Detrimental Role of Nerve Injury-Induced Protein 1 in Myeloid Cells under Intestinal Inflammatory Conditions
Abstract
1. Introduction
2. Results
2.1. Ninj1 Expression Increases under Intestinal Inflammatory Conditions
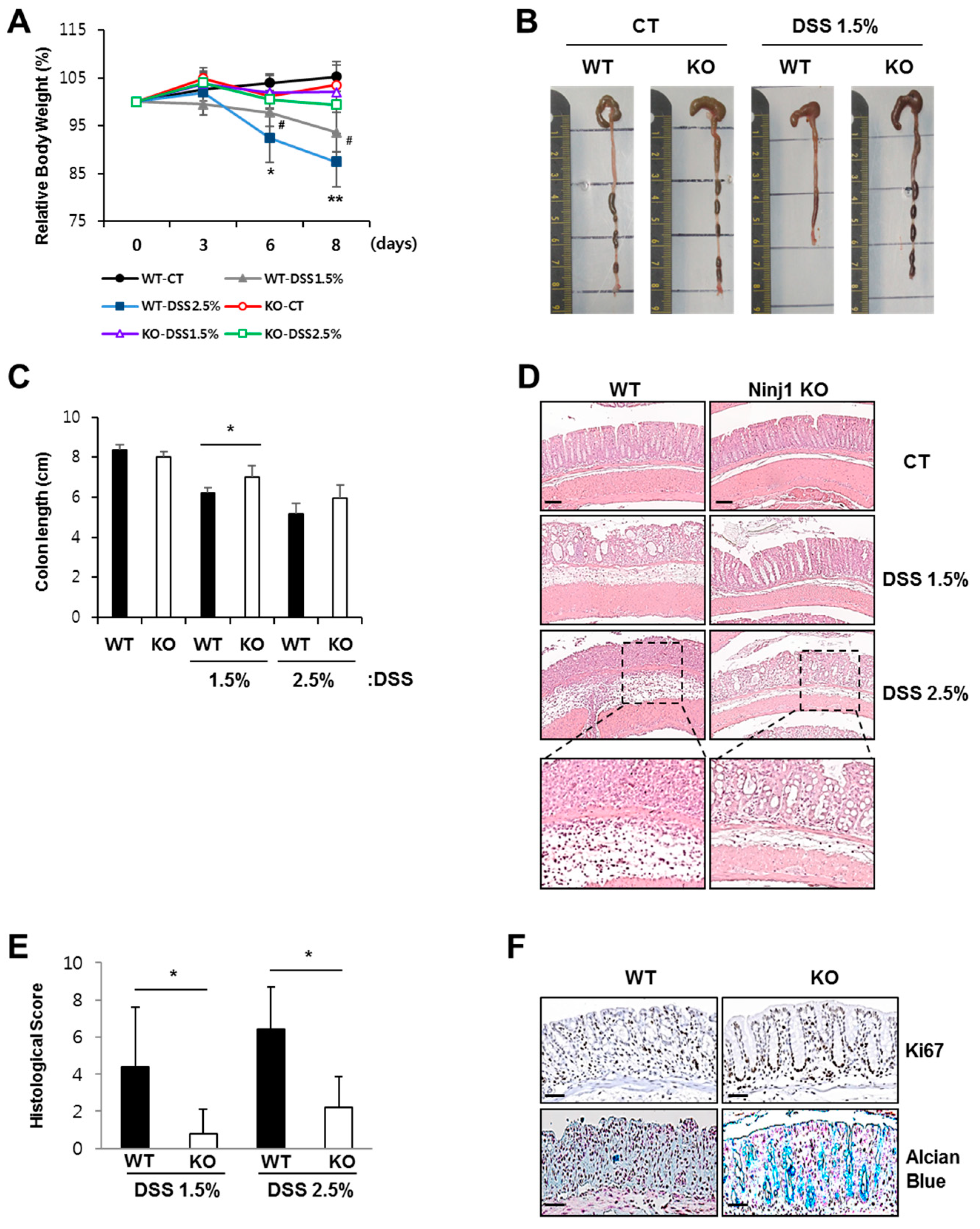
2.2. Ninj1 Deficiency Alleviates Experimental Colitis
2.3. Macrophages during Colitis Development Show Increased Expression of Ninj1
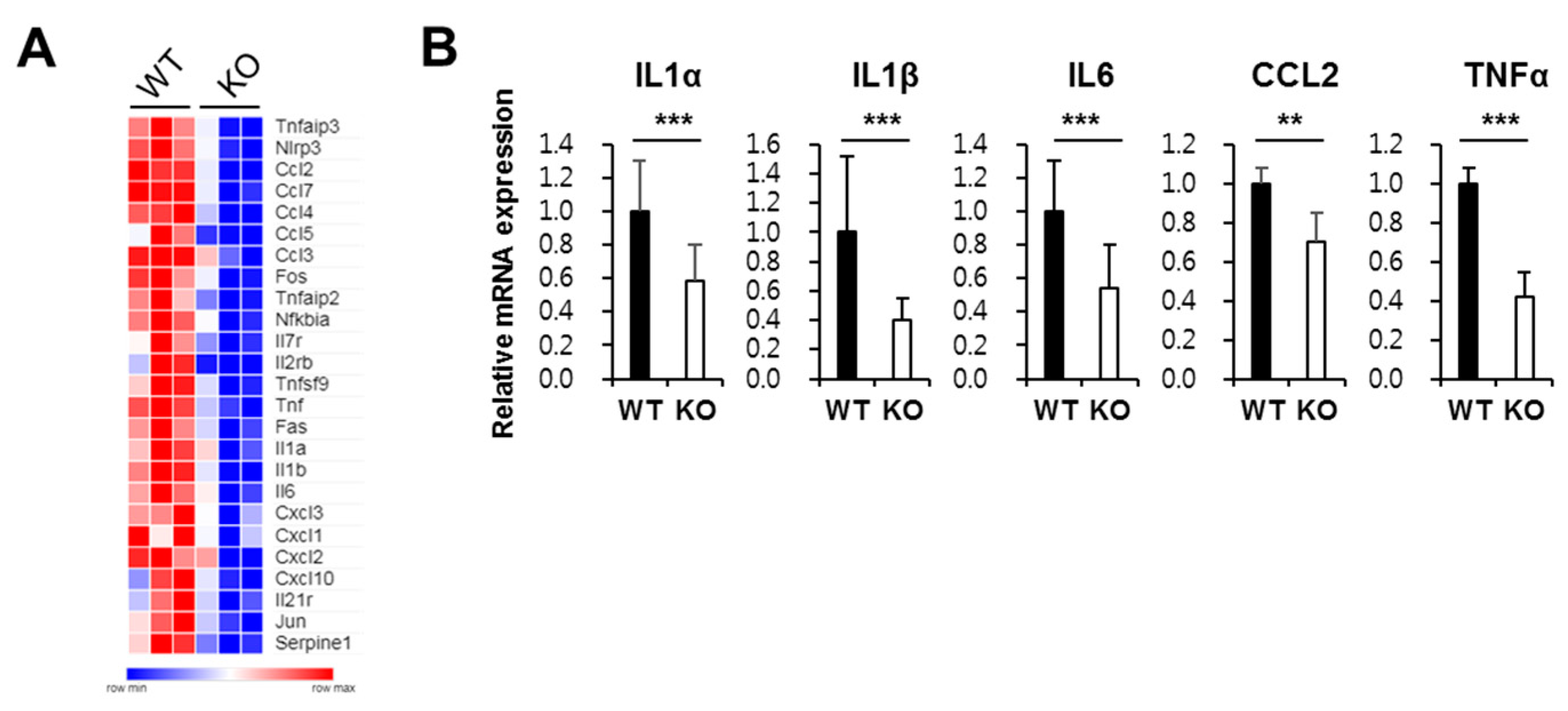
2.4. Ninj1 in Macrophages Enhances Production of Cytokines Modulating Colon Inflammation
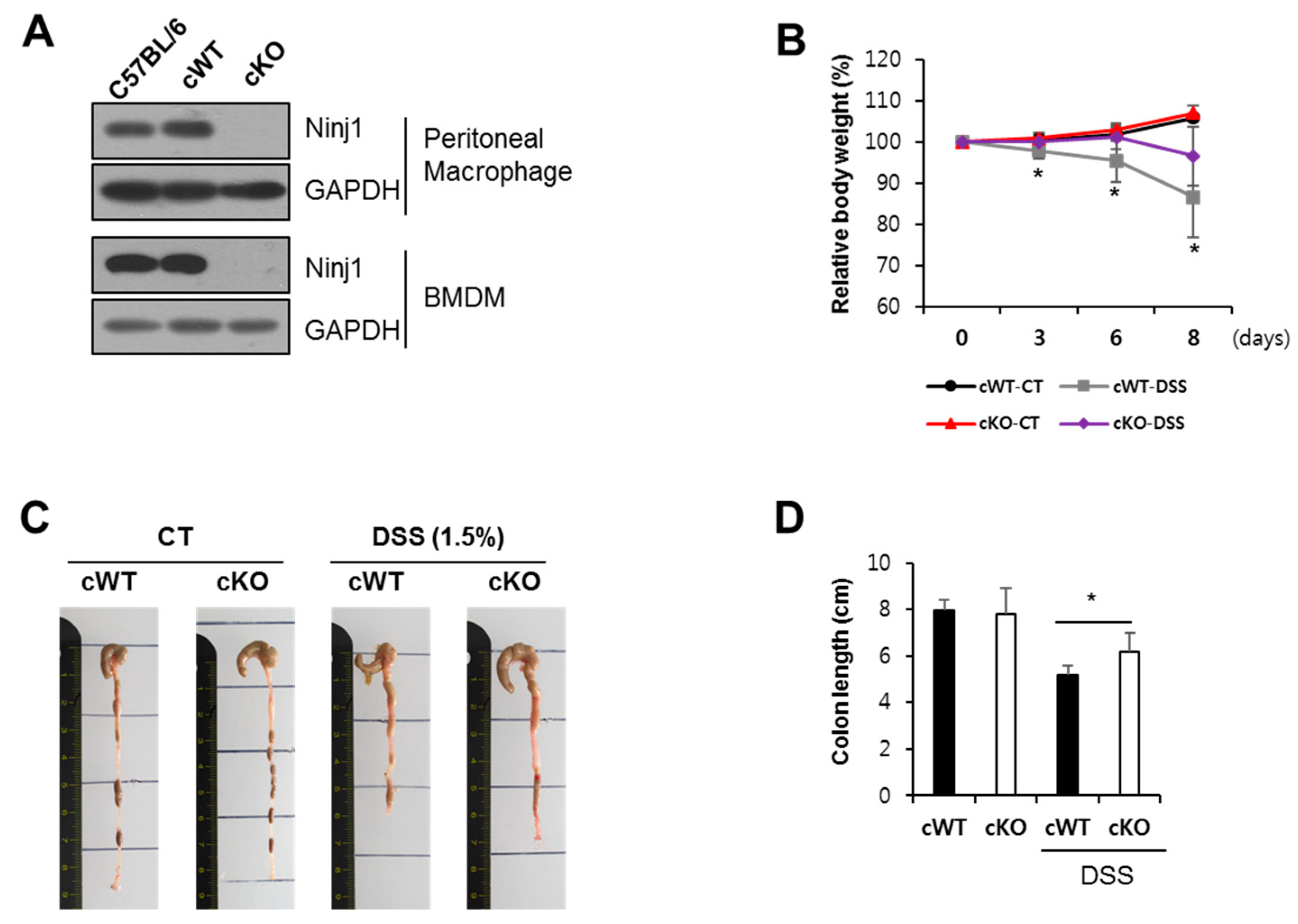
2.5. Ninj1 Deficiency in Myeloid Cells Decreases Susceptibility to Experimental Colitis
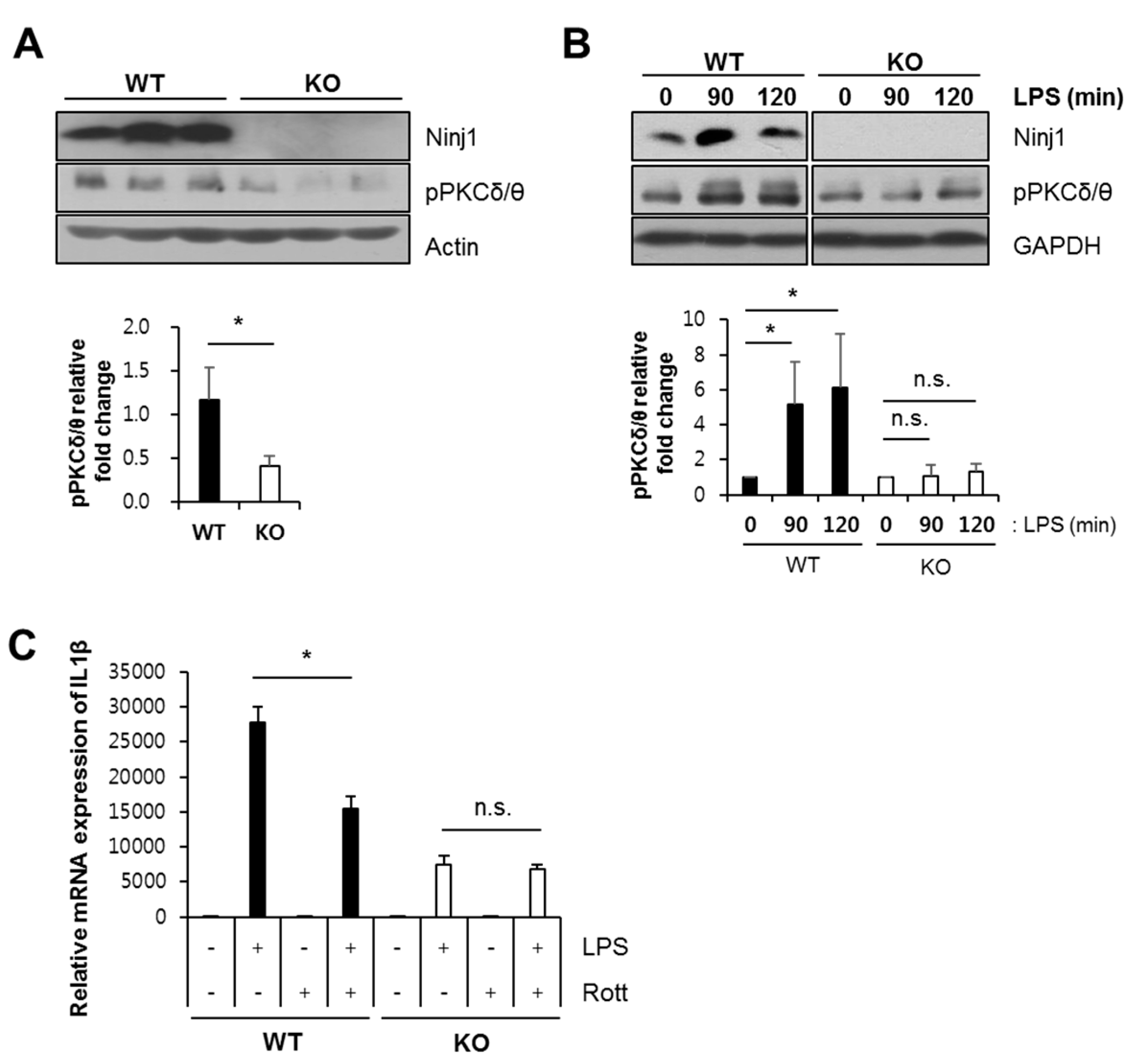
2.6. Ninj1 Modulates PKCδ/θ Activation
3. Discussion
4. Materials and Methods
4.1. Mice and Experimental Colitis
4.2. Cell Lines
4.3. Immunoblotting
4.4. Assessment of Inflammation in Colon
4.5. Immunofluorescence
4.6. Immunohistochemistry
4.7. Purification and Activation of Lymphocytes
4.8. RNA Isolation and Reverse Transcription–Polymerase Chain Reaction (RT-PCR)/Real-Time Quantitative PCR (qRT-PCR) Analysis
4.9. Cell Isolation from Colons
4.10. Flow Cytometric Analysis
4.11. Peritoneal Macrophage Isolation
4.12. Bone Marrow-Derived Macrophage (BMDM) Isolation and Culture
4.13. Microarray Analysis
4.14. Colon Explant and ELISA
4.15. Analysis of Tight Junction Protein Complex in Vitro
4.16. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Araki, T.; Milbrandt, J. Ninjurin, a novel adhesion molecule, is induced by nerve injury and promotes axonal growth. Neuron 1996, 17, 353–361. [Google Scholar] [CrossRef]
- Ahn, B.J.; Le, H.; Shin, M.W.; Bae, S.J.; Lee, E.J.; Wee, H.J.; Cha, J.H.; Lee, H.J.; Lee, H.S.; Kim, J.H.; et al. Ninjurin1 deficiency attenuates susceptibility of experimental autoimmune encephalomyelitis in mice. J. Biol. Chem. 2014, 289, 3328–3338. [Google Scholar] [CrossRef] [PubMed]
