Abstract
Sinisan has been widely used to treat depression. However, its pharmacologically-effective constituents are largely unknown, and the pharmacological effects and clinical efficacies of Sinisan-containing processed medicinal herbs may change. To address these important issues, we developed an ultra-high performance liquid chromatography coupled with electrospray ionization tandem quadrupole-time-of-flight mass spectrometry (UHPLC-Q-TOF/MS) method coupled with multiple statistical strategies to analyze the compound profiles of Sinisan, including individual herb, herb-pair, and complicated Chinese medicinal formula. As a result, 122 different constituents from individual herb, herb-pair, and complicated Chinese medicinal formula were identified totally. Through the comparison of three progressive levels, it suggests that processing herbal medicine and/or altering medicinal formula compatibility could change herbal chemical constituents, resulting in different pharmacological effects. This is also the first report that saikosaponin h/i and saikosaponin g have been identified in Sinisan.
1. Introduction
Chinese medicine processing and Chinese medicinal formula compatibility are two outstanding characteristics in the clinical applications of Chinese medicine. However, current studies often focus on the compatibility mechanism or processing mechanism alone without combining them together organically, and reports discussing Chinese medicine processing mechanisms in Chinese medicinal formulae have been rarely involved. Therefore, the selection of the processed products of Chinese herbal medicines contained in Chinese medicinal formulae has only to rely on the experiences of clinicians without sufficient basis of scientific theories.
Sinisan (SNS), an ancient well-known Chinese medicinal formula consisting of four Chinese herbal medicines—Bupleuri Radix (BR), Paeoniae Radix Alba (PRA), Aurantii Fructus Immaturus (AFI), and Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle (GRM), has been regarded as an effective anti-depression prescription according to the traditional Chinese medicine (TCM) theories of six channels and depression. SNS was initially described by Zhongjing Zhang in ‘Treatise on Febrile Diseases’ as a traditional Chinese herbal formula to cure mental disorders. It has been widely used for thousands of years, and even today it is still the fundamental and essential prescription for the treatment of depression [1,2]. However, the application of processed BR and processed PRA contained in SNS is quite controversial, which is necessary to improve our understanding whether the processing procedures has changed any chemical constituents of the herbal medicine.
At present, ultra-high performance liquid chromatography coupled with electrospray ionization tandem quadrupole/time of flight mass spectrometry (UHPLC-Q-TOF/MS) is a powerful tool for the analysis of complex samples in TCM and possesses high resolution, efficiency, and sensitivity to obtain accurate mass information [3,4,5,6]. Multivariate statistical analysis based on all the available chemical information has made the identification of potential chemical markers possible. In this report, we successfully developed an UHPLC-Q-TOF/MS method coupled with multiple statistical strategy to analyze the compound profiles of SNS.
Based on the theory of TCM, processing with vinegar can enhance the effects of coursing liver and resolving depression [7]. A previous report has suggested that vinegar-processed BR (VPBR) is more effective in the treatment of liver disorders, including hepatitis, cirrhosis, and liver cancer [8]. In this study, we creatively analyzed the compound profiles of individual herb, herb-pair, and complicated Chinese herbal formula according to their representative herbal medicine: BR, PRA, BR-PRA herb-pair, and SNS, respectively, and also systematically compared the changes of chemical constituents of BR, PRA, BR-PRA herb-pair, and SNS before and after processing to reveal the scientific connotation of processing and formula compatibility. We are looking forward to seeking out the common mechanism of processing and formula compatibility of Chinese herbal medicine in order to provide scientific theory for safe clinical applications of Chinese medicine and rational herbal medicine processing in Chinese medicinal formula.
