1. Introduction
Dermatophilosis is a contagious zoonotic skin disease caused by the Gram-positive actinomycete
Dermatophilus congolensis, primarily affecting cattle but also other domestic and wild animals, and occasionally humans [
1]. The disease manifests as exudative dermatitis with characteristic scab formation, crusting, and hair loss, leading to significant animal welfare concerns and economic losses through reduced growth, milk yield, and hide quality [
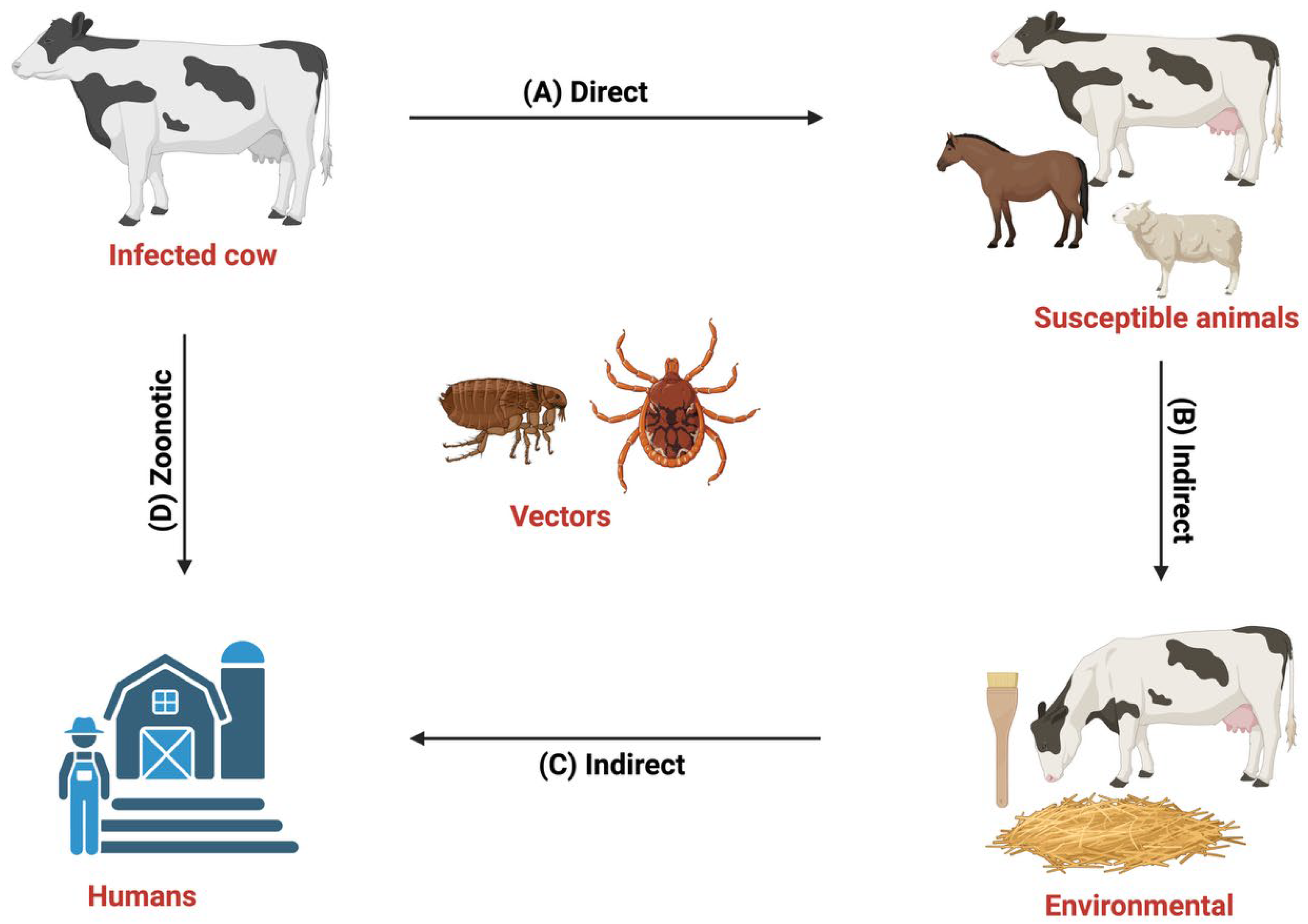
2]. Transmission occurs via direct contact with infected animals or indirectly through fomites and insect vectors [
3], with prevalence highest in warm, humid tropical and subtropical regions where environmental and management factors exacerbate disease impact [
4] (
Figure 1).
