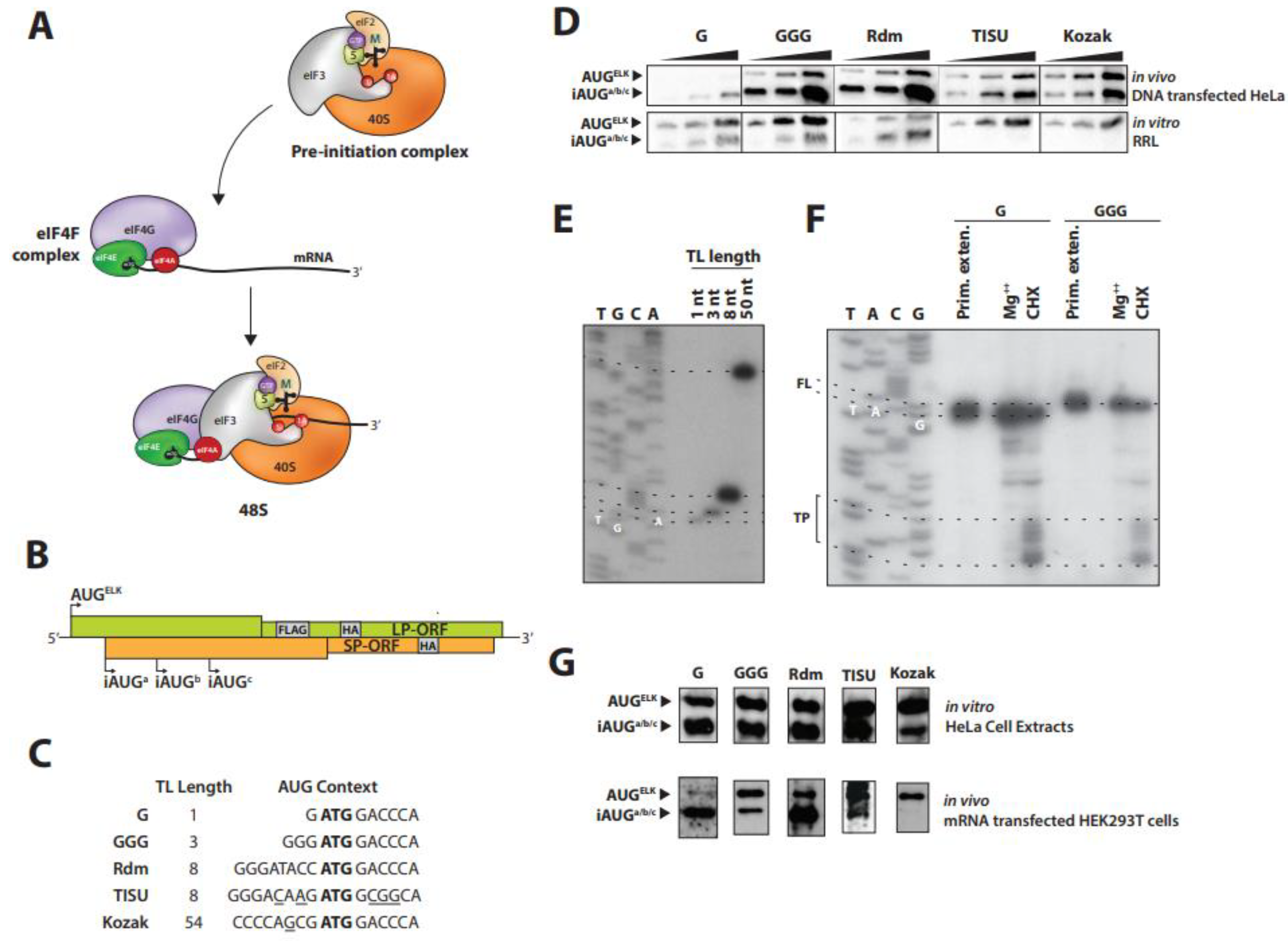
2.1. Translation Initiation Can Occur on Very Short 5′ TLs
2.2. TISU Reduces Leaky Scanning on a Short 5′ TL
2.3. Translation from a Short 5′ TL Is Cap Dependent
- RRLs are a poor in vitro system to monitor 5′ cap dependence of translation, an observation already noted by a number of investigators [34].
