Exploration of Blood Lipoprotein and Lipid Fraction Profiles in Healthy Subjects through Integrated Univariate, Multivariate, and Network Analysis Reveals Association of Lipase Activity and Cholesterol Esterification with Sex and Age
Abstract
1. Introduction
2. Results
2.1. Univariate Analysis: Lipoprotein and Lipid Fraction Concentrations Differ between Sexes and Age Groups
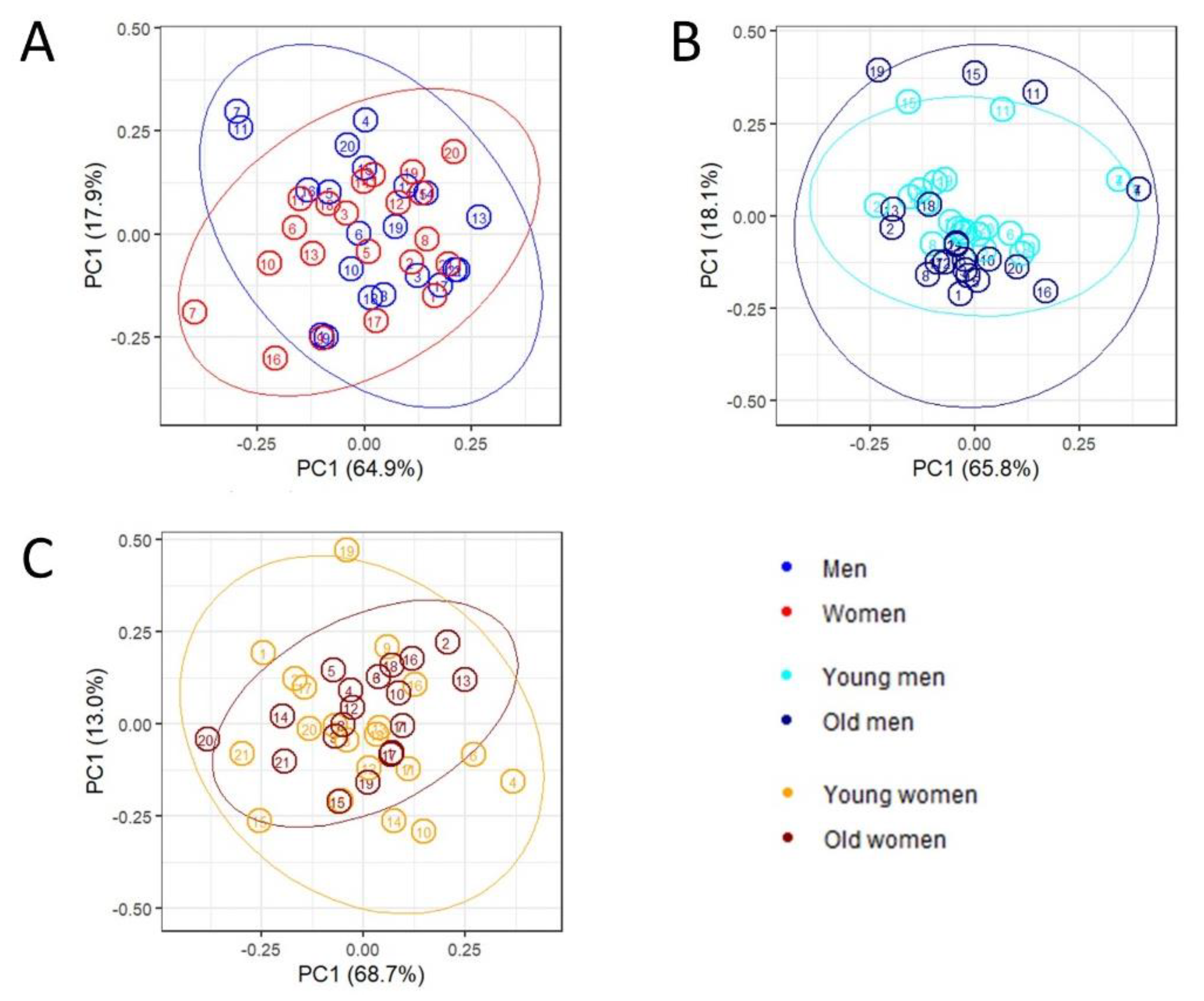
2.2. Multivariate Analysis and Predictive Modeling Indicate the Existence of Sex- and Age-Specific Lipoprotein and Lipid Fraction Profiles
2.3. Network Inference and Analysis
Exploratory Analysis of Lipoprotein and Lipid Fractions Highlights Subtle Remodulation of Correlation Patterns
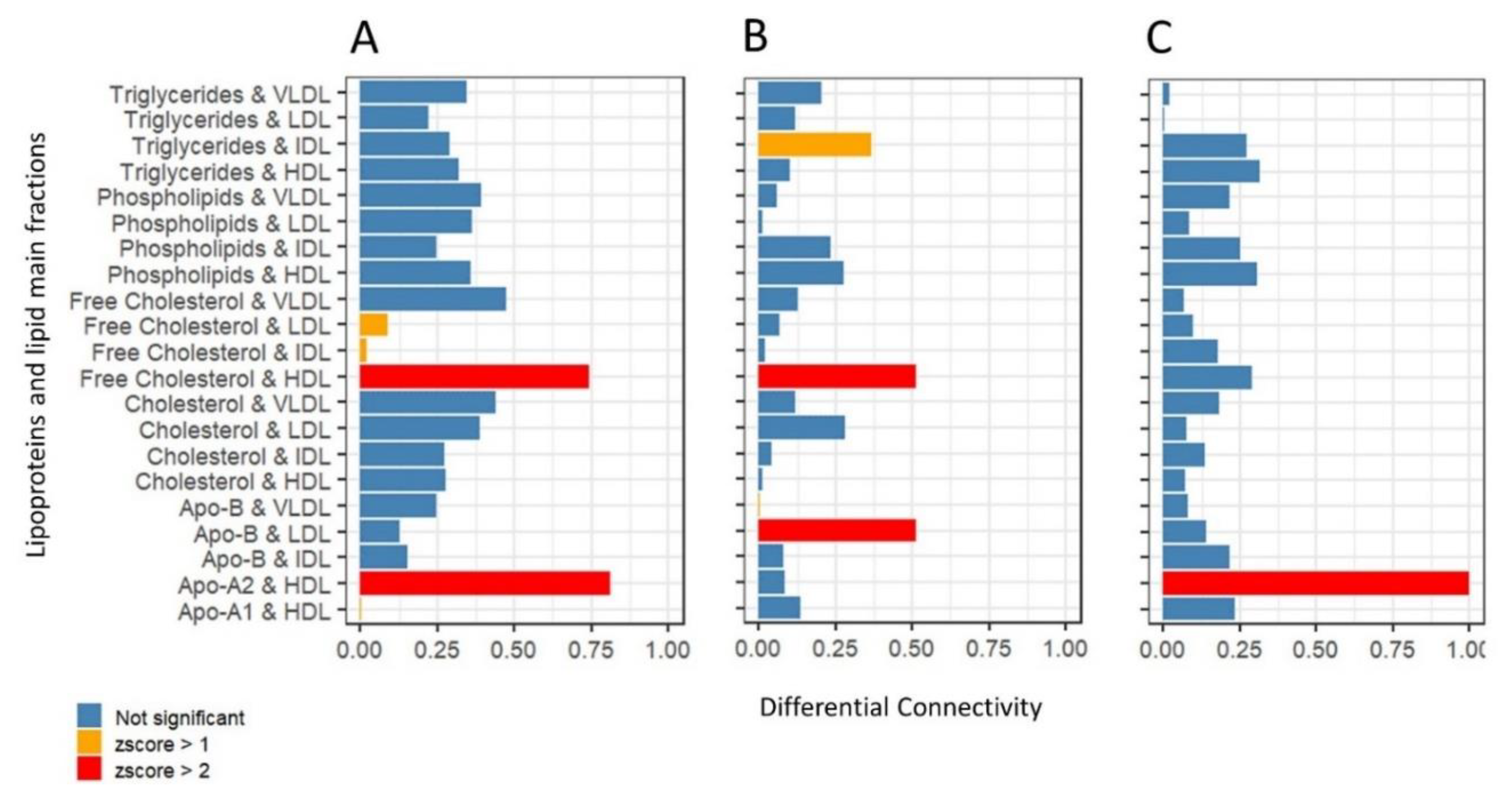
2.4. Differential Network Analysis Indicates Relevant Topological Differences in Lipoprotein and Lipid Fractions Specific to Sex and Age Group
3. Discussion
3.1. Considerations Regarding Confounding Factors
3.2. Considerations Regarding Group Size
3.3. Sex Affects Lipoproteins and Lipid Fraction Profiles in Healthy Subjects
3.4. Consideration Regarding Age Groups
3.5. Age Affects Lipoproteins and Lipid Fraction Profiles in Healthy Subjects
4. Materials and Methods
4.1. Study Population
4.2. Study Data
4.3. Sample Collection and Handling
4.4. NMR Sample Preparation
4.5. NMR Analysis and Lipoprotein Quantification
4.6. Definition of Age Groups
4.7. Statistical Analysis
4.7.1. Data Pre-Processing
4.7.2. Univariate Analysis
4.7.3. ROC Analysis
4.7.4. Multivariate Analysis
Exploratory Analysis
Dimensionality Assessment
Predictive Modeling
4.7.5. Network Analysis
Inference of Association Networks
Gaussian Graphical Modeling
Differential Network Analysis
Network Topology Measures
4.7.6. Covariance Simultaneous Component Analysis
4.7.7. Software
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Quehenberger, O.; Dennis, E.A. The human plasma lipidome. N. Engl. J. Med. 2011, 365, 1812–1823. [Google Scholar] [CrossRef]
- Muro, E.; Atilla-Gokcumen, G.E.; Eggert, U.S. Lipids in cell biology: How can we understand them better? Mol. Biol. Cell 2014, 25, 1819–1823. [Google Scholar] [CrossRef]
- Lee, J.S.; Chang, P.-Y.; Zhang, Y.; Kizer, J.R.; Best, L.G.; Howard, B.V. Triglyceride and hdl-c dyslipidemia and risks of coronary heart disease and ischemic stroke by glycemic dysregulation status: The strong heart study. Diabetes Care 2017, 40, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, P.N.; Heiss, G.; Ellison, R.C.; Province, M.A.; Pankow, J.S.; Eckfeldt, J.H.; Hunt, S.C. Coronary artery disease risk in familial combined hyperlipidemia and familial hypertriglyceridemia: A case-control comparison from the national heart, lung, and blood institute family heart study. Circulation 2003, 108, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Beyene, H.B.; Olshansky, G.; Smith, A.A.T.; Giles, C.; Huynh, K.; Cinel, M.; Mellett, N.A.; Cadby, G.; Hung, J.; Hui, J.; et al. High-coverage plasma lipidomics reveals novel sex-specific lipidomic fingerprints of age and bmi: Evidence from two large population cohort studies. PLoS Biol. 2020, 18, e3000870. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Li, L.; Zhu, B.; Li, Y. Lipidome in colorectal cancer. Oncotarget 2016, 7, 33429. [Google Scholar] [CrossRef] [PubMed]
- Klupczynska, A.; Plewa, S.; Kasprzyk, M.; Dyszkiewicz, W.; Kokot, Z.J.; Matysiak, J. Serum lipidome screening in patients with stage i non-small cell lung cancer. Clin. Exp. Med. 2019, 19, 505–513. [Google Scholar] [CrossRef]
- Bougnoux, P.; Hajjaji, N.; Couet, C. The lipidome as a composite biomarker of the modifiable part of the risk of breast cancer. Prostaglandins Leukot. Essent. Fat. Acids 2008, 79, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, H.; Brieva, L.; Tatzber, F.; Jové, M.; Cacabelos, D.; Cassanyé, A.; Lanau-Angulo, L.; Boada, J.; Serrano, J.C.; González, C.; et al. Lipidome analysis in multiple sclerosis reveals protein lipoxidative damage as a potential pathogenic mechanism. J. Neurochem. 2012, 123, 622–634. [Google Scholar] [CrossRef]
- Souto-Carneiro, M.; Tóth, L.; Behnisch, R.; Urbach, K.; Klika, K.D.; Carvalho, R.A.; Lorenz, H.-M. Differences in the serum metabolome and lipidome identify potential biomarkers for seronegative rheumatoid arthritis versus psoriatic arthritis. Ann. Rheum. Dis. 2020, 79, 499–506. [Google Scholar] [CrossRef]
- Vignoli, A.; Orlandini, B.; Tenori, L.; Biagini, M.R.; Milani, S.; Renzi, D.; Luchinat, C.; Calabrò, A.S. Metabolic signature of primary biliary cholangitis and its comparison with celiac disease. J. Proteome Res. 2018, 18, 1228–1236. [Google Scholar] [CrossRef] [PubMed]
- Palmisano, B.T.; Zhu, L.; Eckel, R.H.; Stafford, J.M. Sex differences in lipid and lipoprotein metabolism. Mol. Metab. 2018, 15, 45–55. [Google Scholar] [CrossRef]
- Barupal, D.K.; Zhang, Y.; Fan, S.; Hazen, S.L.; Tang, W.W.; Cajka, T.; Irvin, M.R.; Arnett, D.K.; Kind, T.; Kaddurah-Daouk, R.; et al. The circulating lipidome is largely defined by sex descriptors in the goldn, genebank and the adni studies. BioRxiv 2019, 731448. [Google Scholar] [CrossRef]
- Wong, M.W.K.; Braidy, N.; Pickford, R.; Vafaee, F.; Crawford, J.; Muenchhoff, J.; Schofield, P.; Attia, J.; Brodaty, H.; Sachdev, P.; et al. Plasma lipidome variation during the second half of the human lifespan is associated with age and sex but minimally with bmi. PLoS ONE 2019, 14, e0214141. [Google Scholar] [CrossRef]
- Slade, E.; Irvin, M.R.; Xie, K.; Arnett, D.K.; Claas, S.A.; Kind, T.; Fardo, D.W.; Graf, G.A. Age and sex are associated with the plasma lipidome: Findings from the goldn study. Lipids Health Dis. 2021, 20, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sales, S.; Graessler, J.; Ciucci, S.; Al-Atrib, R.; Vihervaara, T.; Schuhmann, K.; Kauhanen, D.; Sysi-Aho, M.; Bornstein, S.R.; Bickle, M.; et al. Gender, contraceptives and individual metabolic predisposition shape a healthy plasma lipidome. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Vignoli, A.; Tenori, L.; Luchinat, C.; Saccenti, E. Age and sex effects on plasma metabolite association networks in healthy subjects. J. Proteome Res. 2018, 17, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Vignoli, A.; Ghini, V.; Meoni, G.; Licari, C.; Takis, P.G.; Tenori, L.; Turano, P.; Luchinat, C. High-throughput metabolomics by 1d nmr. Angew. Chem. Int. Ed. 2019, 58, 968–994. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. Nmr spectroscopy for metabolomics research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Rosato, A.; Tenori, L.; Cascante, M.; De Atauri Carulla, P.R.; Martins dos Santos, V.A.P.; Saccenti, E. From correlation to causation: Analysis of metabolomics data using systems biology approaches. Metabolomics 2018, 14, 37. [Google Scholar] [CrossRef]
- Szymańska, E.; Bouwman, J.; Strassburg, K.; Vervoort, J.; Kangas, A.J.; Soininen, P.; Ala-Korpela, M.; Westerhuis, J.; van Duynhoven, J.P.; Mela, D.J.; et al. Gender-dependent associations of metabolite profiles and body fat distribution in a healthy population with central obesity: Towards metabolomics diagnostics. Omics J. Integr. Biol. 2012, 16, 652–667. [Google Scholar] [CrossRef] [PubMed]
- Jansen, J.J.; Szymańska, E.; Hoefsloot, H.C.; Smilde, A.K. Individual differences in metabolomics: Individualised responses and between-metabolite relationships. Metabolomics 2012, 8, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Saccenti, E.; Menichetti, G.; Ghini, V.; Remondini, D.; Tenori, L.; Luchinat, C. Entropy-based network representation of the individual metabolic phenotype. J. Proteome Res. 2016, 15, 3298–3307. [Google Scholar] [CrossRef]