Despite its global distribution and significant burden on livestock production,
D. congolensis remains a neglected pathogen with limited molecular characterization. The bacterium exhibits a complex life cycle involving filamentous hyphae and motile zoospores [
3,
5] and shows genetic diversity with distinct strains reported across geographic locations [
6]. There is currently no fully effective treatment for dermatophilosis in cattle [
3,
7,
8], as clinically recommended topical, parenteral, and local formulations have shown limited success, and a vaccine remains unavailable—largely due to critical gaps that persist in understanding its genomic architecture, virulence mechanisms, and antimicrobial resistance profiles. Notably, rising antibiotic resistance complicates disease control and treatment [
9]. Furthermore, the zoonotic potential of
D. congolensis underscores its importance within a One Health framework, yet studies addressing its public health implications are scarce [
10].
This review aims to synthesize current molecular and genomic knowledge of D. congolensis, focusing on its resistome landscape and virulence factors. We discuss recent advances in genomic sequencing, phylogenetic insights, and antimicrobial resistance mechanisms, highlighting their implications for disease diagnosis, treatment, and management in cattle. By identifying research gaps and emerging trends, this review seeks to inform future investigations and support the development of effective strategies to mitigate the impact of dermatophilosis on animal health and livestock production.
2. Molecular Characterization of D. congolensis
D. congolensis has been characterized by whole-genome sequencing and bioinformatic analysis, exploring the genomic characteristics that provide a clear comprehension of its genetic structure and diversity. The genome is approximately 2.6 Mb in size [
6], where the base composition of
D. congolensis has been analyzed, revealing a guanine plus cytosine (GC) content ranging from 57.4 to 58.7 mol%, which is consistent with its classification within the family
Dermatophilacea [
11]. In contrast to soil-dwelling Actinobacteria like Streptomyces, which frequently have larger genomes and higher GC content to allow complex secondary metabolite production,
D. congolensis’s moderate GC content may represent genomic adaptations to its biological niche as a skin pathogen [
12]. The genome of
D. congolensis typically had 2527 coding sequences (CDS), 49 transfer RNAs (tRNA), apart from BTSK20, BTSK22, and BTSK31, which encode 52 tRNAs, and BTSK5, BTSK28, and BTSK34, which encode 43, 46, and 48 tRNAs, respectively. Each genome also contains three ribosomal RNAs (rRNA) [
13]. A few studies have investigated the genomic characteristics of
D. congolensis to better understand the pathogen’s genetic composition, pathogenesis, and its evolutionary adaptation. Investigating the genome of
D. congolensis is important for the identification of major virulence factors, resistance patterns, and developing control strategies [
14]. Through analysis and better comprehension of this pathogen, a potential vaccine candidate and therapeutic target can be developed [
15]. Previous studies have shown that
D. congolensis is a high GC-content member of the Actinobacteria, with a genome encoding approximately 2527 genes [
6]. Among these, 23 housekeeping genes are associated with antimicrobial susceptibility mechanisms, and some isolates have been found to encode the acquired antimicrobial resistance gene
tet(Z), which is implicated in tetracycline resistance [
6].
To better understand
D. congolensis genetic diversity, evolutionary links, and divergence from related bacteria, phylogenetic analysis has been performed using a variety of molecular approaches. Whole-genome comparisons, single-nucleotide polymorphism (SNP) analysis, and 16S rRNA gene sequencing have been among the primary areas of research. Phylogenetic analysis of
D. congolensis has shown minimal genetic diversity among isolates, indicating structural and functional conservation. A study on Nigerian isolates confirmed
D. congolensis as phylogenetically distinct but more closely related to
Nocardia brasiliensis than
Streptomyces sp. [
16]. Additionally, strain D 363 closely resembled DSM44180, NBRC105199, C2, and D.C16SrRNA, while strain D 362 was similar to S3 and S2 from GenBank [
16]. Moreover, sequence analysis of a
D. congolensis isolate from sheep revealed 99–100% sequence homology with other
D. congolensis isolates in the NCBI database [
17]. The phylogenetic analysis showed a close clustering with isolates from Congo, Nigeria, and Angola, indicating strong genetic relatedness among African strains [
16]. Another study on the phenotypic and genotypic characterization of
D. congolensis isolates from cattle, sheep, and goats in Jos, Nigeria, revealed over 98% sequence similarity among the isolates, with variations observed at specific nucleotide positions [
18]. Phylogenetic analysis further confirmed a 99% identity between the isolates and
D. congolensis DSM 44180 from GenBank. The phylogenetic tree clustered the isolates into two main groups, indicating a close evolutionary relationship across host species. This high degree of genetic similarity suggests that the isolates are likely strains of the same species [
18]. Further investigation of the genetic diversity of
D. congolensis was gained through SNP-based phylogenetic analysis, which identified three major clusters. The first cluster comprised geographically diverse strains, which contained 27–33 CRISPR spacers, reflecting genetic variability. The second cluster consisted of strains from a single location, all of which carried six CRISPR spacers and frequently harbored the
tet(Z) antimicrobial resistance gene, suggesting a localized adaptation to tetracycline exposure. The third cluster comprised strains from two distinct locations, exhibiting 4–5 CRISPR spacers, which represent an intermediate genetic profile [
6].
