Abstract
Fatty acid desaturase 12 (FAD12) is a key enzyme in fatty acid biosynthesis, responsible for converting oleic acid to linoleic acid through desaturase activity. Euglena gracilis (Euglena) is an emerging platform for the industrial production of various metabolites, including lipids. However, a comprehensive understanding of Euglena’s fatty acid biosynthesis pathways remains incomplete, posing a significant barrier to the commercialization of Euglena bioproducts. To address this gap, we employed a bioinformatics approach to identify a Euglena gracilis FAD12 (Eg FAD12). We analyzed the evolutionary relationship of Eg FAD12 with its homologs from other organisms and revealed that the three canonical histidine box motifs are conserved among FAD12s. To characterize EgFAD12, we cloned it into the pEAQ-hyperstrans vector and overexpressed it in Nicotiana benthamiana to take advantage of its endogenous fatty acid pool, which could act as a substrate. The heterologous expression of FAD12 in N. benthamiana led to an increased linoleic acid content, demonstrating the suspected desaturase activity. To further confirm the function of Eg FAD12, we performed CRISPR-Cas9-mediated knockout of Eg FAD12 in Euglena, which resulted in a drastic reduction in linoleic acid (C18:2) without compromising biomass yield or lipid content. This work advances our understanding of fatty acid biosynthesis in Euglena and will aid in its adoption as a platform for producing customized lipids.
1. Introduction
Microorganisms like bacteria, yeasts, fungi, and microalgae produce microbial lipids. They are like traditional plant and animal lipids but offer several benefits when considering them as a lipid source. This includes (i) reduced land and water need, (ii) efficient labour, (iii) climate change resiliency, (iv) faster production cycles, and (v) scalability. Fatty acids (FA) are integral components of lipids and play a substantial role in defining their physical and chemical properties. This, in turn, influences the industrial use of such lipids [1]. As such, developing methods to modify the FA composition of microbial lipids is central, enabling the customization of lipids for specific industrial purposes. This strategy can enhance the applications of microbial lipids, making them more economically viable and attractive for commercial use [2].
Euglena gracilis (Euglena) is an adaptable microalgae capable of growing phototrophically, heterotrophically, or mixotrophically on a variety of substrates and in harsh environments [3,4,5]. Euglena can produce a wide range of commercially relevant bioproducts such as amino acids, pro(vitamins), lipids, and the immunogenic glycopolymer paramylon [1,6]. This makes Euglena an attractive biological platform for food, pharmaceutical, and fuel industries. Besides this, it also has the potential to be used as a biological agent to treat wastewater for sustainable management [7] or heavy metal contaminated sites [8]. There are studies demonstrating its medicinal properties, such as wound healing [9], next-generation prebiotics [10] and a booster of the immune system by activating natural killer cells [11]. In addition, there is a growing interest in its use as a producer of sustainable biofuel [12,13].
Euglena can accumulate lipids comprising up to 37% of its dry weight, with a FA profile that varies considerably depending on environmental growth conditions [14,15]. Among these lipids, oleic acid, linoleic acid, and stearic acid are of particular interest due to their functional applications in biofuels, cosmetics, and nutritional products [16,17]. Despite its industrial potential, relatively little effort has been directed toward targeted modifications of Euglena FA composition to enrich specific FAs [18]. A key limitation is the incomplete understanding of FA biosynthetic pathways in Euglena. Although some enzymes involved in this process have been characterized, such as FA desaturases FAD5 and FAD8 [19,20] many components of these pathways remain unidentified.
A complete understanding of the FA biosynthetic pathway, along with identification of the genes involved, will enable the engineering of Euglena strains to produce lipids with a tailored fatty acid profile, often referred to as customized lipids. Therefore, in this study, we identified the FA desaturase 12 (FAD12) enzyme, which converts oleic acid to linoleic acid in Euglena. We employed a bioinformatics approach to identify a candidate gene, and its function was investigated by overexpression in Nicotiana benthamiana and targeted gene knockout. As expected, EG FAD12 overexpression in tobacco increased linoleic acid content, while CRISPR-mediated knockout in Euglena resulted in a drastic reduction in linoleic acid. This is the first use of CRISPR in Euglena to understand FA biosynthesis and to characterize a novel FAD12 enzyme (Figure 1A). The identification of the enzyme using CRISPR builds upon our knowledge of FA biochemistry in Euglena and will help open the door for customized lipid production.
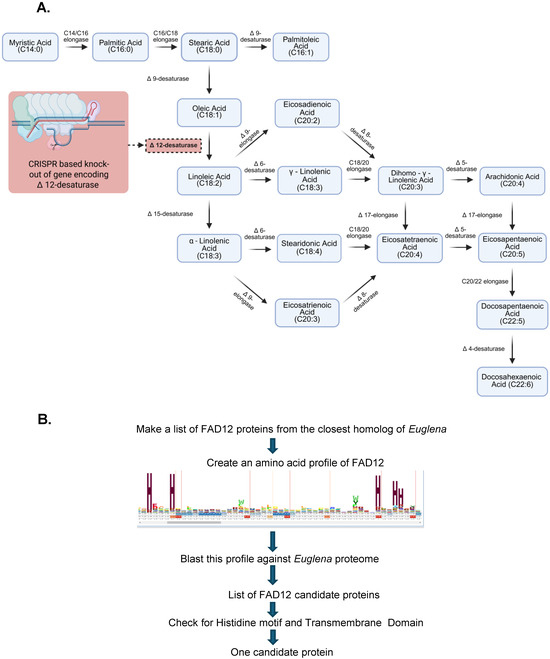
Figure 1.