2. Results and Discussion
2.1. Identification of Compounds
According to the previous reports [9,10,11,12], saponins, terpenoids, and flavones are the main chemical components in BR (VPBR), PRA (VPPRA), AFI, and GRM. These components easily lose a proton under mass spectrum detection, resulting in a better mass response in negative ion mode than in positive one. As shown in Table 1, 101 compounds were identified in negative ion mode and 21 compounds were identified in positive ion mode [13,14,15]. The typical total ion chromatograms (TICs) of BR, VPBR, PRA, vinegar-processed PRA (VPPRA), BR-PRA herb-pair, VPBR-VPPRA herb-pair, SNS, and SNS-containing VPBR and VPPRA in both ion modes are shown in Figure 1.

Table 1.
Identification of chemical compounds by ultrahigh performance liquid chromatography coupled with electrospray ionization tandem quadrupole-time-of-flight mass spectrometry (UHPLC-Q-TOF-MS/MS).
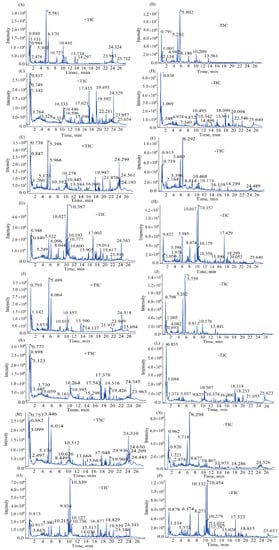
Figure 1.
Typical total ion chromatograms (TICs) in positive ion mode of PRA (A), VPPRA (I), BR (C), VPBR (K), BR-PRA herb-pair (E), VPBR-VPPRA herb-pair (M), SNS (G), and SNS-containing VPBR and VPPRA (O). Typical total ion chromatograms (TICs) in negative ion mode of PRA (B), VPPRA (J), BR (D), VPBR (L), BR-PRA herb-pair (F), VPBR-VPPRA herb-pair (N), SNS (H), and SNS-containing VPBR and VPPRA (P).
2.2. Multivariate Data Analysis
Using MarkerViewTM 1.2.1 data handling software, multivariate data analysis were completed. The principal component analysis (PCA) score plot in negative and positive ion modes were shown in Figure 2. The results showed that all crude and processed samples including individual herb, herb-pair, and complicated Chinese medicinal formula were successfully classified into two categories in both positive and negative ion modes.
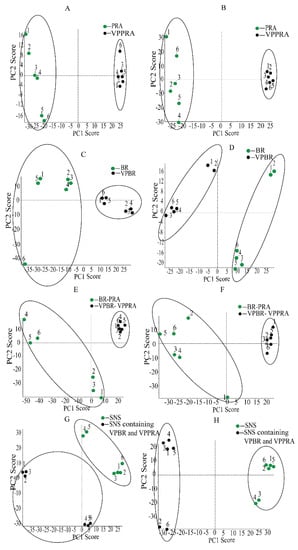
Figure 2.
Principal component analysis (PCA) score plots in positive ion mode of PRA and VPPRA (A), BR and VPBR (C), BR-PRA herb-pair and VPBR-VPPRA herb-pair (E), SNS and SNS containing VPBR and VPPRA (G). PCA score plots in negative ion mode of PRA and VPPRA (B), BR and VPBR (D), BR-PRA herb-pair and VPBR-VPPRA herb-pair (F), and SNS and SNS-containing VPBR and VPPRA (H).
2.3. Compounds Changed after Processing and Formula Compatibility
The variations of components (p < 0.05) in the individual herb, herb-pair, and complicated Chinese herbal formula before and after processing were shown in Table 2 and Table 3. For BR, 22 peaks were shown significant differences after processing. Comparing with BR, the intensity of seven peaks increased in VPBR; the other 15 peaks declined in VPBR. Taking compatibility into consideration, it was interesting to find that 14 peaks contributing to differentiate crude and processed individual herbs disappeared in herb-pair, while three new peaks (isorhamnetin-3-rutinoside, HOSSd, 2′′-O-AcetylSSd) appeared. Additionally, prosaikogenin f decreased in individual herb but increased in herb-pair. Compatibility may be responsible for these changes. On the contrary, adonltol, SSh, SSi, SSg, SSb1, 3′′-O-AcetylSSa, and SSd all showed the same trend after processing of BR in the individual herb and herb-pair. Thus, it was hard to distinguish that the seven components were affected by processing, compatibility, or even their combination. Taking into further account the formula compatibility effect of AFI and GRM, eight peaks showing significant differences in herb-pair vanished in the formula, however seven new peaks (isorhamnetin, buddlejasaponin IV, acetylSSc, 4′′-O-AcetylSSa, SSe, 6′′-O-AcetylSSa, and 6′′-O-AcetylSSd) appeared. Meanwhile SSg, 3′′-O-AcetylSSa, and SSd showed the same tendency and this would result in the unidentifiable problem.