- Vignoli, A.; Tenori, L.; Giusti, B.; Valente, S.; Carrabba, N.; Balzi, D.; Barchielli, A.; Marchionni, N.; Gensini, G.F.; Marcucci, R.; et al. Differential network analysis reveals metabolic determinants associated with mortality in acute myocardial infarction patients and suggests potential mechanisms underlying different clinical scores used to predict death. J. Proteome Res. 2020, 19, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Saccenti, E.; Suarez-Diez, M.; Luchinat, C.; Santucci, C.; Tenori, L. Probabilistic networks of blood metabolites in healthy subjects as indicators of latent cardiovascular risk. J. Proteome Res. 2014, 14, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Vignoli, A.; Tenori, L.; Luchinat, C.; Saccenti, E. Differential network analysis reveals molecular determinants associated with blood pressure and heart rate in healthy subjects. J. Proteome Res. 2020, 19, 64–74. [Google Scholar] [CrossRef]
- Afzal, M.; Saccenti, E.; Madsen, M.B.; Hansen, M.B.; Hyldegaard, O.; Skrede, S.; Martins Dos Santos, V.A.P.; Norrby-Teglund, A.; Svensson, M. Integrated univariate, multivariate, and correlation-based network analyses reveal metabolite-specific effects on bacterial growth and biofilm formation in necrotizing soft tissue infections. J. Proteome Res. 2020, 19, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Jahagirdar, S.; Saccenti, E. Evaluation of single sample network inference methods for metabolomics-based systems medicine. J. Proteome Res. 2021, 20, 932–949. [Google Scholar] [CrossRef]
- Gini, C. Variabilità e Mutuabilità. Contributo Allo Studio Delle Distribuzioni e Delle Relazioni Statistiche; Cuppini: Bologna, Italy, 1912. [Google Scholar]
- Suarez-Diez, M.; Adam, J.; Adamski, J.; Chasapi, S.A.; Luchinat, C.; Peters, A.; Prehn, C.; Santucci, C.; Spyridonidis, A.; Spyroulias, G.A.; et al. Plasma and serum metabolite association networks: Comparability within and between studies using nmr and ms profiling. J. Proteome Res. 2017, 16, 2547–2559. [Google Scholar] [CrossRef]
- Yu, Z.; Kastenmüller, G.; He, Y.; Belcredi, P.; Möller, G.; Prehn, C.; Mendes, J.; Wahl, S.; Roemisch-Margl, W.; Ceglarek, U.; et al. Differences between human plasma and serum metabolite profiles. PLoS ONE 2011, 6, e21230. [Google Scholar] [CrossRef]
- Jahagirdar, S.; Saccenti, E. On the use of correlation and mi as a measure of metabolite—metabolite association for network differential connectivity analysis. Metabolites 2020, 10, 171. [Google Scholar] [CrossRef] [PubMed]
- Jahagirdar, S.; Suarez-Diez, M.; Saccenti, E. Simulation and reconstruction of metabolite-metabolite association networks using a metabolic dynamic model and correlation based-algorithms. J. Proteome Res. 2019, 18, 1099–1113. [Google Scholar] [CrossRef] [PubMed]
- Pearl, J. Causality, 2nd ed.; Cambridge University Press: Cambridge, UK, 2009. [Google Scholar]
- Koller, D.; Friedman, N. Probabilistic Graphical Models: Principles and Techniques; MIT Press: Cambridge, MA, USA, 2009. [Google Scholar]
- Epskamp, S.; Fried, E.I. A tutorial on regularized partial correlation networks. Psychol. Methods 2018, 23, 617. [Google Scholar] [CrossRef] [PubMed]
- Opgen-Rhein, R.; Strimmer, K. From correlation to causation networks: A simple approximate learning algorithm and its application to high-dimensional plant gene expression data. BMC Syst. Biol. 2007, 1, 1–10. [Google Scholar] [CrossRef]
- Junker, B.H.; Schreiber, F. Analysis of Biological Networks; John Wiley & Sons: Hoboken, NJ, USA, 2011; Volume 2. [Google Scholar]
- He, X.; Zhang, J. Why do hubs tend to be essential in protein networks? PLoS Genet 2006, 2, e88. [Google Scholar] [CrossRef]
- Carter, S.; Brechbuhler, C.; Griffin, M.; Bond, A. Gene co-expression network topology provides a framework for molecular characterization of cellular state. Bioinformatics 2004, 20, 2242–2250. [Google Scholar] [CrossRef]
- Stuart, J.; Segal, E.; Koller, D.; Kim, S. A gene-coexpression network for global discovery of conserved genetic modules. Science 2003, 302, 249–255. [Google Scholar] [CrossRef]
- Jeong, H.; Mason, S.; Barabasi, A.; Oltvai, Z. Lethality and centrality in protein networks. Nature 2001, 411, 41–42. [Google Scholar] [CrossRef]
- Rzhetsky, A.; Gomez, S.M. Birth of scale-free molecular networks and the number of distinct DNA and protein domains per genome. Bioinformatics 2001, 17, 988–996. [Google Scholar] [CrossRef]
- Yook, S.H.; Oltvai, Z.N.; Barabási, A.L. Functional and topological characterization of protein interaction networks. Proteomics 2004, 4, 928–942. [Google Scholar] [CrossRef]
- Hartwell, L.; Hopfield, J.; Leibler, S.; Murray, A. From molecular to modular cell biology. Nature 1999, 402, C47–C52. [Google Scholar] [CrossRef]
- Zheng, Y.; Qi, L. Diet and lifestyle interventions on lipids: Combination with genomics and metabolomics. Clin. Lipidol. 2014, 9, 417–427. [Google Scholar] [CrossRef]
- Hegele, R.A. Plasma lipoproteins: Genetic influences and clinical implications. Nat. Rev. Genet. 2009, 10, 109–121. [Google Scholar] [CrossRef]
- Mensink, R.P.; Zock, P.L.; Kester, A.D.; Katan, M.B. Effects of dietary fatty acids and carbohydrates on the ratio of serum total to hdl cholesterol and on serum lipids and apolipoproteins: A meta-analysis of 60 controlled trials. Am. J. Clin. Nutr. 2003, 77, 1146–1155. [Google Scholar] [CrossRef] [PubMed]
- Parks, E.J. Effect of dietary carbohydrate on triglyceride metabolism in humans. J. Nutr. 2001, 131, 2772S–2774S. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.A.; Fensom, G.K.; Rinaldi, S.; Scalbert, A.; Gunter, M.J.; Holmes, M.V.; Key, T.J.; Travis, R.C. Nmr metabolite profiles in male meat-eaters, fish-eaters, vegetarians and vegans, and comparison with ms metabolite profiles. Metabolites 2021, 11, 121. [Google Scholar] [CrossRef] [PubMed]
- Bani, M.; Strepparava, M.; Giussani, B. Gender differences and frequency of whole blood donation in italian donors: Even though I want to, I cannot? Transfus. Apher. Sci. 2014, 50, 81–86. [Google Scholar] [CrossRef]
- Cohen, J. A power primer. Psychol. Bull. 1992, 112, 155. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, B.; Alikhani, A.; Mokhtari, H.; Rasouli, M. The ratio of unesterified/esterified cholesterol is the major determinant of atherogenicity of lipoprotein fractions. Med. Arch. 2018, 72, 103. [Google Scholar] [CrossRef]
- Saku, K.; Zhang, B.; Ohta, T.; Arakawa, K. Quantity and function of high density lipoprotein as an indicator of coronary atherosclerosis. J. Am. Coll. Cardiol. 1999, 33, 436–443. [Google Scholar] [CrossRef]
- Rasouli, M.; Trischuk, T.C.; Lehner, R. Calmodulin antagonist w-7 inhibits de novo synthesis of cholesterol and suppresses secretion of de novo synthesized and preformed lipids from cultured hepatocytes. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2004, 1682, 92–101. [Google Scholar] [CrossRef]
- Steinberg, D. The ldl modification hypothesis of atherogenesis: An update. J. Lipid Res. 2009, 50, S376–S381. [Google Scholar] [CrossRef] [PubMed]
- Abbott, R.D.; Garrison, R.J.; Wilson, P.; Epstein, F.H.; Castelli, W.P.; Feinleib, M.; LaRue, C. Joint distribution of lipoprotein cholesterol classes. The framingham study. Arterioscler. Off. J. Am. Heart Assoc. Inc. 1983, 3, 260–272. [Google Scholar] [CrossRef]
- Cohn, J.S.; McNamara, J.; Cohn, S.; Ordovas, J.; Schaefer, E. Postprandial plasma lipoprotein changes in human subjects of different ages. J. Lipid Res. 1988, 29, 469–479. [Google Scholar] [CrossRef]