- Ifergan, I.; Kebir, H.; Terouz, S.; Alvarez, J.I.; Lecuyer, M.A.; Gendron, S.; Bourbonniere, L.; Dunay, I.R.; Bouthillier, A.; Moumdjian, R.; et al. Role of Ninjurin-1 in the migration of myeloid cells to central nervous system inflammatory lesions. Ann. Neurol. 2011, 70, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Jennewein, C.; Sowa, R.; Faber, A.C.; Dildey, M.; von Knethen, A.; Meybohm, P.; Scheller, B.; Drose, S.; Zacharowski, K. Contribution of Ninjurin1 to Toll-like receptor 4 signaling and systemic inflammation. Am. J. Respir Cell Mol. Biol. 2015, 53, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Woo, J.K.; Jang, Y.-S.; Kang, J.-H.; Hwang, J.-I.; Seong, J.K.; Yoon, Y.S.; Oh, S.H. Ninjurin1 Plays a Crucial Role in Pulmonary Fibrosis by Promoting Interaction between Macrophages and Alveolar Epithelial Cells. Sci. Rep. 2018, 8, 17542. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wang, Y.; Zhang, Z.; He, L.; Zhu, J.; Zhang, M.; He, X.; Cheng, Z.; Ao, Q.; Cao, Y.; et al. Chop Deficiency Protects Mice Against Bleomycin-induced Pulmonary Fibrosis by Attenuating M2 Macrophage Production. Mol. Ther. 2016, 24, 915–925. [Google Scholar] [CrossRef]
- Wynn, T.A.; Vannella, K.M. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity 2016, 44, 450–462. [Google Scholar] [CrossRef]
- Xu, X.R.; Liu, C.Q.; Feng, B.S.; Liu, Z.J. Dysregulation of mucosal immune response in pathogenesis of inflammatory bowel disease. World J. Gastroenterol. 2014, 20, 3255–3264. [Google Scholar] [CrossRef]
- Tamoutounour, S.; Henri, S.; Lelouard, H.; de Bovis, B.; de Haar, C.; van der Woude, C.J.; Woltman, A.M.; Reyal, Y.; Bonnet, D.; Sichien, D.; et al. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur. J. Immunol. 2012, 42, 3150–3166. [Google Scholar] [CrossRef]
- Lv, Z.; Wang, Z.; Luo, L.; Chen, Y.; Han, G.; Wang, R.; Xiao, H.; Li, X.; Hou, C.; Feng, J.; et al. Spliceosome protein Eftud2 promotes colitis-associated tumorigenesis by modulating inflammatory response of macrophage. Mucosal Immunol. 2019, 12, 1164–1173. [Google Scholar] [CrossRef]