Fatty acid biosynthesis pathway and the approach to study in Euglena. (A) Fatty acid desaturase Δ12 is knocked out by CRISPR-Cas9 in this study. The solid arrow lines indicate the direction of fatty acid synthesis, and the dashed arrow indicates the site of CRISPR intervention (brick red box). (B) Flowchart of bioinformatic identification of FAD12 genes in Euglena gracilis. The high-resolution image of the FAD12 profile is in Figure S1.
2. Methods
- Organisms and growth conditions
Euglena gracilis strain Z (UTEX 753) was cultured in sterilized MEGM medium (sodium acetate trihydrate 1 g/L, lab-lemco powder 1 g/L, tryptone 2 g/L, yeast extract 2 g/L, calcium chloride 1 g/L, and glucose 15 g/L) in heterotrophic conditions as previously described [21]. The cells were grown at 28 °C in a shaking incubator in the dark for a week. Then, the cells were harvested by centrifugation and used for downstream process analyses of biomass, dry mass, and lipid.
- 2.
- Bioinformatics Identification of the FAD12 gene in Euglena gracilis
A list of FAD12 proteins from 19 different microorganisms and plants was obtained from the NCBI. The HMMER 3.4 software was then used to analyze the data [22]. First, a unique amino acid profile (hmm profile) of fatty acid desaturase protein was generated by hmmbuild, which uses the principle of the Hidden Markov Model (Figure S1). Using the hmmsearch, the hmm profile was blasted against the translatome of publicly available RNAseq of E. gracilis [23]. All the hits were scanned by hmmscan for the presence of all three histidine boxes in this order (HCEGH, HXXHH, HVXHH, where X is any amino acid). Next, the transmembrane domains, which are the features of FAD12, were computationally predicted using DeepTMHMM-1.0 [24]. Finally, we obtained one candidate protein that fits the previously described profile of FAD12. (Figure S2).
- 3.
- FAD12 gene synthesis and plasmid construction for bacterial transformation
The native FAD12 transcript from Euglena was too long to synthesize by RNA isolation and cDNA synthesis, so we opted to synthesize the gene in-house using BioXP 3250 DNA printer. The synthetic gene sequence was verified by sequencing and agarose gel electrophoresis. The DNA fragment was cloned into the pEAQ-HT vector between Agel and Xmal by the restriction digestion method. The plasmid harbouring the FAD12 gene was expressed in E.coli (DH5a) by the Heat Shock method for amplification. Finally, the plasmid was transformed into Agrobacterium tumefaciens (GV31010) by electroporation.
- 4.
- Heterologous expression of E.G FAD12 in Tobacco
The transient expression of EG FAD12 in Tobacco (Nicotiana benthamiana) was performed as previously described [25]. Briefly, the Agrobacterium harbouring the empty plasmid (control) and the plasmid with the FAD12 coding sequence under the 35S promoter were cultured in LB broth at 28 °C with appropriate antibiotics. The bacterial cultures were centrifuged and resuspended in the infiltration buffer (5 mM 4–Morpholineethanesulfonic acid (MES), 50 µM acetosyringone, and 5 mM MgSO4. Then, it was injected into the ventral side of tobacco leaves. The injected portion of leaves was collected after 3 days for lipid extraction and downstream analysis.
- 5.
- Amino acid alignment and Phylogenetic Tree Analysis
The amino acid sequences of FAD12s from nineteen organisms were retrieved from NCBI. The amino acid alignment of twenty FAD12s and their phylogenetic analysis were conducted in the COBALT program in NCBI [26]. The COBALT works by finding a cluster of pairwise constraints from database searches, sequence similarity, and user input. Then, it merges these pairwise constraints and integrates them into a progressive multiple alignment. The image of the phylogenetic analysis tree was generated from the COBALT program. The amino acid sequences from different organisms with high similarity are clustered in the same node. Those in different nodes exhibit lower similarity and indicate greater evolutionary distance.
- 6.
- Biomass and Lipid Quantification
To determine the Euglena gracilis dry biomass weight, 10 mL of culture broth was centrifuged for 15 min at 2500 rpm. The supernatant was discarded, and the wet cells were washed twice with deionized water (20 mL), dried overnight at 60 °C, and weighed. Lipid was separated using a single-step procedure as previously reported [27]. A 100 mg dried biomass sample was resuspended in a 2:1 ratio of chloroform: methanol. To achieve this, 5.26 mL of chloroform and 2.63 mL of methanol were added, and the tube was manually shaken strongly until the biomass was dispersed entirely. Subsequently, 2.11 mL of 0.73% NaCl solution was added to produce a 2:1:0.8 (v/v/v) system, bringing the total volume to 10 mL in a 15 mL Falcon tube. The mixture was centrifuged at 4000 rpm for 10 min to separate the phases. The bottom chloroform layer containing the isolated lipids was cautiously collected in a pre-weighted vial. The extraction procedure was repeated with a fresh 2:1:0.8 solvent system to increase the total lipid content. The combined chloroform layers were dried under vacuum to remove the solvent. Lipid content was determined gravimetrically from the weight difference.
The lipid from the tobacco plant was extracted as described [28]. The tobacco leaves were freeze-dried and ground into a fine powder. The solvent of a chloroform: methanol mixture (2:1 v:v) was added to the powder, and rotated in a shaker for 4 min. Then, 1:3 v:v NaCl (0.1 M) was added to the mixture, and rotated in a shaker for 4 min. Next, it was centrifuged for 5 min at 14,000 g, and a lower lipid phase was collected and dried with N2 gas.
- 7.