- Jalali, M.T.; Honomaror, A.M.; Rekabi, A.; Latifi, M. Reference ranges for serum total cholesterol, hdl-cholesterol, ldl-cholesterol, and vldl-cholesterol and triglycerides in healthy iranian ahvaz population. Indian J. Clin. Biochem. 2013, 28, 277–282. [Google Scholar] [CrossRef][Green Version]
- Hanai, K.; Babazono, T.; Yoshida, N.; Nyumura, I.; Toya, K.; Hayashi, T.; Bouchi, R.; Tanaka, N.; Ishii, A.; Iwamoto, Y. Gender differences in the association between hdl cholesterol and the progression of diabetic kidney disease in type 2 diabetic patients. Nephrol. Dial. Transplant. 2012, 27, 1070–1075. [Google Scholar] [CrossRef] [PubMed]
- Madsen, C.M.; Varbo, A.; Nordestgaard, B.G. Extreme high high-density lipoprotein cholesterol is paradoxically associated with high mortality in men and women: Two prospective cohort studies. Eur. Heart J. 2017, 38, 2478–2486. [Google Scholar] [CrossRef]
- Kim, H.J.; Park, H.A.; Cho, Y.G.; Kang, J.H.; Kim, K.W.; Kang, J.H.; Kim, N.-R.; Chung, W.-C.; Kim, C.H.; Whang, D.H.; et al. Gender difference in the level of hdl cholesterol in korean adults. Korean J. Fam. Med. 2011, 32, 173. [Google Scholar] [CrossRef]
- Balder, J.W.; de Vries, J.K.; Nolte, I.M.; Lansberg, P.J.; Kuivenhoven, J.A.; Kamphuisen, P.W. Lipid and lipoprotein reference values from 133,450 dutch lifelines participants: Age-and gender-specific baseline lipid values and percentiles. J. Clin. Lipidol. 2017, 11, 1055–1064.e6. [Google Scholar] [CrossRef]
- Wang, X.; Magkos, F.; Mittendorfer, B. Sex differences in lipid and lipoprotein metabolism: It’s not just about sex hormones. J. Clin. Endocrinol. Metab. 2011, 96, 885–893. [Google Scholar] [CrossRef]
- Tian, L.; Fu, M. The relationship between high density lipoprotein subclass profile and plasma lipids concentrations. Lipids Health Dis. 2010, 9, 1–9. [Google Scholar] [CrossRef]
- McLaughlin, T.; Reaven, G.; Abbasi, F.; Lamendola, C.; Saad, M.; Waters, D.; Simon, J.; Krauss, R.M. Is there a simple way to identify insulin-resistant individuals at increased risk of cardiovascular disease? Am. J. Cardiol. 2005, 96, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, F.; Reaven, G.M. Comparison of two methods using plasma triglyceride concentration as a surrogate estimate of insulin action in nondiabetic subjects: Triglycerides× glucose versus triglyceride/high-density lipoprotein cholesterol. Metabolism 2011, 60, 1673–1676. [Google Scholar] [CrossRef]
- Laws, A.; Reaven, G. Evidence for an independent relationship between insulin resistance and fasting plasma hdl-cholesterol, triglyceride and insulin concentrations. J. Intern. Med. 1992, 231, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Vital, P.; Larrieta, E.; Hiriart, M. Sexual dimorphism in insulin sensitivity and susceptibility to develop diabetes in rats. J. Endocrinol. 2006, 190, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Camporez, J.P.; Lyu, K.; Goldberg, E.L.; Zhang, D.; Cline, G.W.; Jurczak, M.J.; Dixit, V.D.; Petersen, K.F.; Shulman, G.I. Anti-inflammatory effects of oestrogen mediate the sexual dimorphic response to lipid-induced insulin resistance. J. Physiol. 2019, 597, 3885–3903. [Google Scholar] [CrossRef]
- Geer, E.B.; Shen, W. Gender differences in insulin resistance, body composition, and energy balance. Gend. Med. 2009, 6, 60–75. [Google Scholar] [CrossRef] [PubMed]
- Grove, K.; Fried, S.; Greenberg, A.; Xiao, X.; Clegg, D. A microarray analysis of sexual dimorphism of adipose tissues in high-fat-diet-induced obese mice. Int. J. Obes. 2010, 34, 989–1000. [Google Scholar] [CrossRef] [PubMed]
- Butruille, L.; Drougard, A.; Knauf, C.; Moitrot, E.; Valet, P.; Storme, L.; Deruelle, P.; Lesage, J. The apelinergic system: Sexual dimorphism and tissue-specific modulations by obesity and insulin resistance in female mice. Peptides 2013, 46, 94–101. [Google Scholar] [CrossRef]
- Orimo, H.; Ito, H.; Suzuki, T.; Araki, A.; Hosoi, T.; Sawabe, M. Reviewing the definition of “elderly”. Geriatr. Gerontol. Int. 2006, 6, 149–158. [Google Scholar] [CrossRef]
- WHO. Women, Ageing and Health: Achieving Health Across the Life Span; World Health Organization: Geneva, Switzerland, 1998. [Google Scholar]
- WHO. Men, Ageing and Health: Achieving Health Across the Life Span; World Health Organization: Geneva, Switzerland, 1999. [Google Scholar]
- Holloszy, J.O. The biology of aging. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2000; pp. S3–S9. [Google Scholar]
- George, A.J.; Ritter, M.A. Thymic involution with ageing: Obsolescence or good housekeeping? Immunol. Today 1996, 17, 267–272. [Google Scholar] [CrossRef]
- Fédération, C.; Schwartz, D.; Mayaux, M. Female fecundity as a function of age: Results of artificial insemination in 2193 nulliparous women with azoospermic husbands. N. Eng. J. Med. 1982, 306, 404–406. [Google Scholar] [CrossRef] [PubMed]
- Stovall, D.W.; Toma, S.K.; Hammond, M.G.; Talbert, L.M. The effect of age on female fecundity. Obstet. Gynecol. 1991, 77, 33–36. [Google Scholar] [CrossRef]
- Attie, A.D. Lipoprotein/cholesterol metabolism. In Encyclopedia of Physical Science and Technology, 3rd ed.; Meyers, R.A., Ed.; Academic Press: New York, NY, USA, 2003; pp. 643–660. [Google Scholar]
- Mead, J.R.; Irvine, S.A.; Ramji, D.P. Lipoprotein lipase: Structure, function, regulation, and role in disease. J. Mol. Med. 2002, 80, 753–769. [Google Scholar] [CrossRef]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011, 3, a004952. [Google Scholar] [CrossRef]
- Mallov, S. Aortic lipoprotein lipase activity in relation to species, age, sex, and blood pressure. Circ. Res. 1964, 14, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Carlile, S.I.; Lacko, A.G. Age-related changes in plasma lipid levels and tissue lipoprotein lipase activities of fischer-344 rats. Arch. Gerontol. Geriatr. 1985, 4, 133–140. [Google Scholar] [CrossRef]
- Perreault, L.; Lavely, J.M.; Kittelson, J.M.; Horton, T.J. Gender differences in lipoprotein lipase activity after acute exercise. Obes. Res. 2004, 12, 241–249. [Google Scholar] [CrossRef]
- Maehira, F.; Miyagi, I.; Eguchi, Y. Sex-and age-related variations in the in vitro heparin-releasable lipoprotein lipase from mononuclear leukocytes in blood. Biochim. Biophys. Acta (BBA) Lipids Lipid Metab. 1990, 1042, 344–351. [Google Scholar] [CrossRef]
- Kiens, B.; Roepstorff, C.; Glatz, J.F.; Bonen, A.; Schjerling, P.; Knudsen, J.; Nielsen, J.N. Lipid-binding proteins and lipoprotein lipase activity in human skeletal muscle: Influence of physical activity and gender. J. Appl. Physiol. 2004, 97, 1209–1218. [Google Scholar] [CrossRef]
- Meschia, M.; Pansini, F.; Modena, A.B.; De Aloysio, D.; Gambacciani, M.; Parazzini, F.; Campagnoli, C.; Maiocchi, G.; Peruzzi, E.; Group, I.S. Determinants of age at menopause in italy: Results from a large cross-sectional study. Maturitas 2000, 34, 119–125. [Google Scholar] [CrossRef]
- Cagnacci, A.; Pansini, F.S.; Bacchi-Modena, A.; Giulini, N.; Mollica, G.; De Aloysio, D.; Vadora, E.; Volpe, A. Season of birth influences the timing of menopause. Hum. Reprod. 2005, 20, 2190–2193. [Google Scholar] [CrossRef] [PubMed]
- Parazzini, F.; Group, P.M.I.S. Determinants of age at menopause in women attending menopause clinics in italy. Maturitas 2007, 56, 280–287. [Google Scholar] [CrossRef]
- Borrás, C.; Gambini, J.; López-Grueso, R.; Pallardó, F.V.; Viña, J. Direct antioxidant and protective effect of estradiol on isolated mitochondria. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2010, 1802, 205–211. [Google Scholar] [CrossRef]