Table 2.
Results of the t-test of 26 peaks from BR showing significant difference in individual herb, herb-pair, and complicated Chinese herbal formula before and after processing (n = 6).

Table 3.
Results of t-test of 22 peaks from PRA showing significant difference in individual herb, herb-pair, and complicated Chinese herbal formula before and after processing (n = 6).
For PRA, 20 peaks showed significant differences after processing. Comparing with PRA, the intensity of 13 peaks enhanced in VPPRA, the other seven peaks decreased in VPPRA. Considering compatibility, 10 of these 20 peaks disappeared in the herb-pair, at the same time, 6-O-β-d-glucopyranosyl lactinolide and benzoylpaeoniflorin appeared. Also, cianidanol enhanced in individual herb but decreased in herb-pair. These changes perhaps resulted from compatibility. Moreover, nine peaks had the same trend after processing of PRA in individual herb and herb-pair, and it was also hard to distinguish as BR. Under further influence of formula compatibility with AFI and GRM, five peaks showing significant differences in herb-pair vanished in formula; oppositely, five new peaks (oxypaeoniflora, 6′-O-β-d-glucopyranosylalbiflorin, galloylpaeoniflorin, lactiflorin, benzoylalbiflorin) appeared. Formula compatibility may be responsible for these changes. In addition, seven peaks (6-O-β-d-glucopyranosyl lactinolide, mudanpioside f, albiflorin, isomaltoalbiflorin, paeoniflorigenone, paeonol, and benzoic acid) displayed an identical trend; this still led to the unidentifiable problem. Figure 3 shows the comparison of the contents of the components identified with significant differences. Processing with vinegar and formula compatibility can both regulate the acidity and alkalinity of the solution and promote changes in chemical composition, such as hydrolysis reaction, isomerization reaction, etc., resulting in increased or decreased dissolution of some components. Finally, we found that processing of BR and PRA also had the impact on AFI and GRM, and the results were shown in Table 4.
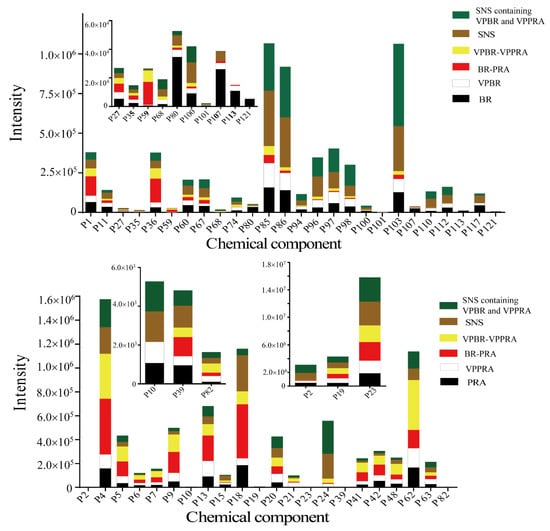
Figure 3.
Contents of components identified with significant differences in individual herb, herb-pair, and complicated Chinese herbal formula of SNS.

Table 4.
Results of t-test of 12 peaks from AFI and GRM showing significant difference (n = 6).