- Fatty acid analysis by Gas Chromatography Flame Ionization Detector (GC-FID)
The Fatty acid methyl esters (FAMEs) of the lipids were prepared using the boron trifluoride (BF3) methylation method as previously reported, with slight modifications [29]. Briefly, glyceryl triundecanoate dissolved in heptane was added to the lipid samples as an internal standard. Heptane was then evaporated under a moderate stream of nitrogen (N2), leaving the internal standard in the sample. Next, the samples were hydrolyzed with alcoholic sodium hydroxide (NaOH), followed by methylation using BF3-methanol solution. FAMEs were extracted with heptane, and a saturated sodium chloride (NaCl) solution was added to assist phase separation. The top heptane layer containing FAMEs was transferred into a new vial and dried over anhydrous sodium sulphate. Gas chromatography-flame ionization detection (GC-FID) was performed using an Agilent 7890B system equipped with an SP2560 column (100 m × 0.25 mm × 0.2 μm) (Santa Clara, CA, USA). Injection volume was 1 μL with a split ratio of 50:1 and a flow rate of 1.2 mL/min. Oven temperature was programmed to hold at 100 °C for 5 min, ramp at 4 °C/min to 240 °C, and hold for 15 min. Peak identification and quantification were carried out using Agilent Technologies 1100 ChemStation software.
- 8.
- Guide RNA (gRNA) design and CRISPR knockout of FAD12
The gRNA for FAD12 was designed using Integrated DNA Technologies software, and the CRISPR knockout experiment was conducted as described [30]. Briefly, crRNA (100 µM) and tracrRNA (100 µM) were mixed to form a complex, and then Cas9 (62 µM) was added. The CRISPR/Cas9 ribonucleoproteins (RNPs) complex was mixed with Euglena cells (1 M), and electroporation was conducted. The cells were grown for 24 h at 28 °C, and single cells were selected for further growth and analysis. The knockout of FAD12 was confirmed by DNA sequencing.
3. Results
3.1. Bioinformatic Identification of FAD12 in Euglena gracilis
To identify FAD12, we used a bioinformatics approach based on sequence homology (Figure 1B). First, we compiled a list of known FAD12 proteins from different organisms available in the National Center for Biotechnology Information (NCBI) database. Then, an amino acid profile of FAD12 (profile HMM, Figure S1) was generated by Hidden Markov Model [22]. The HMM profile displays the most likely amino acid at each position of the protein, which is vital for identifying conserved and non-conserved regions. Since there were no publicly available Euglena proteome data, we took advantage of publicly available Euglena transcriptome data [23]. Using this data, we created a translated protein database of all six possible reading frames (3 Forward, 3 Reverse). The FAD12 HMM profile was searched against the translated protein data to identify candidate genes. Obtained candidates were scanned for the three conserved histidine motifs in this order (HECGH, HXXHH, and HVXHH (X = any amino acids)). Finally, we selected one candidate (532 amino acids, Table S1) with three histidine boxes and multiple transmembrane domains, which are the peculiar characteristics of FAD enzymes. The corresponding FAD12 transcriptome sequence was aligned with our in-house Euglena genome data to identify the FAD12 genomic sequence (Table S2).
3.2. Amino Acid Alignment and Phylogenetic Analysis of FAD12
The amino acid sequences of FAD12 from Euglena gracilis and other organisms were aligned using the Constraint-based Multiple Alignment Tool (COBALT), NCBI [26]. The alignment focuses on conserved domains and local sequence similarity. The compact view alignment with conservative setting 2 bits is shown in Figure 2. The red letter indicates conserved regions, while blue indicates less conserved regions with no gaps. The FAD12 amino acids are vastly conserved across different organisms. The histidine boxes HECGH, HXXHH, HVXHH (X = any amino acids) are found in EG FAD12 along with other organisms. All the FAD12s are below 500 AA.
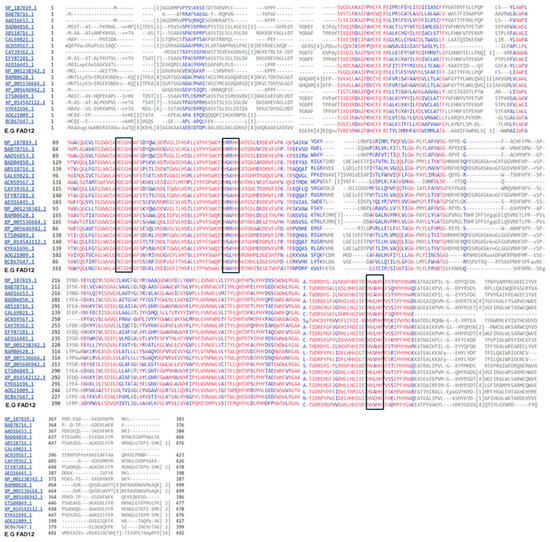
Figure 2.
Amino acids alignment of FAD12 from different organisms. Red color indicates highly conservative regions, and blue color indicates less conserved regions with no gaps. The three histidine boxes are highlighted with black boxes.
To study the evolutionary relationship between EG FAD12 and others, a phylogenetic tree was constructed using the amino acid sequence by the Neighbour-joining method, using the COBALT program (Figure 3). EG FAD12 falls into the same category as Phytophthora. It is evolutionarily nearest to Ascomycete fungi and farthest from Green Algae. FAD12 is highly conserved across species in different kingdoms, so there is high similarity (70–88%) among them.
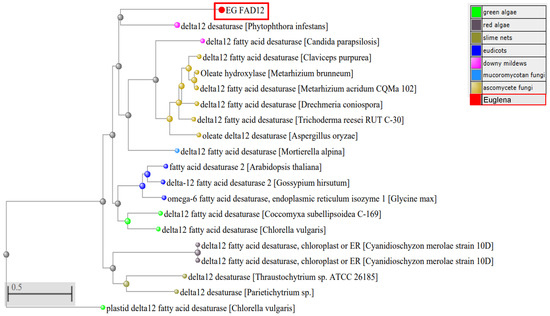
Figure 3.