- Miller, V.M.; Lahr, B.D.; Bailey, K.R.; Heit, J.A.; Harman, S.M.; Jayachandran, M. Longitudinal effects of menopausal hormone treatments on platelet characteristics and cell-derived microvesicles. Platelets 2016, 27, 32–42. [Google Scholar] [CrossRef]
- Guetta, V.; Cannon, R.O., III. Cardiovascular effects of estrogen and lipid-lowering therapies in postmenopausal women. Circulation 1996, 93, 1928–1937. [Google Scholar] [CrossRef]
- Fåhraeus, L. The effects of estradiol on blood lipids and lipoproteins in postmenopausal women. Obstet. Gynecol. 1988, 72, 18S–22S. [Google Scholar] [CrossRef]
- Rasouli, M.; Kiasari, A.M.; Arab, S. Indicators of dehydration and haemoconcentration are associated with the prevalence and severity of coronary artery disease. Clin. Exp. Pharmacol. Physiol. 2008, 35, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Rothblat, G.H.; Phillips, M.C. High-density lipoprotein heterogeneity and function in reverse cholesterol transport. Curr. Opin. Lipidol. 2010, 21, 229. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Zong, C.; Liu, Q.; Si, Y.; Liu, J.; Li, W.; Zhu, P.; Qin, S. Sr-bi associates with abcg1 and inhibits abcg1-mediated cholesterol efflux from cells to high-density lipoprotein 3. Lipids Health Dis. 2012, 11, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Mihajlovic, M.; Gojkovic, T.; Vladimirov, S.; Miljkovic, M.; Stefanovic, A.; Vekic, J.; Zeljkovic, D.; Trifunovic, B.; Kotur-Stevuljevic, J.; Spasojevic-Kalimanovska, V.; et al. Changes in lecithin: Cholesterol acyltransferase, cholesteryl ester transfer protein and paraoxonase-1 activities in patients with colorectal cancer. Clin. Biochem. 2019, 63, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Kiziltunç, A.; Akçay, F.; Polat, F.; Kuşkay, S.; Sahin, Y.N. Reduced lecithin: Cholesterol acyltransferase (lcat) and na+, k+, atpase activity in diabetic patients. Clin. Biochem. 1997, 30, 177–182. [Google Scholar] [CrossRef]
- Dullaart, R.P.; Perton, F.; Sluiter, W.J.; de Vries, R.; van Tol, A. Plasma lecithin: Cholesterol acyltransferase activity is elevated in metabolic syndrome and is an independent marker of increased carotid artery intima media thickness. J. Clin. Endocrinol. Metab. 2008, 93, 4860–4866. [Google Scholar] [CrossRef]
- Kappelle, P.J.; de Boer, J.F.; Perton, F.G.; Annema, W.; de Vries, R.; Dullaart, R.P.; Tietge, U.J. Increased lcat activity and hyperglycaemia decrease the antioxidative functionality of hdl. Eur. J. Clin. Investig. 2012, 42, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Albers, J.J.; Chen, C.H.; Adolphson, J.L. Lecithin: Cholesterol acyltransferase (lcat) mass; its relationship to lcat activity and cholesterol esterification rate. J. Lipid Res. 1981, 22, 1206–1213. [Google Scholar] [CrossRef]
- Bernini, P.; Bertini, I.; Luchinat, C.; Tenori, L.; Tognaccini, A. The cardiovascular risk of healthy individuals studied by nmr metabonomics of plasma samples. J. Proteome Res. 2011, 10, 4983–4992. [Google Scholar] [CrossRef]
- Haug, K.; Salek, R.M.; Conesa, P.; Hastings, J.; de Matos, P.; Rijnbeek, M.; Mahendraker, T.; Williams, M.; Neumann, S.; Rocca-Serra, P.; et al. Metabolights—an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 2013, 41, D781–D786. [Google Scholar] [CrossRef]
- Bernini, P.; Bertini, I.; Luchinat, C.; Nincheri, P.; Staderini, S.; Turano, P. Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. J. Biomol. NMR 2011, 49, 231–243. [Google Scholar] [CrossRef]
- Jimeénez, B.; Holmes, E.; Heude, C.; Tolson, R.F.; Harvey, N.; Lodge, S.L.; Chetwynd, A.J.; Cannet, C.; Fang, F.; Pearce, J.T.; et al. Quantitative lipoprotein subclass and low molecular weight metabolite analysis in human serum and plasma by 1h nmr spectroscopy in a multilaboratory trial. Anal. Chem. 2018, 90, 11962–11971. [Google Scholar] [CrossRef]
- Monsonis Centelles, S.; Hoefsloot, H.C.; Khakimov, B.; Ebrahimi, P.; Lind, M.V.; Kristensen, M.; De Roo, N.; Jacobs, D.M.; Van Duynhoven, J.; Cannet, C.; et al. Toward reliable lipoprotein particle predictions from nmr spectra of human blood: An interlaboratory ring test. Anal. Chem. 2017, 89, 8004–8012. [Google Scholar] [CrossRef]
- Bartlett, M.S. The square root transformation in analysis of variance. Suppl. J. R. Stat. Soc. 1936, 3, 68–78. [Google Scholar] [CrossRef]
- Van Den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; Van Der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Mann, H.B.; Whitney, D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Bonferroni, C.E. Il calcolo delle assicurazioni su gruppi di teste. In Studi in Onore Del Professore Salvatore Ortu Carboni; Bardi: Rome, Italy, 1935; pp. 13–60. [Google Scholar]
- Søreide, K. Receiver-operating characteristic curve analysis in diagnostic, prognostic and predictive biomarker research. J. Clin. Pathol. 2009, 62, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (roc) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef]
- Mason, S.J.; Graham, N.E. Areas beneath the relative operating characteristics (roc) and relative operating levels (rol) curves: Statistical significance and interpretation. Q. J. R. Meteorol. Soc. A J. Atmos. Sci. Appl. Meteorol. Phys. Oceanogr. 2002, 128, 2145–2166. [Google Scholar] [CrossRef]
- Hotelling, H. Analysis of a complex of statistical variables into principal components. J. Educ. Psychol. 1931, 24, 417441. [Google Scholar]
- Pearson, K. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Jolliffe, I. Principal Component Analysis, 2nd ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Johnstone, I.M. On the distribution of the largest eigenvalue in principal components analysis. Ann. Stat. 2001, 29, 295–327. [Google Scholar] [CrossRef]
- Saccenti, E.; Timmerman, M.E. Considering horn’s parallel analysis from a random matrix theory point of view. Psychometrika 2017, 82, 186–209. [Google Scholar] [CrossRef]
- Saccenti, E.; Camacho, J. Determining the number of components in principal components analysis: A comparison of statistical, crossvalidation and approximated methods. Chemom. Intell. Lab. Syst. 2015, 149, 99–116. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Olshen, R.; Stone, C. Classification and regression trees. Brooks/Cole Publishing: Honkong, China, 1984. [Google Scholar]
- Parikh, R.; Mathai, A.; Parikh, S.; Sekhar, G.C.; Thomas, R. Understanding and using sensitivity, specificity and predictive values. Indian J. Ophthalmol. 2008, 56, 45. [Google Scholar] [CrossRef] [PubMed]
- Ishwaran, H. The effect of splitting on random forests. Mach. Learn. 2015, 99, 75–118. [Google Scholar] [CrossRef]
- Faith, J.J.; Hayete, B.; Thaden, J.T.; Mogno, I.; Wierzbowski, J.; Cottarel, G.; Kasif, S.; Collins, J.J.; Gardner, T.S. Large-scale mapping and validation of escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007, 5, e8. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, J.; Opgen-Rhein, R.; Strimmer, K. Reverse engineering genetic networks using the genenet package. Newsl. R Proj. 2006, 6, 50. [Google Scholar]
- Smilde, A.K.; Timmerman, M.E.; Saccenti, E.; Jansen, J.J.; Hoefsloot, H.C.J. Covariances simultaneous component analysis: A new method within a framework for modeling covariances. J. Chemom. 2015, 29, 277–288. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing v3. 1.0; The R Foundation for Statistical Computing: Vienna, Austria; Available online: http://www.R-project.org (accessed on 21 January 2014).