As shown in Table 2, the intensity of paeonol significantly increased after stir-frying with vinegar. According to a previous report [16], adding acid could greatly improve the extraction efficiency of paeonol. Since the boiling point of paeonol is 154 °C, the use of slow fire (130 °C) controlled by infrared radiation thermometer during the processing minimized the loss of paeonol. In addition, acetic acid plays an important role to form intermolecular hydrogen bonds by Van der Waals’ force with paeonol, resulting in the increase of dissolution rate. Modern researches indicate that paeonol has analgesic and antiphlogistic pharmacological activities [17,18] and is consistent with TCM theory that processing of medicinal herbs with vinegar can enhance the effects of promoting blood circulation and relieving pain. As an illustration, Figure 4 revealed the course of deducing fragmentation of paeonol.
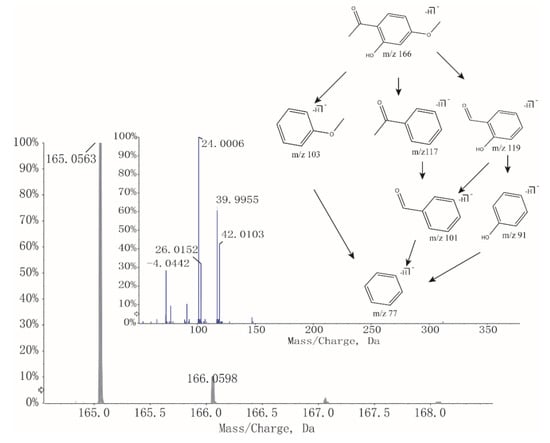
Figure 4.
MS and tandem mass spectroscopy (MS/MS) spectra and fragmentation of Paeonol.
As shown in Table 3, we found that the intensity of SSa and SSd declined but the intensity of SSb2 and SSb1 increased in the BR. SSs, a kind of oleanane type triterpenoid saponin, could be divided into seven types according to their different aglycones. SSa, SSd, and SSc are epoxy-ether saikosaponins (type I), while SSb2 and SSb1 with a different aglycone, form a heterocyclic diene saikosaponin (type II) [19]. The glycosidic bond is very easily hydrolyzed in the acidic conditions or being heated [20,21]. Vinegar processing could promote the hydrolyzation from 13 to 28 allyl oxide linkage to its corresponding heteroannular diene structure, resulting in the aglycone accumulation. As shown in Figure 5, peak No. 94 was clearly observed in VPBR, VPBR-VPPRA herb-pair, SNS-containing VPBR and VPPRA, and SNS, and almost undetectable in BR and BR-PRA herb-pair. According to the fragmentations in both positive and negative ion modes and other reports [22,23,24], we suggested that peak No. 94 is SSg. SSg in SNS could be related to the acidic compounds of herbal formula, such as glycyrrhizic acid. Also, peak No. 68 (SSh/i), as the isomer of SSc, had the same change with SSg. Based on these, we hypothesized that SSa and SSd could be transformed to SSb2, SSb1, and SSg, while SSc could be converted to SSh and SSi after processing and formula compatibility.
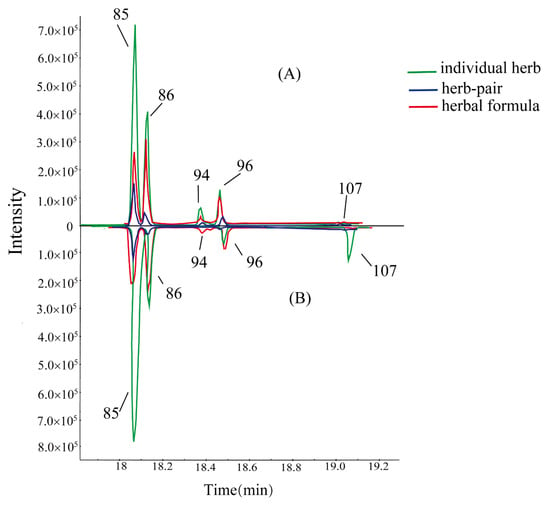
Figure 5.