Phylogenetic analysis of FAD12 from different organisms. The phylogenetic tree was created using COBALT, max seq difference 0.85, Distance- Grishin (Protein). FAD12 from different classes of organisms are shown in different color. The EG FAD12 is highlighted in red color.
3.3. Cloning of EG FAD12 and Heterologous Expression in Tobacco
Once the coding sequence of EG FAD12 was identified using our bioinformatics approach, we synthesized it in-house using a Codex DNA printer. Resultant DNA was cloned into the pEAQ-HT vector through the restriction digestion method and confirmed by sequencing. The size of the DNA was further confirmed using agarose gel electrophoresis (Figure 4A). The control vector and the vector harbouring FAD12 were injected into tobacco leaves, and the leaves were collected after three days for lipid extraction and analysis. Extracted lipids were methylated using the boron trifluoride (BF3) methylation method and were analyzed using GC-FID using the method as previously reported [29]. There was no difference in C18:1 and C18:2 in tobacco injected with the control vector (Figure 4B). However, C18:2 levels increased in all six tobacco plant replicates injected with the FAD12 gene sequence, showing an average rise of 33%. This increase suggests an enhanced delta-12 desaturase activity. The raw data of the lipid profile are shown in Figure 4C. The ratio of C18:2 to 18:1 in the control vector is 0.93, while it increased to 1.25 in the vector expressing FAD12. The t-test between treatments showed that C18:2 significantly increased in plants expressing FAD12 (p = 0.0048, in a two-tailed paired t-test). In addition to the changes in C18:1 and C18:2, a general upregulation of other lipids was observed in FAD12 expressing plants (Figure 4C). Notably, there were increases in upstream saturated fatty acids, C16:0 (palmitic acid) and C18:0 (stearic acid), as well as the downstream product, C18:3 (linolenic acid). This suggests that increased FAD12 activity impacts the broader fatty acid metabolic network. The increase in C18:3 may indicate that the newly synthesized C18:2 is further desaturated by endogenous tobacco enzymes. The corresponding rise in C16:0 and C18:0 could suggest a compensatory positive feedback mechanism, pulling more precursors into the pathway to accommodate for increased flux.
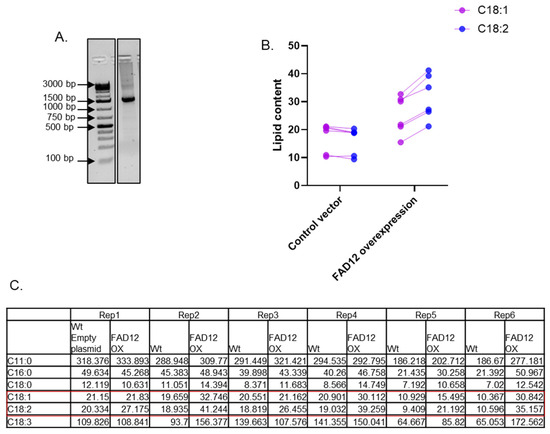
Figure 4.
Cloning of E.G FAD12 and its heterologous expression in Tobacco. (A) The coding sequence of EG FAD12 was synthesized using the Codex system and run on agarose gel to confirm the size (1476 bp), (original image Figure S3). (B) The lipid profile (C18:1 & C18:2) of tobacco injected with control vector or FD12 overexpression vector. (C) The lipid analysis data from tobacco injected with the control or FAD12 OX vector.
3.4. CRISPR-Cas9-Mediated Knockout of FAD12
To further confirm the role of the identified FAD12 candidate in the fatty acid biosynthesis pathway, we used RNP-mediated CRISPR-Cas9 technology to edit this gene in Euglena. First, the FAD12 transcript was aligned with the genomic sequence to identify the gene structure, which contains 10 introns and 11 exons (Figure 5A). Then, a guide RNA was designed in the second exon to knock out the gene (gRNA and related primers are listed in Table S3). The CRISPR-Cas9-mediated knockout was conducted as described [30]. Then, a single cell was isolated from the CRISPR knockout experiment, cultured, and sequenced to confirm the gene knockout (Figure 5B). The DNA sequencing revealed a single nucleotide deletion at position 190, which resulted in a codon shift and the appearance of multiple early stop codons (Table S4). Under the microscope, the CRISPR-edited cell line was similar in morphology and motility to that of wild-type Euglena (Figure 5C). Lipids were extracted from this gene-edited cell line and subjected to GC-FID analysis. As predicted, compared to wild type, there was a drastic reduction in C18:2 in the gene-edited cell line (Figure 5D). The t-test indicates that the C18:2 is significantly reduced in the knockout line compared to the control (p = 0.00047, paired t-test). The raw data of lipid analysis are available in Table S5. Since FAD12 catalyzes the conversion of C18:1 to C18:2, knockout of this gene prevented the formation of C18:2 in Euglena.
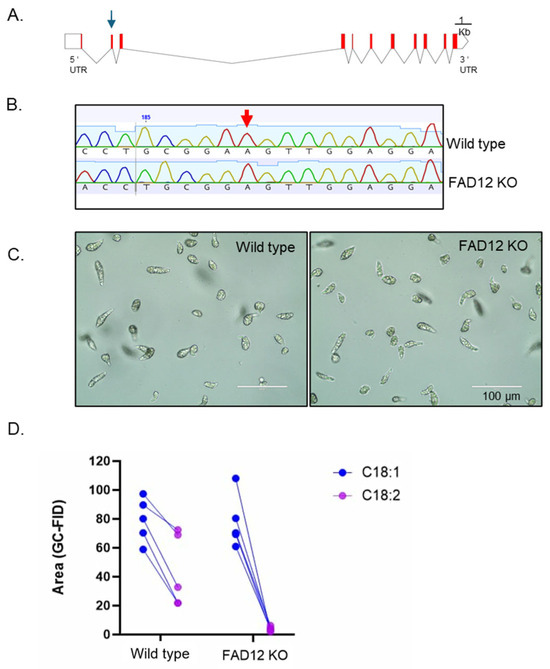
Figure 5.