- Liaw, A.; Wiener, M. Classification and regression by randomforest. R News 2002, 2, 18–22. [Google Scholar]
- Archer, E.; Archer, M.E. Package ’Rfpermute’ . Available online: https://cran.r-project.org/web/packages/rfPermute/index.html (accessed on 21 January 2021).
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. Proc: An open-source package for r and s+ to analyze and compare roc curves. BMC Bioinform. 2011, 12, 1–8. [Google Scholar] [CrossRef]
- Kuhn, M. Building predictive models in r using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.-E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef]
- Dezső, Z.; Barabási, A.-L. Halting viruses in scale-free networks. Phys. Rev. E 2002, 65, 055103. [Google Scholar] [CrossRef]
- Bavelas, A. Communication patterns in task-oriented groups. J. Acoust. Soc. Am. 1950, 22, 725–730. [Google Scholar] [CrossRef]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of small-‘world’ networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Diestel, R.; Schrijver, A.; Seymour, P. Graph theory. Oberwolfach Rep. 2010, 7, 521–580. [Google Scholar] [CrossRef]
- Wasserman, S.; Faust, K. Social Network Analysis: Methods and Applications; Cambridge University Press: Cambridge, UK, 1994. [Google Scholar]
- Maslov, S.; Sneppen, K. Specificity and stability in topology of protein networks. Science 2002, 296, 910–913. [Google Scholar] [CrossRef] [PubMed]
- Shimbel, A. Structural parameters of communication networks. Bull. Math. Biophys. 1953, 15, 501–507. [Google Scholar] [CrossRef]
- Stelzl, U.; Worm, U.; Lalowski, M.; Haenig, C.; Brembeck, F.H.; Goehler, H.; Stroedicke, M.; Zenkner, M.; Schoenherr, A.; Koeppen, S.; et al. A human protein-protein interaction network: A resource for annotating the proteome. Cell 2005, 122, 957–968. [Google Scholar] [CrossRef]
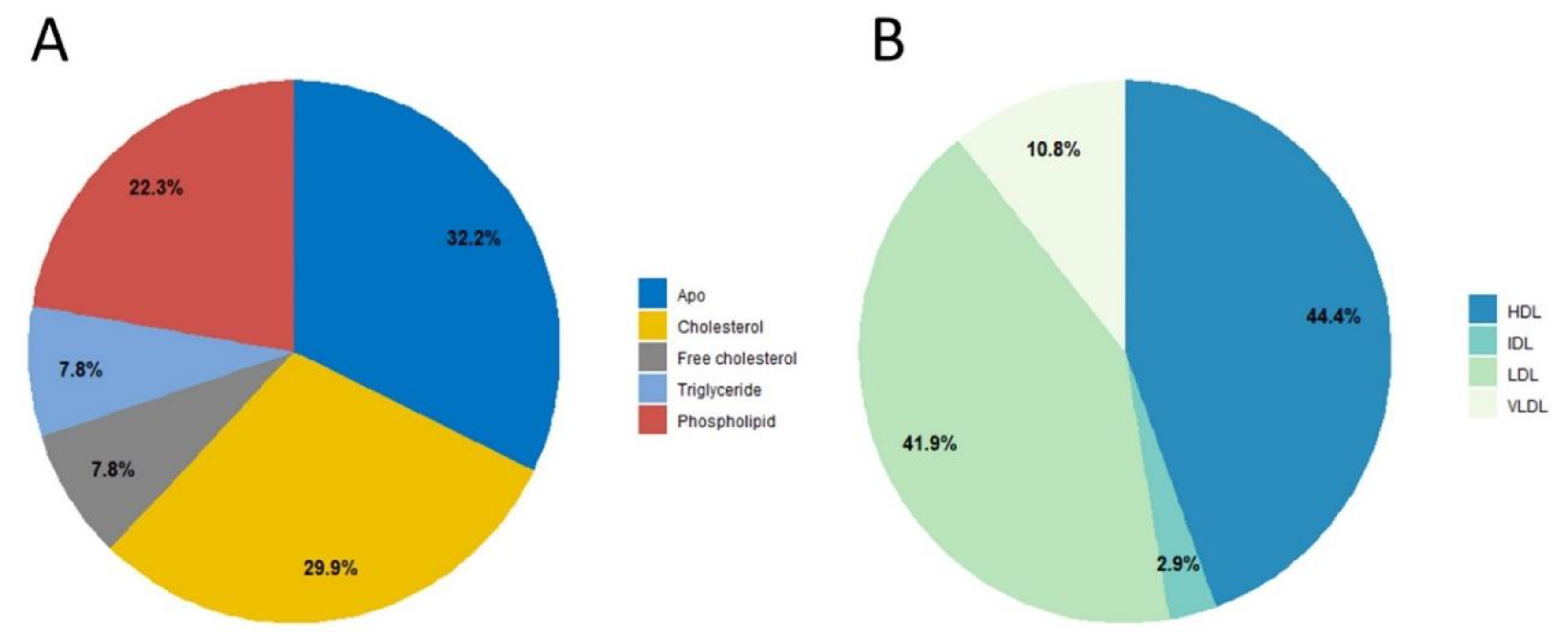
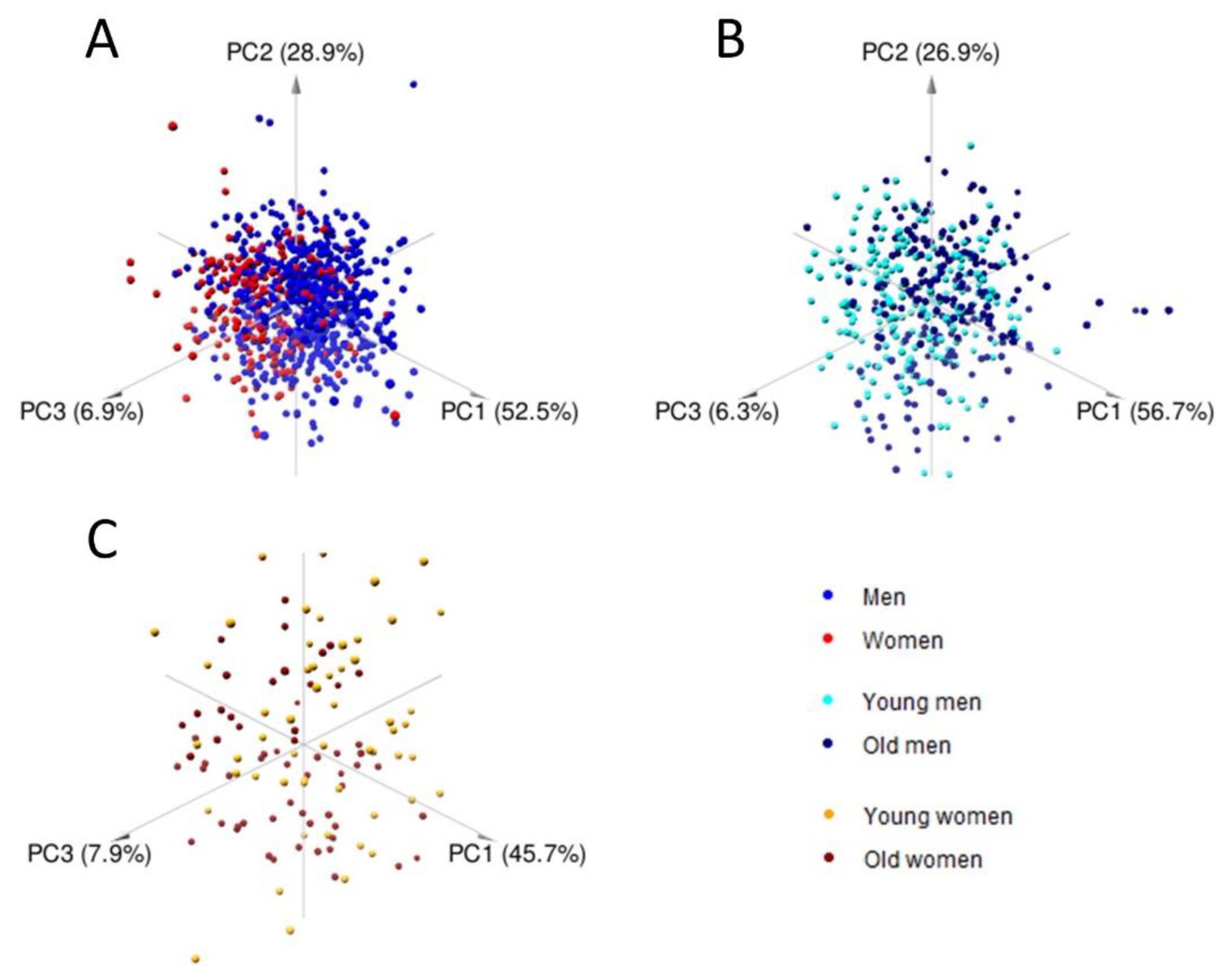
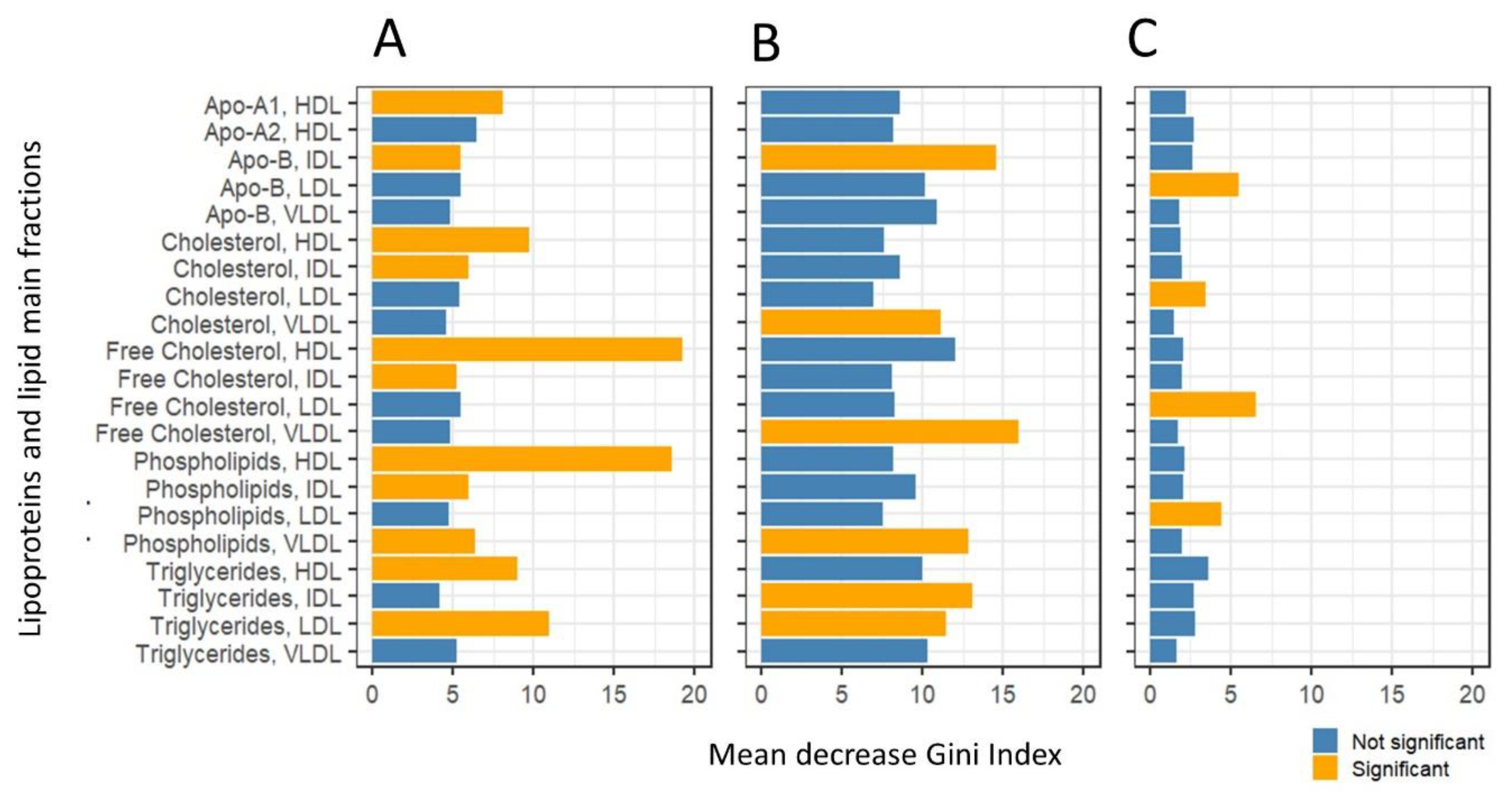

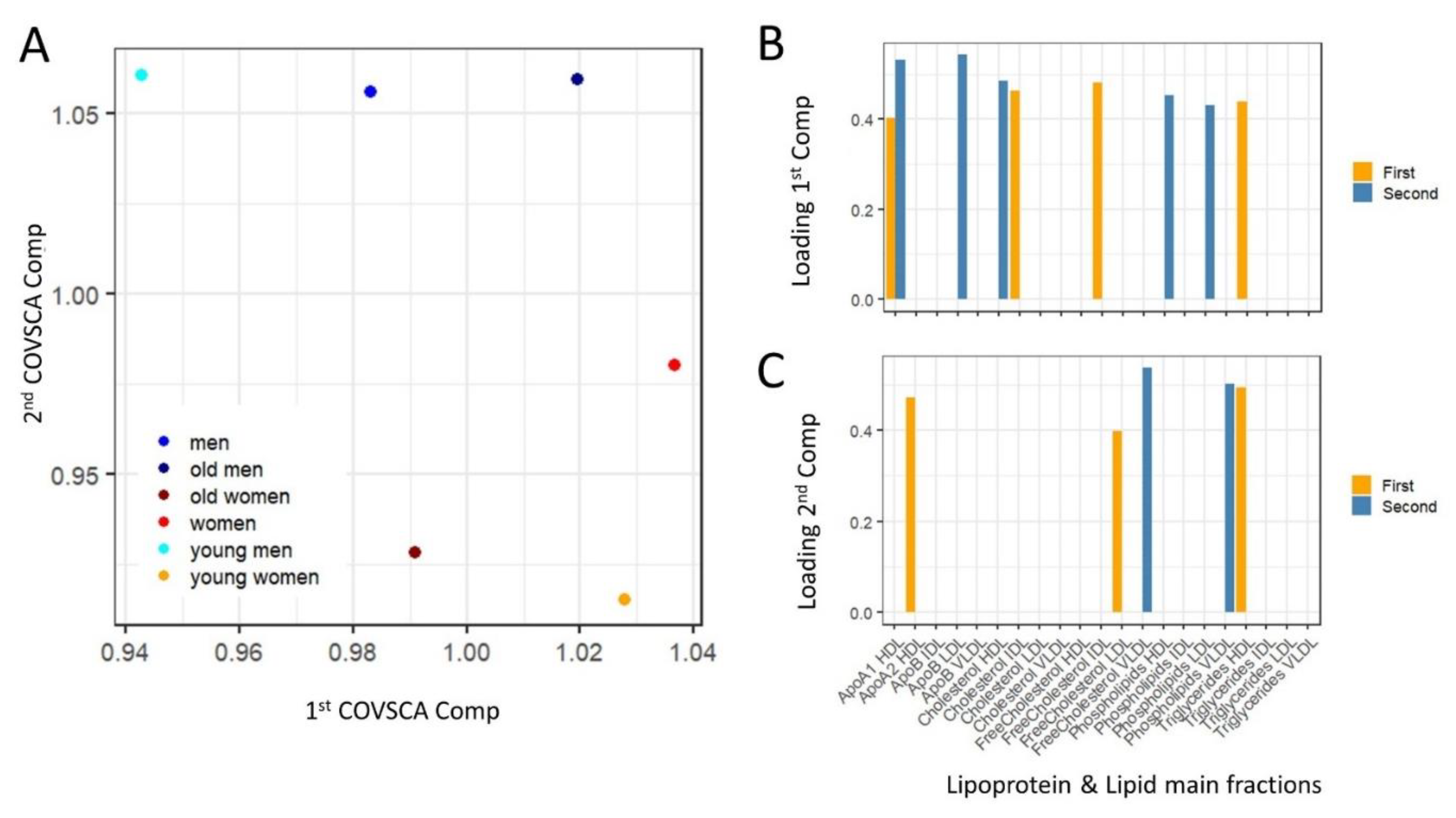
| Group Name | Age Group | Subjects (n) | Median Age (Years) |
|---|---|---|---|
| W | Women (all) | 183 | 43 |
| YW | Young women (<37 yrs) | 56 | 27 |
| OW | Old women (>48 yrs) | 60 | 55 |
| M | Men (all) | 661 | 40 |
| YM | Young men (<35 yrs) | 216 | 29 |
| OM | Old men (>45 yrs) | 213 | 52 |
| Concentration | |||||||
|---|---|---|---|---|---|---|---|
| Lipid/Lipoprotein | Men | Women | Young Men | Old Men | Young Women | Old Women | |
| 1 | Apo-A1, HDL | 156.6 * | 176.8 * | 155.3 | 160.1 | 170.3 | 184.0 |
| 2 | Apo-A2, HDL | 26.8 * | 30.3 * | 26.2 | 28.1 | 28.6 | 32.2 |
| 3 | Apo-B, IDL | 3.1 * | 2.6 * | 2.6 * | 3.9 * | 1.8 * | 3.2 * |
| 4 | Apo-B, LDL | 61.1 | 63.2 | 55.9 * | 67.8 * | 51.9 * | 74.1 * |
| 5 | Apo-B, VLDL | 6.6 * | 5.0 * | 5.4 * | 8.1 * | 4.7 | 5.2 |
| 6 | Cholesterol, HDL | 66.7 * | 76.9 * | 66.4 | 67.7 | 73.8 | 80.1 |
| 7 | Cholesterol, IDL | 6.9 * | 4.7 * | 5.6 * | 8.9 * | 3.1 | 6.0 |
| 8 | Cholesterol, LDL | 154.8 | 146.1 | 153.3 | 159.3 | 124.6 * | 165.7 * |
| 9 | Cholesterol, VLDL | 14.0 * | 9.0 * | 10.6 * | 18.1 * | 8.4 | 8.9 |
| 10 | Free Cholesterol, HDL | 13.4 * | 16.8 * | 12.9 | 13.8 | 16.2 | 17.5 |
| 11 | Free Cholesterol, IDL | 1.9 * | 1.4 * | 1.6 * | 2.6 * | 0.90 | 1.8 |
| 12 | Free Cholesterol, LDL | 40.0 | 38.8 | 38.8 | 41.6 | 33.4 * | 44.2 * |
| 13 | Free Cholesterol, VLDL | 7.7 * | 5.7 * | 5.8 * | 9.7 * | 5.5 | 5.9 |
| 14 | Phospholipids, HDL | 74.7 * | 93.3 * | 73.2 | 77.0 | 91.0 | 96.3 |
| 15 | Phospholipids, IDL | 5.2 * | 3.9 * | 4.5 * | 6.435 * | 2.9 | 4.7 |