Comparison on intensity of five isomers of Saikosaponins in BR, BR-PRA herb-pair, and SNS (B). Comparison on intensity of five isomers of Saikosaponins in VPBR, VPBR-VPPRA herb-pair, and SNS-containing VPBR and VPPRA (A).
3. Materials and Methods
3.1. Materials and Reagents
Acetonitrile (Merck, Darmstadt, Germany) and formic acid from Anaqua Chemical Supply (ACS, Houston, TX, USA) of HPLC/MS-grade were purchased for UHPLC-Q-TOF/MS analysis. Deionized water was prepared using a Milli-Q system (Millipore, Molsheim, France). SPE columns (LC-C18, 500 mg/mL) were purchased from ANPLE Scientific Instrument (Shanghai, China). Other reagents of analytical grade were purchased from Nanjing Chemical Reagent Co., Ltd. (Jiangsu, China).
BR, PRA, AFI, and GRM were obtained from different Chinese pharmacies and pharmaceutical factories, and authenticated by Professor Hao Cai. The quality of all collected samples was strictly evaluated and consistent with the regulations of Chinese Pharmacopoeia (Edition 2015, Part One). VPBR and VPPRA were prepared according to the processing standards described in Chinese Pharmacopoeia (Edition 2015, Part Four). The voucher specimens were deposited in School of Pharmacy, Nanjing University of Chinese Medicine (Nanjing, China).
3.2. Sample Preparation
The decoction of BR was prepared as follows. Eight grams of BR were extracted twice in a reflux water heating mantle in 48 mL and 32 mL of deionized water for 1.5 h and 1 h of reflux, respectively. The mixed solution was filtered through a four-layer mesh following the reflux. One milliliter of the solution was loaded onto a C18 RP SPE column and the gradient elution was performed as the following sequence. One milliliter of 20% acetonitrile in water (20:80, v/v), 1 mL of 40% acetonitrile in water (40:60, v/v), 1 mL of 60% acetonitrile in water (60:40, v/v), 1 mL of 80% acetonitrile in water (80:20, v/v), and 1 mL of acetonitrile. After the sequent elution, the collected eluent was eddied for 2 min and centrifuged at 13,000 rpm for 5 min. Finally, the supernatant was collected as the injection solution. The decoctions of VPBR, PRA, and VPPRA were prepared according to the same procedures above.
The decoction of BR-PRA herb-pair consisted of 4 g of BR and 4 g of PRA, and prepared as the same procedures as individual herb described above. The decoction of VPBR-VPPRA herb-pair was prepared using the same procedures as the decoction of BR-PRA herb-pair. The decoction of SNS was consist of 2 g of BR, 2 g of PRA, 2 g of AFI, and 2 g of GRM, and prepared using the same procedures as individual herb. The decoction of SNS containing VPBR and VPPRA was prepared using the same procedures as the decoction of SNS.
3.3. Chromatographic Separation
Chromatographic analysis was performed using a UHPLC system (Shimadzu, Kyoto, Japan) consisting of an LC-30AD binary pump, an autosampler (Model SIL-30SD), an online degasser (DGU-20A5R), and a temperature controller for columns (CTO-30A). Separation was carried out on an extended C18 Column (2.1 mm × 100 mm, 1.8 μm; Agilent, Palo Alto, CA, USA) at 30 °C and the flow rate was 0.3 mL/min. The optimal mobile phase consisted of A (HCOOH/H2O, 0.1:100, v/v) and B (C2H3N). The optimized UHPLC elution conditions were as follows 0–2 min, 3–15% B; 2–7 min, 15–20% B;7–8 min, 20% B; 8–9 min, 20–30% B; 9–13 min, 30–32% B; 13–21 min; 32–54% B; 21–23 min, 54–100% B; 23–27 min, 100–3% B; and 27–28 min, 3% B. The injection volume was 2 μL.