- The expression profiles in both systems reveals no evident difference in the behavior of the short and long 5′ TLs with regards to cap-dependence. Translation from the short 5′ TL is consequently cap-dependent.
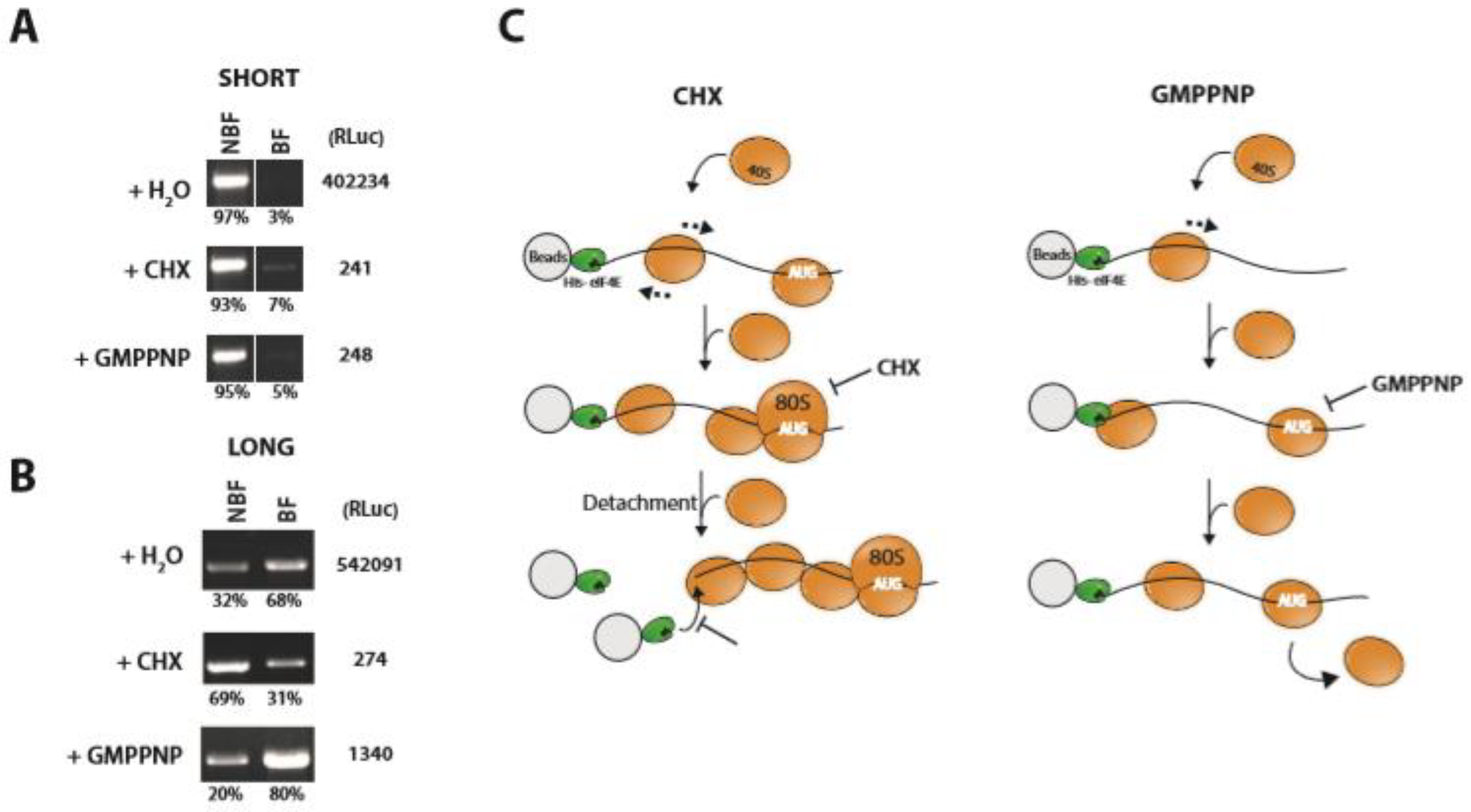
2.4. PIC Loading
2.5. Future Directions
3. Materials and Methods
- G: 5′TAATACGACTCACTATAGATGGACCCATCTGTG
- GGG: 5′TAATACGACTCACTATAGGGATGACTTCGAAAGTTTATGA
- Rdm:5′CGCTAGCTAATACGACTCACTATAGGGAGAAAATGACTTCGAAAGTTTAT
- TISU: 5′-TAATACGACTCACTATAGGGACAAGATGGCGGCATCTGTGACGCTGTGG
- Reverse: 5′-TCAGCGAGCTCTAGCATTTAGGTG
- (-VE): GAACACCACGGTAGGCTGCGAAATG
- (+VE) GATCAAAGCAATAGTTCACGCTGAAAGTGTAG
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Buttgereit, F.; Brand, M.D. A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 1995, 312, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Szamecz, B.; Rutkai, E.; Cuchalová, L.; Munzarová, V.; Herrmannová, A.; Nielsen, K.H.; Burela, L.; Hinnebusch, A.G.; Valášek, L. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRN.A. Genes Dev. 2008, 22, 2414–2425. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. MMBR 2011, 75, 434–467. [Google Scholar] [CrossRef]
- Merrick, W.C. eIF4F: A Retrospective. J. Biol. Chem. 2015, 290, 24091–24099. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987, 15, 8125–8148. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.P.; Firth, A.E.; Michel, A.M.; Atkins, J.F.; Baranov, P.V. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 2011, 39, 4220–4234. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Marintchev, A.; Kolupaeva, V.G.; Unbehaun, A.; Veryasova, T.; Lai, S.-C.; Hong, P.; Wagner, G.; Hellen, C.U.T.; Pestova, T.V. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res. 2009, 37, 5167–5182. [Google Scholar] [CrossRef]
- Weisser, M.; Voigts-Hoffmann, F.; Rabl, J.; Leibundgut, M.; Ban, N. The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A. Nat. Struct. Mol. Biol. 2013, 20, 1015–1017. [Google Scholar] [CrossRef]
- Aitken, C.E.; Lorsch, J.R. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012, 19, 568–576. [Google Scholar] [CrossRef]
- Passmore, L.A.; Schmeing, T.M.; Maag, D.; Applefield, D.J.; Acker, M.G.; Algire, M.A.; Lorsch, J.R.; Ramakrishnan, V. The Eukaryotic Translation Initiation Factors eIF1 and eIF1A Induce an Open Conformation of the 40S Ribosome. Mol. Cell 2007, 26, 41–50. [Google Scholar] [CrossRef]
- Algire, M.A.; Maag, D.; Lorsch, J.R. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell 2005, 20, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1991, 1, 111–115. [Google Scholar]
- Ingolia, N.T. Chapter 6—Genome-Wide Translational Profiling by Ribosome Footprinting. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2010; pp. 119–142. [Google Scholar]