- Wang, S.; Ye, Q.; Zeng, X.; Qiao, S. Functions of Macrophages in the Maintenance of Intestinal Homeostasis. J. Immunol Res. 2019, 2019, 1512969. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zikusoka, M.; Trindade, A.; Dassopoulos, T.; Harris, M.L.; Bayless, T.M.; Brant, S.R.; Chakravarti, S.; Kwon, J.H. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology 2008, 135, 1624–1635. [Google Scholar] [CrossRef] [PubMed]
- Deusch, K.; Reich, K. Immunological aspects of inflammatory bowel disease. Endoscopy 1992, 24, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Arango Duque, G.; Descoteaux, A. Macrophage cytokines: Involvement in immunity and infectious diseases. Front. Immunol. 2014, 5, 491. [Google Scholar] [CrossRef] [PubMed]
- Mazzucchelli, L.; Hauser, C.; Zgraggen, K.; Wagner, H.E.; Hess, M.W.; Laissue, J.A.; Mueller, C. Differential in situ expression of the genes encoding the chemokines MCP-1 and RANTES in human inflammatory bowel disease. J. Pathol. 1996, 178, 201–206. [Google Scholar] [CrossRef]
- Ludwiczek, O.; Vannier, E.; Borggraefe, I.; Kaser, A.; Siegmund, B.; Dinarello, C.A.; Tilg, H. Imbalance between interleukin-1 agonists and antagonists: Relationship to severity of inflammatory bowel disease. Clin. Exp. Immunol. 2004, 138, 323–329. [Google Scholar] [CrossRef]
- Cominelli, F.; Nast, C.C.; Clark, B.D.; Schindler, R.; Lierena, R.; Eysselein, V.E.; Thompson, R.C.; Dinarello, C.A. Interleukin 1 (IL-1) gene expression, synthesis, and effect of specific IL-1 receptor blockade in rabbit immune complex colitis. J. Clin. Invest. 1990, 86, 972–980. [Google Scholar] [CrossRef]
- Su, D.; Coudriet, G.M.; Hyun Kim, D.; Lu, Y.; Perdomo, G.; Qu, S.; Slusher, S.; Tse, H.M.; Piganelli, J.; Giannoukakis, N.; et al. FoxO1 links insulin resistance to proinflammatory cytokine IL-1beta production in macrophages. Diabetes 2009, 58, 2624–2633. [Google Scholar] [CrossRef]
- Li, S.; Gallup, M.; Chen, Y.T.; McNamara, N.A. Molecular mechanism of proinflammatory cytokine-mediated squamous metaplasia in human corneal epithelial cells. Invest. Ophthalmol. Vis. Sci. 2010, 51, 2466–2475. [Google Scholar] [CrossRef]