3.4. MS and MS/MS Experiments
A triple TOF 5600+ System (AB Sciex, Concord, CA, USA) equipped with an electrospray ionization (ESI) source was performed. The MS was operated in both positive and negative ion modes. Parameters were set as follows: ion spray voltage of +4500/−4500 V; turbo spray temperature of 550 °C; declustering potential (DP) of +60/−60 V; collision energy of +35/−45 V; nebulizer gas (gas 1) of 55 psi; heater gas (gas 2) of 55 psi and curtain gas of 35 psi. TOF MS and TOF MS/MS were scanned with the mass ranges of m/z 100–2000 and 50–1000, respectively. The experiments were run with 200 ms accumulation time for TOF MS and 80 ms accumulation time for TOF MS/MS. Continuous recalibration was performed at the intervals of 3 h. Dynamic background subtraction and information-dependent acquisition techniques were applied to reduce the impact of matrix interference and increase the efficiency of analysis.
3.5. MS and MS/MS Data Processing and Analysis
The raw data were obtained by the Analyst TF 1.6 software (AB Sciex, Concord, CA, USA). Before data processing, a database about chemical components of medicinal herbs in SNS, including names, molecular formulas, chemical structures, and accurate molecular weights, was established by searching relevant reported literature and database websites, including PubMed and SciFinder. The data were analyzed by using PeakViewTM 1.2 software (AB Sciex, Concord, CA, USA) for a perfect match with the information in the established database, according to fragmentations of the different peaks. The main parameters used were set as follows: retention time range of 0–28 min, mass range of 100 to 2000 Da, and mass tolerance of 10 ppm. By using the method of PCA with MarkerViewTM 1.2.1 software (AB Sciex, Concord, CA, USA) to check for outliers and variation trend, the gathered data were more intuitionistic. The Student’s t-test was performed to find out a list of peaks that were finally defined as the main contributors to the significant difference between raw and processed medicinal herbs (p < 0.05).
4. Conclusions
A total of 122 constituents had been identified by creative global analysis in individual herb, herb-pair, and complicated Chinese herbal formula of SNS. Taking BR as an example, 29 kinds of SSs had been identified, including some new discoveries in recent years, such as SSq, SSm, and so forth. Monoterpene glycosides (oxypaeoniflora, mudanpioside f, paeoniflorigenone, etc) showed a marked increase after processing of PRA. This is the first report of SSh/i and SSg being identified in SNS. Through three progressive levels of comparison, it suggests that processing herbal medicine and/or changing medicinal formula compatibility could alter herbal chemical constituents, resulting in different pharmaceutical effects. Herbal formula has always been the predicament of Chinese medicine research, and some scholars only employed SSd and paeoniflorin (the main components of BR and PRA) for research [25], whereas the effects between individual components and herbal formula containing individual components are quite different. We hope that the thoughts of this article would be some helpful for further research of herbal formula.
Author Contributions
J.Z., H.C., and Y.D. contributed to the design of the study. J.Z., S.T., Y.D., K.P., Y.X., J.L., and L.S. performed the laboratory experiments and data analysis. J.Z., H.C., Y.D., M.N., and Y.Z. wrote the manuscript. H.C., S.T., and Q.Z. revised the final manuscript. All authors reviewed the manuscript.