CRISPR-Cas9-Mediated Knockout of FAD12 dramatically reduced linoleic acid production. (A) FAD12 gene structure showing introns (line break), exons (red box) and guideRNA location (blue arrow). (B) DNA sequencing of FAD12 knockdown Euglena showing a single nucleotide deletion (Adenosine) highlighted by a red arrow. (C) Culture of FAD12 knockdown cells selected from a single cell. (D) Lipid profile (C18:1 & C18:2) of FAD12 knockdown cells with wild-type cells.
3.5. Characterization of the FAD12 kd Cell Line for Commercial Production
The FAD12 knockout cell line was further evaluated for use in commercial production. First, Euglena cells were cultured in standard growth media to compare with the wild-type strain [21]. Both looked similar in color, texture, flavor, and aroma (Figure 6A). There was also no difference in total biomass and dry biomass between the two strains (Figure 6B,C). Although the lipid profile was different, there was no difference in total lipid content (Figure 6D). Finally, it was tested for glucose consumption, and both strains exhibited a similar pattern of glucose consumption over time (Figure 6E). The t-test shows no significant differences in total biomass, dry biomass, lipid content and glucose consumption rate between wild type and FAD12 KO Euglena cell line (p = 0.42, 0.66, 0.18 and 0.79, respectively). Overall, there was no yield penalty for biomass or lipids in the FAD12 knockout strain.
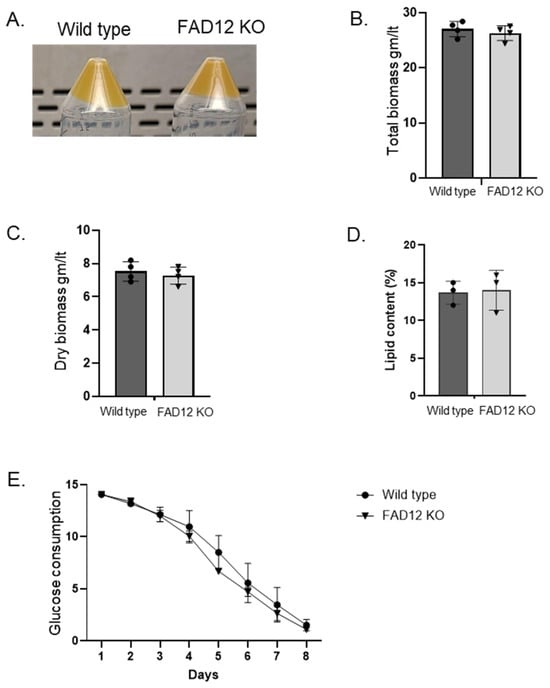
Figure 6.
Characterization of FAD12 knockdown Euglena. (A): Biomass of wildtype and FAD12Δ Euglena. (B): Total biomass comparison between wildtype and FAD12Δ Euglena. (C): Dry biomass comparison between wildtype and FAD12Δ Euglena. (D): Lipid content between wildtype and FAD12Δ Euglena. (E): Glucose consumption between wildtype and FAD12Δ Euglena.
4. Discussion
Euglena has emerged as a promising platform for producing high-value compounds, including tailored lipids and metabolites utilized in cosmetic, food, and nutraceutical applications [31]. Over the last decade, the major hindrance to studying Euglena has been the unavailability of high-quality genomic data. However, there have been some notable accomplishments recently, such as the publication of a draft genome, transcriptome, and proteome [32]. Also, a chromosome-level genome assembly of Euglena was published last year [33]. The availability of these next-gen data is expected to expedite the understanding of several major biochemical pathways, including the fatty acid biosynthesis pathway.
To enhance lipid production in oleaginous microorganisms, including Euglena, researchers have employed both endogenous (such as pathway engineering) and exogenous strategies (including media optimization and external elicitor applications) [34,35]. There has also been an attempt to co-culture Euglena with other microalgae to boost biomass and lipid production [36]. Recent efforts have particularly focused on increasing the production of specific fatty acid types in oil-producing microbes to generate customized lipids with significant industrial potential [37,38,39]. The fatty acid desaturase FAD12 represents a critical enzyme in the lipid biosynthesis pathway of oleaginous microorganisms, catalyzing the conversion of oleic acid (C18:1) to linoleic acid (C18:2). Understanding and manipulating this enzyme’s activity is of considerable interest, as it enables redirection of carbon flux to either increase or decrease specific fatty acid production (C18:1 and C18:2), ultimately facilitating customized lipid synthesis.
In this study, we employed bioinformatics, forward, and reverse genetics approaches to identify and characterize the FAD12 enzyme, a key regulator of fatty acid biosynthesis in Euglena. All characterized FAD12 proteins, including that in Euglena, possess three essential histidine domains that are crucial for desaturase activity (Figure S2). These domains interact with iron ligands to catalyze the desaturation reaction, as previously documented [40]. The domain analysis using DeepTMHMM-1.0 [24] predicted that Euglena FAD12 (EGFAD12) contains five transmembrane domains (Figure S4), a characteristic feature shared among desaturase enzymes across various organisms. The subcellular localization prediction using DeepLoc2.0 [41] indicated that EGFAD12 localizes to the endoplasmic reticulum (Table S6). This localization pattern is consistent with FAD12 enzymes from other organisms, which typically reside in the endoplasmic reticulum, the Golgi apparatus, or plastids [19,20,42]. The FAD12 ER localization is expected, as the ER is one of the primary cellular sites for lipid biosynthesis.