- Xu, J.; Zhou, L.; Ji, L.; Chen, F.; Fortmann, K.; Zhang, K.; Liu, Q.; Li, K.; Wang, W.; Wang, H.; et al. The REGgamma-proteasome forms a regulatory circuit with IkappaBvarepsilon and NFkappaB in experimental colitis. Nat. Commun. 2016, 7, 10761. [Google Scholar] [CrossRef]
- Cuschieri, J.; Billigren, J.; Maier, R.V. Endotoxin tolerance attenuates LPS-induced TLR4 mobilization to lipid rafts: A condition reversed by PKC activation. J. Leukoc. Biol. 2006, 80, 1289–1297. [Google Scholar] [CrossRef] [PubMed]
- Koff, J.L.; Shao, M.X.; Kim, S.; Ueki, I.F.; Nadel, J.A. Pseudomonas lipopolysaccharide accelerates wound repair via activation of a novel epithelial cell signaling cascade. J. Immunol. 2006, 177, 8693–8700. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Chen, L.Y.; Doerner, A.M.; Pan, W.W.; Smith, L.; Huang, S.; Papadimos, T.J.; Pan, Z.K. An atypical protein kinase C (PKC zeta) plays a critical role in lipopolysaccharide-activated NF-kappa B in human peripheral blood monocytes and macrophages. J. Immunol. 2009, 182, 5810–5815. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Jang, J.H.; Lin, H.; Choi, S.W.; Kim, H.R.; Shin, D.H.; Nam, J.H.; Zhang, Y.H.; Kim, S.J. Rise and Fall of Kir2.2 Current by TLR4 Signaling in Human Monocytes: PKC-Dependent Trafficking and PI3K-Mediated PIP2 Decrease. J. Immunol. 2015, 195, 3345–3354. [Google Scholar] [CrossRef] [PubMed]
- Slomiany, B.L.; Slomiany, A. Helicobacter pylori LPS-induced gastric mucosal spleen tyrosine kinase (Syk) recruitment to TLR4 and activation occurs with the involvement of protein kinase Cdelta. Inflammopharmacology 2018, 26, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, K.H.; Pandey, R.K.; Dahiya, Y.; Sodhi, A. Protein kinase Cdelta and protein tyrosine kinase regulate peptidoglycan-induced nuclear factor-kappaB activation and inducible nitric oxide synthase expression in mouse peritoneal macrophages in vitro. Mol. Immunol. 2010, 47, 861–870. [Google Scholar] [CrossRef] [PubMed]
- Kontny, E.; Kurowska, M.; Szczepanska, K.; Maslinski, W. Rottlerin, a PKC isozyme-selective inhibitor, affects signaling events and cytokine production in human monocytes. J. Leukoc. Biol. 2000, 67, 249–258. [Google Scholar] [CrossRef]