Funding
This research was financially supported by the National Natural Science Foundation of China (No. 81673600).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AFI | Aurantii Fructus Immaturus |
| BR | Bupleuri Radix |
| GRM | Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle |
| PRA | Paeoniae Radix Alba; SNS, Sinisan |
| SS | saikosaponin |
| TCM | traditional Chinese medicine |
| UHPLC-Q-TOF/MS | ultrahigh performance liquid chromatography coupled with electrospray ionization tandem quadrupole-time-of-flight mass spectrometry |
| VPBR | vinegar-processed Bupleuri Radix |
| VPPRA | vinegar-processed Paeoniae Radix Alba |
| PCA | principal component analysis |
References
- Shen, X.; Zhao, Z.Y.; Luo, X.; Wang, H.; Hu, B.X.; Guo, Z.H. Systems pharmacology based study of the molecular mechanism of SiNiSan formula for application in nervous and mental diseases. Evid.-Based Complement. Altern. Med. 2016, 2016, 9146378. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.D.; Tang, T.; Lin, X.P.; Yang, Z.Y.; Yang, S.; Xia, Z.A.; Wang, Y.; Zheng, P.; Wang, Y.; Zhang, C.H. Nine traditional Chinese herbal formulas for the treatment of depression: An ethnopharmacology, phytochemistry, and pharmacology review. Neuropsych. Dis. Treat 2016, 12, 2387–2402. [Google Scholar]
- Li, Y.B.; Zhang, T.J.; Zhang, X.L.; Xu, H.Y.; Liu, C.X. Chemical figerprint analysis of Phellodendri Amurensis Cortex by ultra performance LC/Q-TOF-MS methods combined with chemometrics. J. Sep. Sci. 2015, 33, 3347–3353. [Google Scholar] [CrossRef] [PubMed]
- Pei, K.; Duan, Y.; Cai, H.; Tu, S.C.; Qiao, F.X.; Song, X.Q.; Liu, X.; Cao, G.; Fan, K.L.; Cai, B.C. Ultra-high-performance liquid chromatography quadrupole/time of flight mass spectrometry combined with statistical analysis for rapidly revealing the influence of sulfur-fumigated Paeoniae Radix Alba on the chemical constituents of Si Wu Tang. Anal. Methods 2015, 7, 9442–9451. [Google Scholar] [CrossRef]
- Guo, S.; Duan, J.A.; Qian, D.W.; Wang, H.Q.; Tang, Y.P.; Qian, Y.F.; Wu, D.W.; Su, S.L.; Shang, E.X. Hydrophilic interaction ultra-high performance liquid chromatography coupled with triple quadrupole mass spectrometry for determination of nucleotides, nucleosides and nucleobases in Ziziphus plants. J. Chromatogr. A 2013, 1301, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Qi, L.W.; Li, P. Correlative analysis of metabolite profiling of Danggui Buxue Tang in rat biological fluids by rapid resolution LC-TOF/MS. J. Pharm. Biomed. 2011, 55, 146–160. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.M. Materia Medica Companion; People’s Medical Publishing House: Beijing, China, 1988. [Google Scholar]
- Chen, X.Z.; Yu, T.Y.; Chen, Z.X.; Zhao, R.Z.; Mao, S.R. Effect of saikosaponins and extracts of vinegar-baked Bupleuri Radix on the activity of β-glucuronidase. Xenobiotica 2014, 44, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Liang, Z.T.; Yi, T.; Ma, Y.; Zhao, Z.Z.; Guo, B.L.; Zhang, J.Y.; Chen, H.B. Comparison of chemical profiles between the root and aerial parts from three Bupleurum species based on a UHPLC-QTOF-MS metabolomics approach. BMC Complement. Altern. Med. 2017, 17, 305. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, L.; Fan, C.R.; Li, H.; Huang, M.Q.; Xiang, Q.; Xu, W.; Xu, W.; Chu, K.D.; Lin, Y. Qualitative and quantitative analysis of major constituents of Paeoniae Radix Alba and Paeoniae Radix Rubra by HPLC-DAD-Q-TOF-MS/MS. China J. Chin. Mater. Med. 2015, 40, 1762–1770. [Google Scholar]