- Pisarev, A.V.; Kolupaeva, V.G.; Yusupov, M.M.; Hellen, C.U.; Pestova, T.V. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008, 27, 1609–1621. [Google Scholar] [CrossRef] [PubMed]
- Curran, J.A.; Weiss, B. What Is the Impact of mRNA 5′ TL Heterogeneity on Translational Start Site Selection and the Mammalian Cellular Phenotype? Front. Genet. 2016, 7, 156. [Google Scholar] [CrossRef] [PubMed]
- Kawaji, H.; Kasukawa, T.; Fukuda, S.; Katayama, S.; Kai, C.; Kawai, J.; Carninci, P.; Hayashizaki, Y. CAGE Basic/Analysis Databases: The CAGE resource for comprehensive promoter analysis. Nucleic Acids Res. 2006, 34, D632–D636. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Sandelin, A.; Lenhard, B.; Katayama, S.; Shimokawa, K.; Ponjavic, J.; Semple, C.; Taylor, M.; Engström, P.; Frith, M.; et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006, 38, 626–635. [Google Scholar] [CrossRef]
- Elfakess, R.; Sinvani, H.; Haimov, O.; Svitkin, Y.; Sonenberg, N.; Dikstein, R. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res. 2011, 39, 7598–7609. [Google Scholar] [CrossRef]
- Elfakess, R.; Dikstein, R. A translation initiation element specific to mRNAs with very short 5′UTR that also regulates transcription. PLoS ONE 2008, 3, e3094. [Google Scholar] [CrossRef]
- Pestova, T.V.; Kolupaeva, V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002, 16, 2906–2922. [Google Scholar] [CrossRef]
- Kumar, P.; Hellen, C.U.; Pestova, T.V. Toward the mechanism of eIF4F-mediated ribosomal attachment to mammalian capped mRNAs. Genes Dev. 2016, 30, 1573–1588. [Google Scholar] [CrossRef]
- Dikstein, R. Transcription and translation in a package deal: The TISU paradigm. Gene 2012, 491, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Legrand, N.; Araud, T.; Conne, B.; Kuijpers, O.; Jaquier-Gubler, P.; Curran, J. An AUG codon conserved for protein function rather than translational initiation: The story of the protein sElk1. PLoS ONE 2014, 9, e102890. [Google Scholar] [CrossRef] [PubMed]
- Rahim, G.; Araud, T.; Jaquier-Gubler, P.; Curran, J. Alternative splicing within the elk-1 5′ untranslated region serves to modulate initiation events downstream of the highly conserved upstream open reading frame 2. Mol. Cell Biol. 2012, 32, 1745–1756. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Weston, B.F.; Kuzmine, I.; Martin, C.T. Positioning of the start site in the initiation of transcription by bacteriophage T7 RNA polymerase. J. Mol. Biol. 1997, 272, 21–30. [Google Scholar] [CrossRef][Green Version]
- Fuerst, T.R.; Earl, P.L.; Moss, B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol. Cell Biol. 1987, 7, 2538–2544. [Google Scholar]