| 16 | Phospholipids, LDL | 77.7 | 76.3 | 75.6 | 81.1 | 66.1 * | 86.0 * |
| 17 | Phospholipids, VLDL | 19.6 * | 14.0 * | 15.7 * | 24.1 * | 13.6 | 14.2 |
| 18 | Triglycerides, HDL | 7.3 * | 9.2 * | 6.4 * | 8.2 * | 9.5 | 9.1 |
| 19 | Triglycerides, IDL | 7.5 | 5.2 | 4.8 * | 10.7 * | 4.4 | 5.5 |
| 20 | Triglycerides, LDL | 5.49 * | 8.1 * | 4.0 * | 7.5 * | 6.2 | 9.5 |
| 21 | Triglycerides, VLDL | 45.4 * | 29.7 * | 31.5 * | 61.1 * | 27.1 | 29.8 |
| Lipid/Lipoprotein | AUC | CI AUC Lower | CI AUC Upper | Threshold | Accuracy | Specificity | Sensitivity | p-Value | Adjusted p-Value | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Apo-A1, HDL | 0.689 | 0.644 | 0.735 | 166.3 | 0.674 | 0.681 | 0.650 | <0.001 | <0.001 |
| 2 | Apo-A2, HDL | 0.656 | 0.612 | 0.701 | 28.6 | 0.633 | 0.634 | 0.628 | <0.001 | <0.001 |
| 3 | Apo-B, IDL | 0.605 | 0.557 | 0.652 | 1.8 | 0.684 | 0.755 | 0.426 | <0.001 | 0.001 |
| 4 | Apo-B, LDL | 0.524 | 0.477 | 0.572 | 65.4 | 0.582 | 0.622 | 0.437 | 0.315 | 1.000 |
| 5 | Apo-B, VLDL | 0.640 | 0.595 | 0.685 | 4.6 | 0.642 | 0.670 | 0.541 | <0.001 | <0.001 |
| 6 | Cholesterol, HDL | 0.715 | 0.672 | 0.758 | 76.6 | 0.755 | 0.820 | 0.519 | <0.001 | <0.001 |
| 7 | Cholesterol, IDL | 0.644 | 0.599 | 0.690 | 5.3 | 0.600 | 0.585 | 0.650 | <0.001 | <0.001 |
| 8 | Cholesterol, LDL | 0.554 | 0.507 | 0.601 | 153.6 | 0.524 | 0.501 | 0.607 | 0.026 | 1.000 |
| 9 | Cholesterol, VLDL | 0.648 | 0.605 | 0.692 | 12.4 | 0.533 | 0.469 | 0.765 | <0.001 | <0.001 |
| 10 | Free Cholesterol, HDL | 0.784 | 0.747 | 0.820 | 15.5 | 0.763 | 0.793 | 0.656 | <0.001 | <0.001 |
| 11 | Free Cholesterol, IDL | 0.637 | 0.591 | 0.683 | 1.5 | 0.597 | 0.585 | 0.639 | <0.001 | <0.001 |
| 12 | Free Cholesterol, LDL | 0.539 | 0.490 | 0.588 | 35.7 | 0.620 | 0.676 | 0.415 | 0.108 | 1.000 |
| 13 | Free Cholesterol, VLDL | 0.628 | 0.585 | 0.671 | 7.7 | 0.514 | 0.439 | 0.787 | <0.001 | <0.001 |
| 14 | Phospholipids, HDL | 0.793 | 0.756 | 0.830 | 85.1 | 0.763 | 0.785 | 0.683 | <0.001 | <0.001 |
| 15 | Phospholipids, IDL | 0.616 | 0.571 | 0.661 | 4.2 | 0.592 | 0.581 | 0.634 | <0.001 | <0.001 |
| 16 | Phospholipids, LDL | 0.524 | 0.476 | 0.571 | 73.1 | 0.552 | 0.569 | 0.492 | 0.325 | 1.000 |
| 17 | Phospholipids, VLDL | 0.666 | 0.623 | 0.709 | 16.8 | 0.604 | 0.576 | 0.705 | <0.001 | <0.001 |
| 18 | Triglycerides, HDL | 0.623 | 0.577 | 0.669 | 6.8 | 0.532 | 0.486 | 0.699 | <0.001 | <0.001 |
| 19 | Triglycerides, IDL | 0.578 | 0.535 | 0.622 | 7.7 | 0.444 | 0.342 | 0.814 | 0.001 | 0.071 |
| 20 | Triglycerides, LDL | 0.622 | 0.577 | 0.667 | 5.1 | 0.582 | 0.570 | 0.623 | <0.001 | <0.001 |
| 21 | Triglycerides, VLDL | 0.627 | 0.584 | 0.670 | 42.8 | 0.527 | 0.449 | 0.809 | <0.001 | <0.001 |
| Lipid/Lipoprotein | AUC | CI AUC Lower | CI AUC Upper | Threshold | Accuracy | Specificity | Sensitivity | p-Value | Adjusted p-Value | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Apo-A1, HDL | 0.553 | 0.498 | 0.607 | 178.4 | 0.562 | 0.230 | 0.889 | 0.059 | 1.000 |
| 2 | Apo-A2, HDL | 0.581 | 0.527 | 0.635 | 27.3 | 0.578 | 0.549 | 0.606 | 0.004 | 0.227 |
| 3 | Apo-B, IDL | 0.710 | 0.660 | 0.759 | 2.8 | 0.681 | 0.765 | 0.597 | <0.001 | <0.001 |
| 4 | Apo-B, LDL | 0.665 | 0.614 | 0.716 | 65.1 | 0.636 | 0.563 | 0.708 | <0.001 | <0.001 |
| 5 | Apo-B, VLDL | 0.720 | 0.672 | 0.768 | 6.9 | 0.671 | 0.577 | 0.764 | <0.001 | <0.001 |
| 6 | Cholesterol, HDL | 0.530 | 0.475 | 0.585 | 62.7 | 0.534 | 0.653 | 0.417 | 0.281 | 1.000 |
| 7 | Cholesterol, IDL | 0.688 | 0.638 | 0.739 | 7.7 | 0.650 | 0.559 | 0.741 | <0.001 | <0.001 |