- Kilpatrick, L.E.; Standage, S.W.; Li, H.; Raj, N.R.; Korchak, H.M.; Wolfson, M.R.; Deutschman, C.S. Protection against sepsis-induced lung injury by selective inhibition of protein kinase C-delta (delta-PKC). J. Leukoc. Biol. 2011, 89, 3–10. [Google Scholar] [CrossRef]
- Shukla, A.; Lounsbury, K.M.; Barrett, T.F.; Gell, J.; Rincon, M.; Butnor, K.J.; Taatjes, D.J.; Davis, G.S.; Vacek, P.; Nakayama, K.I.; et al. Asbestos-induced peribronchiolar cell proliferation and cytokine production are attenuated in lungs of protein kinase C-delta knockout mice. Am. J. Pathol. 2007, 170, 140–151. [Google Scholar] [CrossRef]
- Zanin-Zhorov, A.; Ding, Y.; Kumari, S.; Attur, M.; Hippen, K.L.; Brown, M.; Blazar, B.R.; Abramson, S.B.; Lafaille, J.J.; Dustin, M.L. Protein kinase C-theta mediates negative feedback on regulatory T cell function. Science 2010, 328, 372–376. [Google Scholar] [CrossRef]
- Costello, C.M.; Mah, N.; Hasler, R.; Rosenstiel, P.; Waetzig, G.H.; Hahn, A.; Lu, T.; Gurbuz, Y.; Nikolaus, S.; Albrecht, M.; et al. Dissection of the inflammatory bowel disease transcriptome using genome-wide cDNA microarrays. PLoS Med. 2005, 2, e199. [Google Scholar] [CrossRef] [PubMed]
- Fang, K.; Bruce, M.; Pattillo, C.B.; Zhang, S.; Stone, R., 2nd; Clifford, J.; Kevil, C.G. Temporal genomewide expression profiling of DSS colitis reveals novel inflammatory and angiogenesis genes similar to ulcerative colitis. Physiol. Genom. 2011, 43, 43–56. [Google Scholar] [CrossRef]
- Ahn, B.J.; Lee, H.J.; Shin, M.W.; Choi, J.H.; Jeong, J.W.; Kim, K.W. Ninjurin1 is expressed in myeloid cells and mediates endothelium adhesion in the brains of EAE rats. Biochem. Biophys Res. Commun. 2009, 387, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Eissa, N.; Hussein, H.; Kermarrec, L.; Grover, J.; Metz-Boutigue, M.E.; Bernstein, C.N.; Ghia, J.E. Chromofungin Ameliorates the Progression of Colitis by Regulating Alternatively Activated Macrophages. Front. Immunol. 2017, 8, 1131. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, X.; Zheng, D.; Zhang, D.; Huang, S.; Zhang, X.; Ai, F.; Wang, X.; Ma, J.; Xiong, W.; et al. Dynamic changes of peritoneal macrophages and subpopulations during ulcerative colitis to metastasis of colorectal carcinoma in a mouse model. Inflamm. Res. 2013, 62, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.J.; Rossi, A.; Jung, Y.S.; Yan, W.; Liu, G.; Zhang, J.; Zhang, M.; Chen, X. Ninjurin1, a target of p53, regulates p53 expression and p53-dependent cell survival, senescence, and radiation-induced mortality. Proc. Natl. Acad. Sci. USA 2013, 110, 9362–9367. [Google Scholar] [CrossRef]
- Woo, J.K.; Jang, Y.S.; Kang, J.H.; Hwang, J.I.; Seong, J.K.; Lee, S.J.; Jeon, S.; Oh, G.T.; Lee, H.Y.; Oh, S.H. Ninjurin1 inhibits colitis-mediated colon cancer development and growth by suppression of macrophage infiltration through repression of FAK signaling. Oncotarget 2016, 7, 29592–29604. [Google Scholar] [CrossRef]
- Duff, M.D.; Mestre, J.; Maddali, S.; Yan, Z.P.; Stapleton, P.; Daly, J.M. Analysis of gene expression in the tumor-associated macrophage. J. Surg. Res. 2007, 142, 119–128. [Google Scholar] [CrossRef]
- Condeelis, J.; Pollard, J.W. Macrophages: Obligate partners for tumor cell migration, invasion, and metastasis. Cell 2006, 124, 263–266. [Google Scholar] [CrossRef]
- Tang, A.; Li, N.; Li, X.; Yang, H.; Wang, W.; Zhang, L.; Li, G.; Xiong, W.; Ma, J.; Shen, S. Dynamic activation of the key pathways: Linking colitis to colorectal cancer in a mouse model. Carcinogenesis 2012, 33, 1375–1383. [Google Scholar] [CrossRef]
- Yang, H.J.; Zhang, J.; Yan, W.; Cho, S.J.; Lucchesi, C.; Chen, M.; Huang, E.C.; Scoumanne, A.; Zhang, W.; Chen, X. Ninjurin 1 has two opposing functions in tumorigenesis in a p53-dependent manner. Proc. Natl. Acad. Sci. USA 2017, 114, 11500–11505. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.S.; Kang, J.H.; Woo, J.K.; Kim, H.M.; Hwang, J.I.; Lee, S.J.; Lee, H.Y.; Oh, S.H. Ninjurin1 suppresses metastatic property of lung cancer cells through inhibition of interleukin 6 signaling pathway. Int. J. Cancer 2016, 139, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Ahn, B.J.; Le, H.; Shin, M.W.; Bae, S.J.; Lee, E.J.; Lee, S.Y.; Yang, J.H.; Wee, H.J.; Cha, J.H.; Seo, J.H.; et al. Ninjurin1 enhances the basal motility and transendothelial migration of immune cells by inducing protrusive membrane dynamics. J. Biol. Chem. 2014, 289, 21926–21936. [Google Scholar] [CrossRef] [PubMed]
- Ingalls, R.R.; Heine, H.; Lien, E.; Yoshimura, A.; Golenbock, D. Lipopolysaccharide recognition, CD14, and lipopolysaccharide receptors. Infect. Dis. Clin. North. Am. 1999, 13, 341–353. [Google Scholar] [CrossRef]
- Dang, S.C.; Wang, H.; Zhang, J.X.; Cui, L.; Jiang, D.L.; Chen, R.F.; Qu, J.G.; Shen, X.Q.; Chen, M.; Gu, M. Are gastric mucosal macrophages responsible for gastric injury in acute pancreatitis? World J. Gastroenterol. 2015, 21, 2651–2657. [Google Scholar] [CrossRef]
- Burczynski, M.E.; Peterson, R.L.; Twine, N.C.; Zuberek, K.A.; Brodeur, B.J.; Casciotti, L.; Maganti, V.; Reddy, P.S.; Strahs, A.; Immermann, F.; et al. Molecular classification of Crohn’s disease and ulcerative colitis patients using transcriptional profiles in peripheral blood mononuclear cells. J. Mol. Diagn. 2006, 8, 51–61. [Google Scholar] [CrossRef]
- Zenlea, T.; Peppercorn, M.A. Immunosuppressive therapies for inflammatory bowel disease. World J. Gastroenterol. 2014, 20, 3146–3152. [Google Scholar] [CrossRef]
- Van Dullemen, H.M.; van Deventer, S.J.; Hommes, D.W.; Bijl, H.A.; Jansen, J.; Tytgat, G.N.; Woody, J. Treatment of Crohn’s disease with anti-tumor necrosis factor chimeric monoclonal antibody (cA2). Gastroenterology 1995, 109, 129–135. [Google Scholar] [CrossRef]
- Colombel, J.F.; Schwartz, D.A.; Sandborn, W.J.; Kamm, M.A.; D’Haens, G.; Rutgeerts, P.; Enns, R.; Panaccione, R.; Schreiber, S.; Li, J.; et al. Adalimumab for the treatment of fistulas in patients with Crohn’s disease. Gut 2009, 58, 940–948. [Google Scholar] [CrossRef]