- Dai, A.; Cao, S.; Dhungel, P.; Luan, Y.; Liu, Y.; Xie, Z.; Yang, Z. Ribosome Profiling Reveals Translational Upregulation of Cellular Oxidative Phosphorylation mRNAs during Vaccinia Virus-Induced Host Shutoff. J. Virol. 2017, 91, e01858-16. [Google Scholar] [CrossRef]
- Szczerba, M.; Subramanian, S.; Trainor, K.; McCaughan, M.; Kibler, K.V.; Jacobs, B.L. Small Hero with Great Powers: Vaccinia Virus E3 Protein and Evasion of the Type I IFN Response. Biomedicines 2022, 10, 235. [Google Scholar] [CrossRef]
- Elroy-Stein, O.; Moss, B. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc. Natl. Acad. Sci. USA 1990, 87, 6743–6747. [Google Scholar] [CrossRef]
- Shenvi, C.L.; Dong, K.C.; Friedman, E.M.; Hanson, J.A.; Cate, J.H. Accessibility of 18S rRNA in human 40S subunits and 80S ribosomes at physiological magnesium ion concentrations--implications for the study of ribosome dynamics. RNA 2005, 11, 1898–1908. [Google Scholar] [CrossRef]
- ten Asbroek, A.L.; van Groenigen, M.; Nooij, M.; Baas, F. The involvement of human ribonucleases H1 and H2 in the variation of response of cells to antisense phosphorothioate oligonucleotides. Eur. J. Biochem. 2002, 269, 583–592. [Google Scholar] [CrossRef]
- Genolet, R.; Rahim, G.; Gubler-Jaquier, P.; Curran, J. The translational response of the human mdm2 gene in HEK293T cells exposed to rapamycin: A role for the 5′-UTRs. Nucleic Acids Res. 2011, 39, 989–1003. [Google Scholar] [CrossRef] [PubMed]
- Andreev, D.E.; Dmitriev, S.E.; Terenin, I.M.; Prassolov, V.S.; Merrick, W.C.; Shatsky, I.N. Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res. 2009, 37, 6135–6147. [Google Scholar] [CrossRef] [PubMed]
- Svitkin, Y.V.; Ovchinnikov, L.P.; Dreyfuss, G.; Sonenberg, N. General RNA binding proteins render translation cap dependent. EMBO J. 1996, 15, 7147–7155. [Google Scholar] [CrossRef]
- Berthelot, K.; Muldoon, M.; Rajkowitsch, L.; Hughes, J.; McCarthy, J.E. Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol. Microbiol. 2004, 51, 987–1001. [Google Scholar] [CrossRef] [PubMed]
- Abaeva, I.S.; Pestova, T.V.; Hellen, C.U. Attachment of ribosomal complexes and retrograde scanning during initiation on the Halastavi árva virus IRE.S. Nucleic Acids Res. 2016, 44, 2362–2377. [Google Scholar] [CrossRef]
- Haimov, O.; Sinvani, H.; Martin, F.; Ulitsky, I.; Emmanuel, R.; Tamarkin-Ben-Harush, A.; Vardy, A.; Dikstein, R. Efficient and Accurate Translation Initiation Directed by TISU Involves RPS3 and RPS10e Binding and Differential Eukaryotic Initiation Factor 1A Regulation. Mol. Cell Biol. 2017, 37, e00150-17. [Google Scholar] [CrossRef]
- Sinvani, H.; Haimov, O.; Svitkin, Y.; Sonenberg, N.; Tamarkin-Ben-Harush, A.; Viollet, B.; Dikstein, R. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab. 2015, 21, 479–492. [Google Scholar] [CrossRef]