| 8 | Cholesterol, LDL | 0.546 | 0.491 | 0.601 | 172.1 | 0.562 | 0.437 | 0.685 | 0.100 | 1.000 |
| 9 | Cholesterol, VLDL | 0.686 | 0.636 | 0.736 | 16.2 | 0.650 | 0.502 | 0.796 | <0.001 | <0.001 |
| 10 | Free Cholesterol, HDL | 0.586 | 0.533 | 0.640 | 11.4 | 0.573 | 0.789 | 0.361 | 0.002 | 0.126 |
| 11 | Free Cholesterol, IDL | 0.684 | 0.633 | 0.734 | 2.4 | 0.646 | 0.498 | 0.792 | <0.001 | <0.001 |
| 12 | Free Cholesterol, LDL | 0.591 | 0.537 | 0.645 | 41.7 | 0.592 | 0.507 | 0.676 | 0.001 | 0.069 |
| 13 | Free Cholesterol, VLDL | 0.733 | 0.686 | 0.780 | 7.3 | 0.695 | 0.676 | 0.713 | <0.001 | <0.001 |
| 14 | Phospholipids, HDL | 0.575 | 0.521 | 0.629 | 74.9 | 0.566 | 0.535 | 0.597 | 0.007 | 0.438 |
| 15 | Phospholipids, IDL | 0.659 | 0.607 | 0.710 | 5.3 | 0.634 | 0.587 | 0.681 | <0.001 | <0.001 |
| 16 | Phospholipids, LDL | 0.591 | 0.537 | 0.644 | 79.6 | 0.590 | 0.563 | 0.616 | 0.001 | 0.074 |
| 17 | Phospholipids, VLDL | 0.719 | 0.671 | 0.768 | 20.0 | 0.688 | 0.638 | 0.736 | <0.001 | <0.001 |
| 18 | Triglycerides, HDL | 0.674 | 0.623 | 0.724 | 7.2 | 0.639 | 0.620 | 0.657 | <0.001 | <0.001 |
| 19 | Triglycerides, IDL | 0.720 | 0.672 | 0.768 | 2.7 | 0.678 | 0.854 | 0.505 | <0.001 | <0.001 |
| 20 | Triglycerides, LDL | 0.683 | 0.633 | 0.734 | 6.4 | 0.662 | 0.563 | 0.759 | <0.001 | <0.001 |
| 21 | Triglycerides, VLDL | 0.713 | 0.664 | 0.761 | 43.1 | 0.676 | 0.634 | 0.718 | <0.001 | <0.001 |
| Lipid/Lipoprotein | AUC | CI AUC Lower | CI AUC Upper | Threshold | Accuracy | Specificity | Sensitivity | p-Value | Adjusted p-Value | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Apo-A1, HDL | 0.621 | 0.516 | 0.727 | 171.6 | 0.655 | 0.733 | 0.571 | 0.024 | 1.000 |
| 2 | Apo-A2, HDL | 0.665 | 0.565 | 0.764 | 25.4 | 0.647 | 0.950 | 0.321 | 0.002 | 0.143 |
| 3 | Apo-B, IDL | 0.704 | 0.608 | 0.799 | 1.7 | 0.672 | 0.750 | 0.589 | <0.001 | 0.010 |
| 4 | Apo-B, LDL | 0.793 | 0.710 | 0.875 | 55.5 | 0.750 | 0.867 | 0.625 | <0.001 | <0.001 |
| 5 | Apo-B, VLDL | 0.560 | 0.454 | 0.666 | 4.1 | 0.586 | 0.633 | 0.536 | 0.264 | 1.000 |
| 6 | Cholesterol, HDL | 0.608 | 0.505 | 0.711 | 70.1 | 0.621 | 0.800 | 0.429 | 0.045 | 1.000 |
| 7 | Cholesterol, IDL | 0.682 | 0.585 | 0.779 | 3.5 | 0.690 | 0.700 | 0.679 | 0.001 | 0.042 |
| 8 | Cholesterol, LDL | 0.755 | 0.667 | 0.844 | 127.5 | 0.716 | 0.850 | 0.571 | <0.001 | <0.001 |
| 9 | Cholesterol, VLDL | 0.473 | 0.366 | 0.579 | 1.3 | 0.517 | 0.150 | 0.911 | 0.613 | 1.000 |
| 10 | Free Cholesterol, HDL | 0.603 | 0.499 | 0.707 | 13.6 | 0.612 | 0.950 | 0.250 | 0.056 | 1.000 |
| 11 | Free Cholesterol, IDL | 0.683 | 0.586 | 0.780 | 1.0 | 0.681 | 0.700 | 0.661 | 0.001 | 0.040 |
| 12 | Free Cholesterol, LDL | 0.802 | 0.721 | 0.883 | 34.4 | 0.767 | 0.900 | 0.625 | <0.001 | <0.001 |
| 13 | Free Cholesterol, VLDL | 0.544 | 0.438 | 0.650 | 3.7 | 0.560 | 0.733 | 0.375 | 0.414 | 1.000 |
| 14 | Phospholipids, HDL | 0.560 | 0.453 | 0.667 | 78.4 | 0.612 | 0.883 | 0.321 | 0.269 | 1.000 |
| 15 | Phospholipids, IDL | 0.681 | 0.584 | 0.779 | 4.7 | 0.664 | 0.517 | 0.821 | 0.001 | 0.049 |
| 16 | Phospholipids, LDL | 0.782 | 0.698 | 0.866 | 77.1 | 0.733 | 0.683 | 0.786 | <0.001 | <0.001 |
| 17 | Phospholipids, VLDL | 0.538 | 0.432 | 0.645 | 8.6 | 0.578 | 0.767 | 0.375 | 0.478 | 1.000 |
| 18 | Triglycerides, HDL | 0.506 | 0.397 | 0.615 | 6.7 | 0.569 | 0.817 | 0.304 | 0.916 | 1.000 |
| 19 | Triglycerides, IDL | 0.634 | 0.530 | 0.737 | 1.9 | 0.672 | 0.867 | 0.464 | 0.013 | 0.820 |
| 20 | Triglycerides, LDL | 0.674 | 0.575 | 0.772 | 3.7 | 0.664 | 0.867 | 0.446 | 0.001 | 0.079 |