We verified the FAD12 activity by overexpressing it in tobacco and found a 33% increase in FAD12 compared to the control vector. The FAD12 expression may also have affected lipids like C18:0, C18:1 and others and affected the whole lipid metabolic flux. Additionally, as previously indicated, its overexpression in tobacco not only increased C18:2 but also could have caused a broader shift in the lipid profile, including increases in C16:0, C18:0, and C18:3 (Figure 4C). This suggests that the introduction of Eg FAD12 activity stimulates a pull on the entire biosynthetic pathway, possibly through a positive feedback loop, and that its product (C18:2) is available for further processing by native desaturases into C18:3. To further validate the function of this newly identified gene, we performed a knockout of FAD12 in Euglena using CRISPR-Cas9. As hypothesized, the linoleic acid level was increased after overexpression, and the oleic acid level was increased after knockout. This complementary approach of functional gene study shows that our FAD12 candidate gene has desaturase activity. To improve its activity in the future, researchers can take the path of enzyme engineering; e.g., one can add an extra histidine motif, or replace the iron ligand with copper or other metals. Furthermore, one could optimize the enzyme expression in specific organelles for easy harvesting and downstream processing, which are critical for commercial success. All these desired enzyme traits can be obtained by directed evolution, rational design, computational design or de novo design. Since the FAD12 KO cell line does not incur a yield penalty from altering its lipid profile, it can be further utilized for knockout or knocking in other genes and pathways.
This paper demonstrates the successful CRISPR-Cas9-mediated knockout of EgFAD12 and establishes Euglena gracilis as a promising platform for rational lipid engineering and underscores the potential of this organism for industrial bioproduction. Redirecting fatty acid flux toward monounsaturated oleic acid (C18:1) is advantageous for applications such as biodiesel, oleochemicals, and food-grade oils, where higher oxidative stability and improved properties are desirable. Beyond single-gene knockouts, future work could focus on developing more versatile genetic tools to enable multiplex or sequential editing, thereby facilitating the genetic stacking of complementary traits to optimize lipid yield and composition. Although targeted gene upregulation remains challenging in Euglena, the creation of tunable expression systems and promoter libraries would enhance the ability to fine-tune lipid biosynthetic pathways. Given that Euglena accumulates wax esters under anaerobic conditions, strategies such as disrupting wax ester synthase could redirect carbon flux away from storage waxes and toward triacylglycerol or other neutral lipid pools. Integrating such genetic interventions with process optimization under heterotrophic and mixotrophic growth conditions will be necessary for achieving scalable, high-titer lipid production. This study provides a foundation for building an advanced genetic toolkit in Euglena and positions this organism as a viable chassis for the sustainable production of customized fatty acids and lipid-derived bioproducts. Overall, these findings are expected to have significant implications for the biotechnological production of valuable lipids in microalgae for industrial applications. In particular, the production of Euglena-derived lipids enriched in oleic acid is industrially attractive due to oleic acid’s enhanced oxidative stability and its wide-ranging applications in food, cosmetics, and bio-based materials.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/bioresourbioprod1020008/s1. Supplementary Figure S1: Amino acid profile (HMM) of Fatty acid desaturase. Supplementary Figure S2: Computational prediction of the 3D structure of EG FAD12. Supplementary Figure S3: The original DNA gel electrophoresis image showing different fatty acid desaturase enzymes from Euglena. Supplementary Figure S4: Prediction of transmembrane domains (TMD) in EG FAD12 by TMHMM-2.0. Supplementary Table S1: EG FAD12 amino acids. Supplementary Table S2: Genomic sequence of EG FAD12. Supplementary Table S3: List of Oligos. Supplementary Table S4: CRISPR genome and protein sequence. Supplementary Table S5: FAD12 CRISPR line and lipid profile. Supplementary Table S6: Prediction of subcellular localization of EG FAD12.
Author Contributions
Conceptualization, S.C.F., R.J.N.E., B.K.U. and R.K.T.; methodology, S.C.F., R.K.T. and B.K.U.; investigation, R.K.T. and B.K.U.; resources, S.C.F. and R.J.N.E.; data curation, R.K.T. and S.C.F.; writing—original draft preparation, R.K.T.; writing, review, and editing, R.K.T., B.K.U., R.J.N.E. and S.C.F.; supervision, R.J.N.E. and S.C.F.; project administration, R.J.N.E. and S.C.F.; funding acquisition, R.J.N.E. and S.C.F. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by MITACS to R.J.N.E. and Noblegen.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data from this article are available in the Supplementary Files.
Acknowledgments
This work was funded by Noblegen and MITACS. Raj Kumar Thapa and Bijaya Kumar Uprety were recipients of the MITACS Accelerate Industrial Post-doctoral award. We would like to thank the Cell & Systems Biology team from Noblegen and the Emery lab for experimental assistance and helpful discussions.