- DiTursi, M.K.; Cha, J.; Newman, M.R.; Dordick, J.S. Simultaneous in vitro protein synthesis using solid-phase DNA template. Biotechnol. Prog. 2004, 20, 1705–1709. [Google Scholar] [CrossRef]
- Ivanov, I.P.; Shin, B.-S.; Loughran, G.; Tzani, I.; Young-Baird, S.K.; Cao, C.; Atkins, J.F.; Dever, T.E. Polyamine Control of Translation Elongation Regulates Start Site Selection on Antizyme Inhibitor mRNA via Ribosome Queuing. Mol. Cell 2018, 70, 254–264.e6. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, J.; Lewdorowicz, M.; Zuberek, J.; Grudzien-Nogalska, E.; Bojarska, E.; Stepinski, J.; Rhoads, R.E.; Darzynkiewicz, E.; Davis, R.E.; Jemielity, J. Synthesis and characterization of mRNA cap analogs containing phosphorothioate substitutions that bind tightly to eIF4E and are resistant to the decapping pyrophosphatase DcpS. RNA 2008, 14, 1119–1131. [Google Scholar] [CrossRef]
- Bednarek, S.; Madan, V.; Sikorski, P.J.; Bartenschlager, R.; Kowalska, J.; Jemielity, J. mRNAs biotinylated within the 5′ cap and protected against decapping: New tools to capture RNA–protein complexes. Philos. Trans. B 2018, 373, 20180167. [Google Scholar] [CrossRef] [PubMed]
- de Breyne, S.; Monney, R.S.; Curran, J. Proteolytic Processing and Translation Initiation: Two Independent Mechanisms for The Expression of The Sendai Virus Y Proteins. J. Biol. Chem. 2004, 279, 16571–16580. [Google Scholar] [CrossRef] [PubMed]
- Terenin, I.M.; Andreev, D.E.; Dmitriev, S.E.; Shatsky, I.N. A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent. Nucleic Acids Res. 2013, 41, 1807–1816. [Google Scholar] [CrossRef] [PubMed]

| LEAKINESS | ||||
|---|---|---|---|---|
| VacT7/HeLa | mRNA/HEK293T | RRL | HeLa Extract | |
| G | 95 | 95 | 36 | 68 |
| GGG | 30 | 30 | 22 | 43 |
| Rdm | 85 | 85 | 22 | 32 |
| TISU | 34 | 34 | 0 | 19 |
| Kozak | 5 | 5 | 0 | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weiss, B.; Jaquier-Gubler, P.; Curran, J.A. Preinitiation Complex Loading onto mRNAs with Long versus Short 5′ TLs. Int. J. Mol. Sci. 2022, 23, 13369. https://doi.org/10.3390/ijms232113369
Weiss B, Jaquier-Gubler P, Curran JA. Preinitiation Complex Loading onto mRNAs with Long versus Short 5′ TLs. International Journal of Molecular Sciences. 2022; 23(21):13369. https://doi.org/10.3390/ijms232113369
Chicago/Turabian StyleWeiss, Benjamin, Pascale Jaquier-Gubler, and Joseph Alphonsus Curran. 2022. "Preinitiation Complex Loading onto mRNAs with Long versus Short 5′ TLs" International Journal of Molecular Sciences 23, no. 21: 13369. https://doi.org/10.3390/ijms232113369
APA StyleWeiss, B., Jaquier-Gubler, P., & Curran, J. A. (2022). Preinitiation Complex Loading onto mRNAs with Long versus Short 5′ TLs. International Journal of Molecular Sciences, 23(21), 13369. https://doi.org/10.3390/ijms232113369