| 21 | Triglycerides, VLDL | 0.566 | 0.460 | 0.671 | 18.9 | 0.586 | 0.650 | 0.518 | 0.223 | 1.000 |
| Random Forest Model | Accuracy (p-Value) | Specificity (p-Value) | Sensitivity (p-Value) | AUC (p-Value) |
|---|---|---|---|---|
| Women vs men | 0.776 (0.001) | 0.761 (0.001) | 0.780 (0.001) | 0.826 (0.001) |
| Young vs old men | 0.746 (0.001) | 0.741 (0.001) | 0.751 (0.001) | 0.810 (0.001) |
| Young vs old women | 0.716 (0.001) | 0.679 (0.002) | 0.750 (0.001) | 0.762 (0.005) |
| Men vs Women | Young Men vs Old Men | Young Women vs Old Women | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lipid/Lipoprotein | U | RF | D | C | T | U | RF | D | C | T | U | RF | D | C | T | |
| 1 | Apo-A1 HDL | * | * | * | * | 4 | * | 1 | * | 1 | ||||||
| 2 | Apo-A2 HDL | * | * | 2 | 0 | * | 1 | |||||||||
| 3 | Apo-B IDL | * | * | 2 | * | * | 2 | * | 1 | |||||||
| 4 | Apo-B LDL | 0 | * | * | * | 3 | * | * | * | 3 | ||||||
| 5 | Apo-B VLDL | * | 1 | * | * | 2 | 0 | |||||||||
| 6 | Cholesterol HDL | * | * | 2 | * | 1 | * | 1 | ||||||||
| 7 | Cholesterol IDL | * | * | 2 | * | 1 | 0 | |||||||||
| 8 | Cholesterol LDL | 0 | 0 | * | 1 | |||||||||||
| 9 | Cholesterol VLDL | * | 1 | * | * | 2 | 0 | |||||||||
| 10 | Free cholesterol HDL | * | * | * | 3 | * | * | 2 | * | 1 | ||||||
| 11 | Free cholesterol IDL | * | * | * | * | 4 | * | 1 | 0 | |||||||
| 12 | Free cholesterol LDL | * | 1 | 0 | * | * | 2 | |||||||||
| 13 | Free cholesterol VLDL | * | * | 2 | * | * | 2 | 0 | ||||||||
| 14 | Phospholipids HDL | * | * | 2 | * | 1 | * | 1 | ||||||||
| 15 | Phospholipids IDL | * | * | 2 | * | 1 | 0 | |||||||||
| 16 | Phospholipids LDL | 0 | * | 1 | * | * | * | 3 | ||||||||
| 17 | Phospholipids VLDL | * | * | 2 | * | * | * | 3 | * | 1 | ||||||
| 18 | Triglycerides HDL | * | * | 2 | * | 1 | 0 | |||||||||
| 19 | Triglycerides IDL | 0 | * | * | 2 | 0 | ||||||||||
| 20 | Triglycerides LDL | * | * | 2 | * | * | 2 | 0 | ||||||||
| 21 | Triglycerides VLDL | * | * | 2 | * | 1 | 0 | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balder, Y.; Vignoli, A.; Tenori, L.; Luchinat, C.; Saccenti, E. Exploration of Blood Lipoprotein and Lipid Fraction Profiles in Healthy Subjects through Integrated Univariate, Multivariate, and Network Analysis Reveals Association of Lipase Activity and Cholesterol Esterification with Sex and Age. Metabolites 2021, 11, 326. https://doi.org/10.3390/metabo11050326
Balder Y, Vignoli A, Tenori L, Luchinat C, Saccenti E. Exploration of Blood Lipoprotein and Lipid Fraction Profiles in Healthy Subjects through Integrated Univariate, Multivariate, and Network Analysis Reveals Association of Lipase Activity and Cholesterol Esterification with Sex and Age. Metabolites. 2021; 11(5):326. https://doi.org/10.3390/metabo11050326
Chicago/Turabian StyleBalder, Yasmijn, Alessia Vignoli, Leonardo Tenori, Claudio Luchinat, and Edoardo Saccenti. 2021. "Exploration of Blood Lipoprotein and Lipid Fraction Profiles in Healthy Subjects through Integrated Univariate, Multivariate, and Network Analysis Reveals Association of Lipase Activity and Cholesterol Esterification with Sex and Age" Metabolites 11, no. 5: 326. https://doi.org/10.3390/metabo11050326
APA StyleBalder, Y., Vignoli, A., Tenori, L., Luchinat, C., & Saccenti, E. (2021). Exploration of Blood Lipoprotein and Lipid Fraction Profiles in Healthy Subjects through Integrated Univariate, Multivariate, and Network Analysis Reveals Association of Lipase Activity and Cholesterol Esterification with Sex and Age. Metabolites, 11(5), 326. https://doi.org/10.3390/metabo11050326