Conflicts of Interest
All Authors were employed by the company Noblegen Inc. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Uprety, B.K.; Morrison, E.N.; Emery, R.J.N.; Farrow, S.C. Customizing lipids from oleaginous microbes: Leveraging exogenous and endogenous approaches. Trends Biotechnol. 2022, 40, 482–508. [Google Scholar] [CrossRef] [PubMed]
- Ochsenreither, K.; Glück, C.; Stressler, T.; Fischer, L.; Syldatk, C. Production Strategies and Applications of Microbial Single Cell Oils. Front. Microbiol. 2016, 7, 2016. [Google Scholar] [CrossRef]
- Farjallah; Fillion, M.; Guéguen, C. Metabolic responses of Euglena gracilis under photoheterotrophic and heterotrophic conditions. Protist 2024, 175, 126035. [Google Scholar] [CrossRef]
- Kuhne, A.M.; Morrison, E.N.; Sultana, T.; Kisiala, A.B.; Horlock-Roberts, K.; Noble, A. Cultivation of heterotrophic Euglena gracilis: The effects of recycled media on culture growth and associations with growth-regulating phytohormone profiles. J. Appl. Phycol. 2023, 35, 2161–2175. [Google Scholar] [CrossRef]
- Lewis; Guéguen, C. How growth conditions of Euglena gracilis cells influence cellular composition as evidenced by Fourier transform infrared spectroscopy and direct infusion high-resolution mass spectrometry. J. Appl. Phycol. 2020, 32, 153–163. [Google Scholar] [CrossRef]
- Rodríguez-Zavala, J.S.; Ortiz-Cruz, M.A.; Mendoza-Hernández, G.; Moreno-Sánchez, R. Increased synthesis of α-tocopherol, paramylon and tyrosine by Euglena gracilis under conditions of high biomass production. J. Appl. Microbiol. 2010, 109, 2160–2172. [Google Scholar] [CrossRef]
- Mahapatra, D.M.; Chanakya, H.N.; Ramachandra, T.V. Euglena sp. as a suitable source of lipids for potential use as biofuel and sustainable wastewater treatment. J. Appl. Phycol. 2013, 25, 855–865. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Nguyen, Q.T.; Dang, D.H.; Emery, R.J.N. Phytohormones enhance heavy metal responses in Euglena gracilis: Evidence from uptake of Ni, Pb and Cd and linkages to hormonomic and metabolomic dynamics. Environ. Pollut. 2023, 320, 121094. [Google Scholar] [CrossRef]
- Ko, Y.; Baek, H.; Hwang, J.-H.; Kim, Y.; Lim, K.-M.; Kim, J.; Kim, J.W. Nonanimal Euglena gracilis-Derived Extracellular Vesicles Enhance Skin-Regenerative Wound Healing. Adv. Mater. Interfaces 2023, 10, 2202255. [Google Scholar] [CrossRef]
- Dai, J.; He, J.; Chen, Z.; Qin, H.; Du, M.; Lei, A.; Zhao, L.; Wang, J. Euglena gracilis Promotes Lactobacillus Growth and Antioxidants Accumulation as a Potential Next-Generation Prebiotic. Front. Nutr. 2022, 9, 864565. [Google Scholar] [CrossRef]
- Park, S.-y.; Kim, K.J.; Jo, S.M.; Jeon, J.-Y.; Kim, B.-R.; Hwang, J.E.; Kim, J.Y. Euglena gracilis (Euglena) powder supplementation enhanced immune function through natural killer cell activity in apparently healthy participants: A randomized, double-blind, placebo-controlled trial. Nutr. Res. 2023, 119, 90–97. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, Y.; Zhang, H.; Qin, H.; He, J.; Zheng, Z.; Zhao, L.; Lei, A.; Wang, J. Evaluation of Euglena gracilis 815 as a New Candidate for Biodiesel Production. Front. Bioeng. Biotechnol. 2022, 10, 827513. [Google Scholar] [CrossRef]
- Kim, S.; Im, H.; Yu, J.; Kim, K.; Kim, M.; Lee, T. Biofuel production from Euglena: Current status and techno-economic perspectives. Bioresour. Technol. 2023, 371, 128582. [Google Scholar] [CrossRef]
- Shibata, S.; Arimura, S.I.; Ishikawa, T.; Awai, K. Alterations of Membrane Lipid Content Correlated With Chloroplast and Mitochondria Development in Euglena gracilis. Front. Plant Sci. 2018, 9, 370. [Google Scholar] [CrossRef]
- Wang, Y.; Seppänen-Laakso, T.; Rischer, H.; Wiebe, M.G. Euglena gracilis growth and cell composition under different temperature, light and trophic conditions. PLoS ONE 2018, 13, e0195329. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.; Cordeiro, N. Microalgae as Sustainable Biofactories to Produce High-Value Lipids: Biodiversity, Exploitation, and Biotechnological Applications. Mar. Drugs 2021, 19, 573. [Google Scholar] [CrossRef]
- Khoo, K.S.; Ahmad, I.; Chew, K.W.; Iwamoto, K.; Bhatnagar, A.; Show, P.L. Enhanced microalgal lipid production for biofuel using different strategies including genetic modification of microalgae: A review. Prog. Energy Combust. Sci. 2023, 96, 101071. [Google Scholar] [CrossRef]
- Bedard, S.; Roxborough, E.; O’Neill, E.; Mangal, V. The biomolecules of Euglena gracilis: Harnessing biology for natural solutions to future problems. Protist 2024, 175, 126044. [Google Scholar] [CrossRef]
- Pollak, D.W.; Bostick, M.W.; Yoon, H.; Wang, J.; Hollerbach, D.H.; He, H.; Damude, H.G.; Zhang, H.; Yadav, N.S.; Hong, S.-P.; et al. Isolation of a Δ5 Desaturase Gene from Euglena gracilis and Functional Dissection of Its HPGG and HDASH Motifs. Lipids 2012, 47, 913–926. [Google Scholar] [CrossRef] [PubMed]
