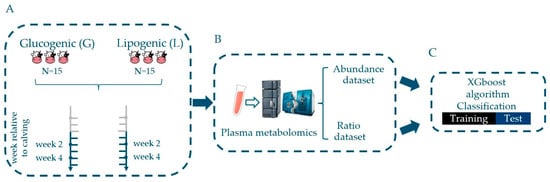
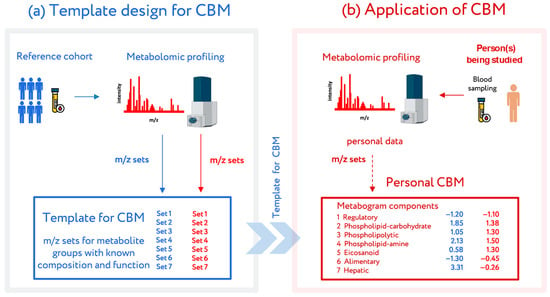
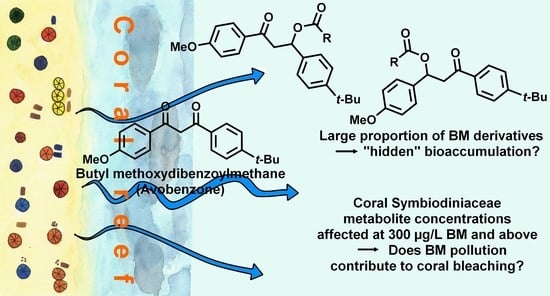
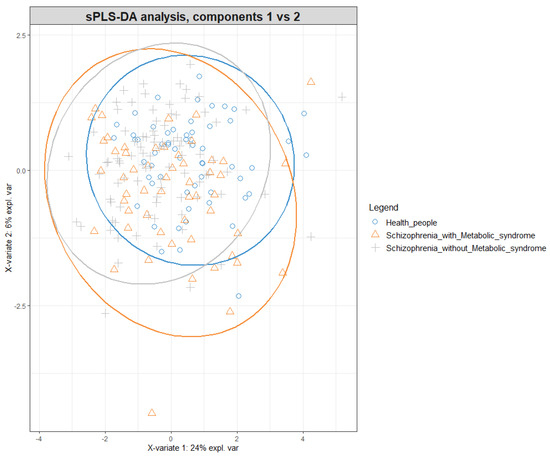
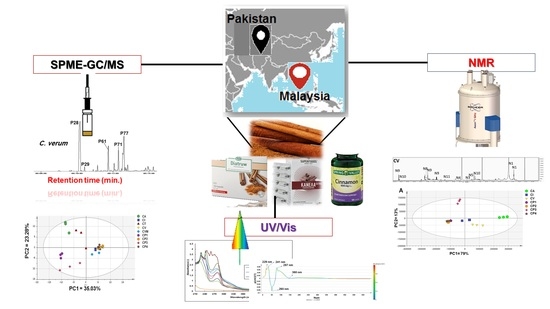
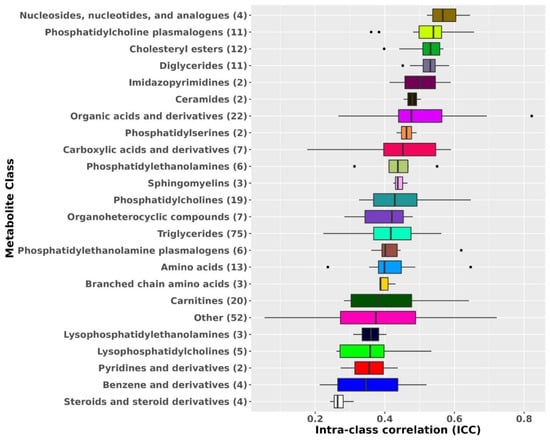
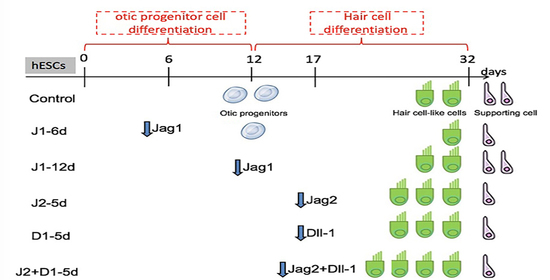
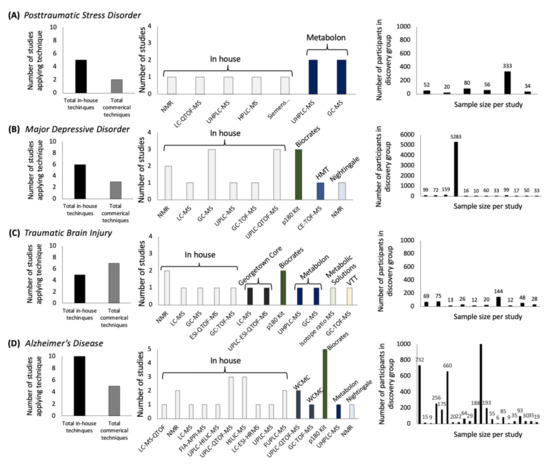
Advances in Metabolomics
A topical collection in Metabolites (ISSN 2218-1989). This collection belongs to the section "Advances in Metabolomics".
Viewed by 47293Editors
2. Department of Chemistry “Ugo Schiff”, University of Florence, 50019 Sesto Fiorentino, Italy
3. Consorzio Interuniversitario Risonanze Magnetiche MetalloProteine (CIRMMP), 50019 Sesto Fiorentino, Italy
Interests: applications of NMR-based metabolomics in biomedicine and in food science; NMR fingerprinting and profiling of biological samples; development of new analytical approaches for NMR metabolomics; development of new tools for NMR data analysis
Special Issues, Collections and Topics in MDPI journals
Interests: mathematics; statistics; biostatistics; chemometrics; comparative genomics; data analysis; data mining; mathematical models; metabolomics; multivariate analysis; genomics; proteomics; statistical analysis; transcriptomics; health; systems biology; statistical sampling techniques; metagenomics; big data
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
At present, metabolomics is a mature research field with a considerable track-record of successful applications, especially in the domains of biomedical and agricultural sciences. During the last decade, it has become a valuable instrument for providing insights into the molecular mechanisms of numerous phenomena, thus contributing to the characterization of the biological functioning of living systems. Albeit no more in its infancy stage, metabolomics still needs constant technological improvement to be exploited to its full potential. For this reason, this Topical Collection aims to publish high-quality research papers or systematic reviews discussing bleeding edge innovative approaches that will boost metabolomics to the very vanguard of its technological advancement.
We make a call for articles from all the various fields of metabolomics applications ranging from biomedicine, pharmacology, biochemistry to nutrition, food and agricultural sciences. Without restrictions to any particular field of application or analytical platform, innovative aspects can be related, for instance, to 1) pre-analytical and analytical protocols, 2) instrumental development, 3) sample collection and data acquisition strategies, 4) data analysis algorithms, tools, and software, 4) multi-omic and multi-platform studies, and 5) groundbreaking applications with a high technology readiness level.
Dr. Leonardo Tenori
Dr. Edoardo Saccenti
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Metabolites is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- metabolomics
- technological advancements
- analytical platforms
- nuclear magnetic resonance
- mass spectrometry
- clinical applications
- food science and nutrition
- metabolite annotation and quantification
- data mining and artificial intelligence
- high technology readiness level