- Targan, S.R.; Feagan, B.G.; Fedorak, R.N.; Lashner, B.A.; Panaccione, R.; Present, D.H.; Spehlmann, M.E.; Rutgeerts, P.J.; Tulassay, Z.; Volfova, M.; et al. Natalizumab for the treatment of active Crohn’s disease: Results of the ENCORE Trial. Gastroenterology 2007, 132, 1672–1683. [Google Scholar] [CrossRef]
- Chelakkot, C.; Ghim, J.; Ryu, S.H. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 2018, 50, 103. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; Song, K.S.; Son, Y.K.; Seong, J.K.; Kim, S.Y.; Oh, S.H. 1,7-Bis(4-hydroxyphenyl)-4-hepten-3-one from Betula platyphylla induces apoptosis by suppressing autophagy flux and activating the p38 pathway in lung cancer cells. Phytother Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; Kang, J.H.; Choi, S.; Son, Y.K.; Lee, K.R.; Seong, J.K.; Kim, S.Y.; Oh, S.H. Pharbitis Nil (PN) induces apoptosis and autophagy in lung cancer cells and autophagy inhibition enhances PN-induced apoptosis. J. Ethnopharmacol. 2017, 208, 253–263. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, H.J.; Kang, J.-H.; Pak, S.; Lee, K.; Seong, J.K.; Oh, S.H. Detrimental Role of Nerve Injury-Induced Protein 1 in Myeloid Cells under Intestinal Inflammatory Conditions. Int. J. Mol. Sci. 2020, 21, 614. https://doi.org/10.3390/ijms21020614
Jung HJ, Kang J-H, Pak S, Lee K, Seong JK, Oh SH. Detrimental Role of Nerve Injury-Induced Protein 1 in Myeloid Cells under Intestinal Inflammatory Conditions. International Journal of Molecular Sciences. 2020; 21(2):614. https://doi.org/10.3390/ijms21020614
Chicago/Turabian StyleJung, Hyun Jin, Ju-Hee Kang, Seongwon Pak, Keunwook Lee, Je Kyung Seong, and Seung Hyun Oh. 2020. "Detrimental Role of Nerve Injury-Induced Protein 1 in Myeloid Cells under Intestinal Inflammatory Conditions" International Journal of Molecular Sciences 21, no. 2: 614. https://doi.org/10.3390/ijms21020614
APA StyleJung, H. J., Kang, J.-H., Pak, S., Lee, K., Seong, J. K., & Oh, S. H. (2020). Detrimental Role of Nerve Injury-Induced Protein 1 in Myeloid Cells under Intestinal Inflammatory Conditions. International Journal of Molecular Sciences, 21(2), 614. https://doi.org/10.3390/ijms21020614