- Wallis, J.G.; Browse, J. The Δ8-Desaturase of Euglena gracilis: An Alternate Pathway for Synthesis of 20-Carbon Polyunsaturated Fatty Acids. Arch. Biochem. Biophys. 1999, 365, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.R.; Morrison, E.N.; Noble, A.J.; Farrow, S.C. A simple and effective cryopreservation protocol for the industrially important and model organism, Euglena gracilis. STAR Protoc. 2022, 3, 101043. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef]
- Mangal, V.; Donaldson, M.E.; Lewis, A.; Saville, B.J.; Guéguen, C. Identifying Euglena gracilis Metabolic and Transcriptomic Adaptations in Response to Mercury Stress. Front. Environ. Sci. 2022, 10, 836732. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Thapa, R.K.; Tian, G.; Lu, Q.S.M.; Yu, Y.; Shu, J.; Chen, C.; Song, J.; Xie, X.; Shan, B.; Nguyen, V.; et al. NUCLEOPORIN1 mediates proteasome-based degradation of ABI5 to regulate Arabidopsisseed germination. bioRxiv 2023. [Google Scholar] [CrossRef]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef]
- Axelsson, M.; Gentili, F. A Single-Step Method for Rapid Extraction of Total Lipids from Green Microalgae. PLoS ONE 2014, 9, e89643. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, K.B.; Taylor, M.C.; Zhou, X.-R.; Vanhercke, T.; Wood, C.C.; Blanchard, C.L.; Singh, S.P.; Petrie, J.R. Metabolic engineering of medium-chain fatty acid biosynthesis in Nicotiana benthamiana plant leaf lipids. Front. Plant Sci. 2015, 6, 164. [Google Scholar] [CrossRef] [PubMed]
- Araujo, P.; Nguyen, T.-T.; Frøyland, L.; Wang, J.; Kang, J.X. Evaluation of a rapid method for the quantitative analysis of fatty acids in various matrices. J. Chromatogr. A 2008, 1212, 106–113. [Google Scholar] [CrossRef]
- Nomura, T.; Yoshikawa, M.; Suzuki, K.; Mochida, K. Highly Efficient CRISPR-Associated Protein 9 Ribonucleoprotein-Based Genome Editing in Euglena gracilis. STAR Protoc. 2020, 1, 100023. [Google Scholar] [CrossRef]
- Ebenezer, T.E.; Low, R.S.; O’Neill, E.C.; Huang, I.; DeSimone, A.; Farrow, S.C.; Field, R.A.; Ginger, M.L.; Guerrero, S.A.; Hammond, M.; et al. Euglena International Network (EIN): Driving euglenoid biotechnology for the benefit of a challenged world. Biol. Open 2022, 11, bio059561. [Google Scholar] [CrossRef]
- Ebenezer, T.E.; Zoltner, M.; Burrell, A.; Nenarokova, A.; Novák Vanclová, A.M.G.; Prasad, B.; Soukal, P.; Santana-Molina, C.; O’Neill, E.; Nankissoor, N.N.; et al. Transcriptome, proteome and draft genome of Euglena gracilis. BMC Biol. 2019, 17, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Dong, Y.; Duan, S.; He, J.; Qin, H.; Bian, C.; Chen, Z.; Liu, C.; Zheng, C.; Du, M.; et al. A chromosome-level genome assembly for the paramylon-producing microalga Euglena gracilis. Sci. Data 2024, 11, 780. [Google Scholar] [CrossRef] [PubMed]
- Matouk, A.M.; Abu-Elreesh, G.M.; Abdel-Rahman, M.A.; Desouky, S.E.; Hashem, A.H. Response surface methodology and repeated-batch fermentation strategies for enhancing lipid production from marine oleaginous Candida parapsilosis Y19 using orange peel waste. Microb. Cell Factories 2025, 24, 16. [Google Scholar] [CrossRef]
- Uprety, B.K.; Rakshit, S.K. Use of Essential Oils From Various Plants to Change the Fatty Acids Profiles of Lipids Obtained From Oleaginous Yeasts. J. Am. Oil Chem. Soc. 2018, 95, 135–148. [Google Scholar] [CrossRef]
- Toyama, T.; Hanaoka, T.; Yamada, K.; Suzuki, K.; Tanaka, Y.; Morikawa, M.; Mori, K. Enhanced production of biomass and lipids by Euglena gracilis via co-culturing with a microalga growth-promoting bacterium, Emticicia sp. EG3. Biotechnol. Biofuels 2019, 12, 205. [Google Scholar] [CrossRef]
- Du, F.; Zhang, F.; Hang, Y.; Jing, H.; Zheng, Y.; Ma, W.; Sun, X.; Huang, H. Advances in production of customized functional unsaturated fatty acids in Yarrowia lipolytica. Agric. Prod. Process. Storage 2025, 1, 14. [Google Scholar] [CrossRef]
- Duman-Özdamar, Z.E.; Martins dos Santos, V.A.P.; Hugenholtz, J.; Suarez-Diez, M. Tailoring and optimizing fatty acid production by oleaginous yeasts through the systematic exploration of their physiological fitness. Microb. Cell Factories 2022, 21, 228. [Google Scholar] [CrossRef]
- Wang, J.; Yu, X.; Wang, K.; Lin, L.; Liu, H.-H.; Ledesma-Amaro, R.; Ji, X.-J. Reprogramming the fatty acid metabolism of Yarrowia lipolytica to produce the customized omega-6 polyunsaturated fatty acids. Bioresour. Technol. 2023, 383, 129231. [Google Scholar] [CrossRef] [PubMed]
- Douglas, R.G.; Rousseau, D.L. Hydrogen bonding of iron-coordinated histidine in heme proteins. J. Struct. Biol. 1992, 109, 13–17. [Google Scholar] [CrossRef]
- Thumuluri, V.; Almagro Armenteros, J.J.; Johansen, A.R.; Nielsen, H.; Winther, O. DeepLoc 2.0: Multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 2022, 50, W228–W234. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.A.; Choudhury, A.R.; Kancharla, P.K.; Arumugam, N. The FAD2 Gene in Plants: Occurrence, Regulation, and Role. Front. Plant Sci. 2017, 8, 1789. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).