- Li, P.; Zeng, S.L.; Duan, L.; Ma, X.D.; Dou, L.L.; Wang, L.J.; Li, P.; Bi, Z.M.; Liu, E.H. Comparison of Aurantii Fructus Immaturus and Aurantii Fructus based on multiple chromatographic analysis and chemometrics methods. J. Chromatogr. A 2016, 1469, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Ota, M.; Xu, F.; Li, Y.L.; Shang, M.Y.; Makino, T.; Cai, S.Q. Comparison of chemical constituents among licorice, roasted licorice, and roasted licorice with honey. J. Nat. Med. 2017, 72, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Y.; Tang, Y.Z.; Fan, C.L.; Luo, H.T.; Guo, P.R.; Chen, J.X. Identification and determination of the saikosaponins in Radix bupleuri by accelerated solvent extraction combined with rapid-resolution LC-MS. J. Sep. Sci. 2010, 33, 1933–1945. [Google Scholar] [CrossRef] [PubMed]
- Ni, F.Y.; Song, Y.L.; Liu, L.; Zhao, Y.W.; Huang, W.Z.; Wang, Z.Z.; Xiao, W. Preparation technology of isochlorogenic acids A, B, and C. Chin. Tradit. Herb. Drugs 2015, 46, 369–373. [Google Scholar]
- Zhu, M.; Duan, J.A.; Tang, Y.P.; Guo, J.M.; Shang, E.X.; Zhu, Z.H. Identification of chemical constituents in SiWu decoction by UHPLC-DAD-TOF/MS. Acta Chromatogr. 2014, 26, 517–537. [Google Scholar] [CrossRef]
- Gong, C.B.; Kang, H.X. Study on extraction of Paeonol from Cortex Moutan by steam distillation with the addition of acid and inorganic salt. J. Shandong Univ. TCM 2013, 37, 162–163. [Google Scholar]
- Jin, X.; Wang, J.; Xia, Z.M.; Shang, C.H.; Chao, Q.L.; Liu, Y.R.; Fan, H.Y.; Chen, D.Q.; Qiu, F.; Zhao, F. Anti-inflammatory and anti-oxidative activities of Paeonol and its metabolites through blocking MAPK/ERK/p38 signaling pathway. Inflammation 2016, 39, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.J.; Wang, M.X.; Xie, X.J.; Di, T.T.; Zhao, J.X.; Lin, Y.; Xu, X.L.; Li, N.F.; Zhai, Y.T.; Wang, Y.; et al. Paeonol ameliorates imiquimod-induced psoriasis-like skin lesions in BALB/c mice by inhibiting the maturation and activation of dendritic cells. Int. J. Mol. Med. 2017, 39, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.Y.; Chiou, C.Y.; Chiou, S.J. Putative genes involved in saikosaponin biosynthesis in Bupleurum species. Int. J. Mol. Sci. 2013, 14, 12806–12826. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jiang, H.; Zhang, Y.P.; Zhang, Q.; Xu, Y. Content change of saikosaponins during preparation of saiko decoction. Chin. J. Exp. Tradit. Med. Formulae 2012, 18, 155–158. [Google Scholar]
- Li, J.; Shi, R.B.; Liu, B.; Jiang, H. Influence of different component combinations of Sini Powder on decocting quantity of saikosaponin-a and b2. J. Beijing Univ. Tradit. Chin. Med. 2007, 30, 115–120. [Google Scholar]
- Li, J.; Xu, Q.; Jiang, H. Identification and characterization of two new degradation products of saikosaponin A under acid hydrolytic conditions. J. Lumin. 2016, 171, 131–137. [Google Scholar] [CrossRef]
- Yu, P.; Qiu, H.; Wang, M.; Tian, Y.; Zhang, Z.J.; Song, R. In vitro metabolism study of saikosaponin d and its derivatives in rat liver microsomes. Xenobiotica 2016, 47, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.B.; Wang, L.; Yin, L.S.; Sun, R. Research on biotransformation of saikosaponin A in vitro based on HPLC-DAD-MSn. Chin. Tradit. Herb. Drugs 2017, 48, 333–338. [Google Scholar]
- Liang, G.W.; Chen, Y.C.; Wang, Y.; Wang, H.M.; Pan, X.Y.; Chen, P.H.; Niu, Q.X. Interaction between saikosaponin D, paeoniflorin, and human serum albumin. Molecules 2018, 23, 249. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of Bupleuri Radix, Paeoniae Radix Alba, Aurantii Fructus Immaturus, and Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle are available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).