In our recent phylogenetic analysis (
Figure 2) we examined 28 16S rRNA gene sequences of
Dermatophilus congolensis obtained from diverse geographic regions. The phylogenetic tree revealed tight clustering of sequences from individual countries, such as Nigeria (2009) and Japan (2022), which exhibited short branch lengths, indicating circulation of closely related or identical strains within those regions. Based on the topology, the tree can be divided into three major clusters (Cluster I, II, and III), representing distinct phylogenetic lineages. Sequences from Japan and Iraq formed separate, well-supported clades and were notably distant from other groups, highlighting significant genetic divergence and suggesting unique evolutionary trajectories in these regions. The pairwise sequence identity within these two countries ranged from 71% to 83%, whereas sequences from other countries displayed higher similarity, with identities ranging from 88% to 100%.
Compared to other cutaneous Actinobacteria such as Nocardia brasiliensis and Corynebacterium species,
D. congolensis exhibits a more compact genome and moderate GC content [
6], reflecting adaptation to its niche as a skin pathogen. Unlike Streptomyces spp., which encode diverse secondary metabolite pathways,
D. congolensis maintains a focused set of virulence-associated genes, highlighting its specialized pathogenic strategy [
14]. Therefore, the genomic and phylogenetic analyses of
D. congolensis provide insight into genetic composition, diversity, and evolutionary adaptations. This knowledge has significant implications for disease control, antimicrobial resistance monitoring, and the development of targeted therapeutic interventions.
D. congolensis employs several virulent factors to establish infection and evade host defenses, though their functional roles remain only partially characterized. Among these are proteolytic enzymes, particularly the nasp (nuclear antigenic serine protease) gene, which encodes an extracellular subtilisin-like serine protease that plays a crucial role in epidermal invasion [
21]. This extracellular enzyme degrades host tissues, enabling the bacterium to penetrate deeper layers of the skin and form lesions [
22]. However, protease activity varies between isolates, with some lacking detectable serine protease genes, indicating functional diversity in virulence strategies [
23]. Besides these enzymatic factors, the agac (alkaline ceramidase) gene, which encodes an enzyme involved in hydrolyzing ceramides that are critical to skin barrier integrity, also serves as a conserved molecular marker widely used for species-specific detection of
D. congolensis. The agac gene forms the basis of a highly specific SYBR Green real-time PCR (RT-PCR) assay, developed to rapidly identify
D. congolensis DNA in clinical samples. This assay demonstrates exceptional sensitivity, with a detection limit of 1 pg DNA per reaction and no cross-reactivity with closely related or other common bacterial species, supporting its diagnostic precision. All clinical isolates tested yielded positive results, confirming the reliability of agac-based molecular detection methods. Although the agac gene’s role in virulence is less directly characterized compared to proteolytic enzymes, its stable presence across
D. congolensis strains underpins its value for laboratory confirmation, while also revealing minimal sequence variability among isolates tested [
24]. In addition to enzymatic degradation,
D. congolensis utilizes structural invasion mechanisms such as hyphae and zoospores. Hyphae are filamentous structures that actively penetrate the epidermis, triggering acute inflammation and leading to repeated cycles of tissue invasion [
23,
25]. Zoospores, which are motile and flagellated, facilitate the spread of infection to new sites, especially in the presence of moisture and skin damage [
26]. Understanding the diversity and function of these virulence factors can inform targeted interventions. For example, knowledge of nasp-mediated proteolysis may guide the development of topical inhibitors or vaccines that block epidermal invasion, while agac-based diagnostics allow rapid identification of infected animals, improving early containment and reducing disease spread.
3. Resistome Profile of D. congolensis
Antibiotics are the major drugs used for the treatment of dermatophilosis; historically, drugs such as amoxicillin, ampicillin, chloramphenicol, erythromycin, novobiocin, penicillin G, spiramycin, streptomycin, and tetracyclines have been effective [
27,
28]. However,
D. congolensis has been reported to have developed resistance to certain antimicrobial agents, although comprehensive data on its antibiotic resistance profile remains limited. Tetracycline is the only antibiotic for which the resistant mechanism and transmission have been studied to some extent. A recent comprehensive genomic study reported the spread of
tet(Z) gene within the population of
D. congolensis isolated from animal origin [
6]. This gene has been seen to be associated with tetracycline resistance.
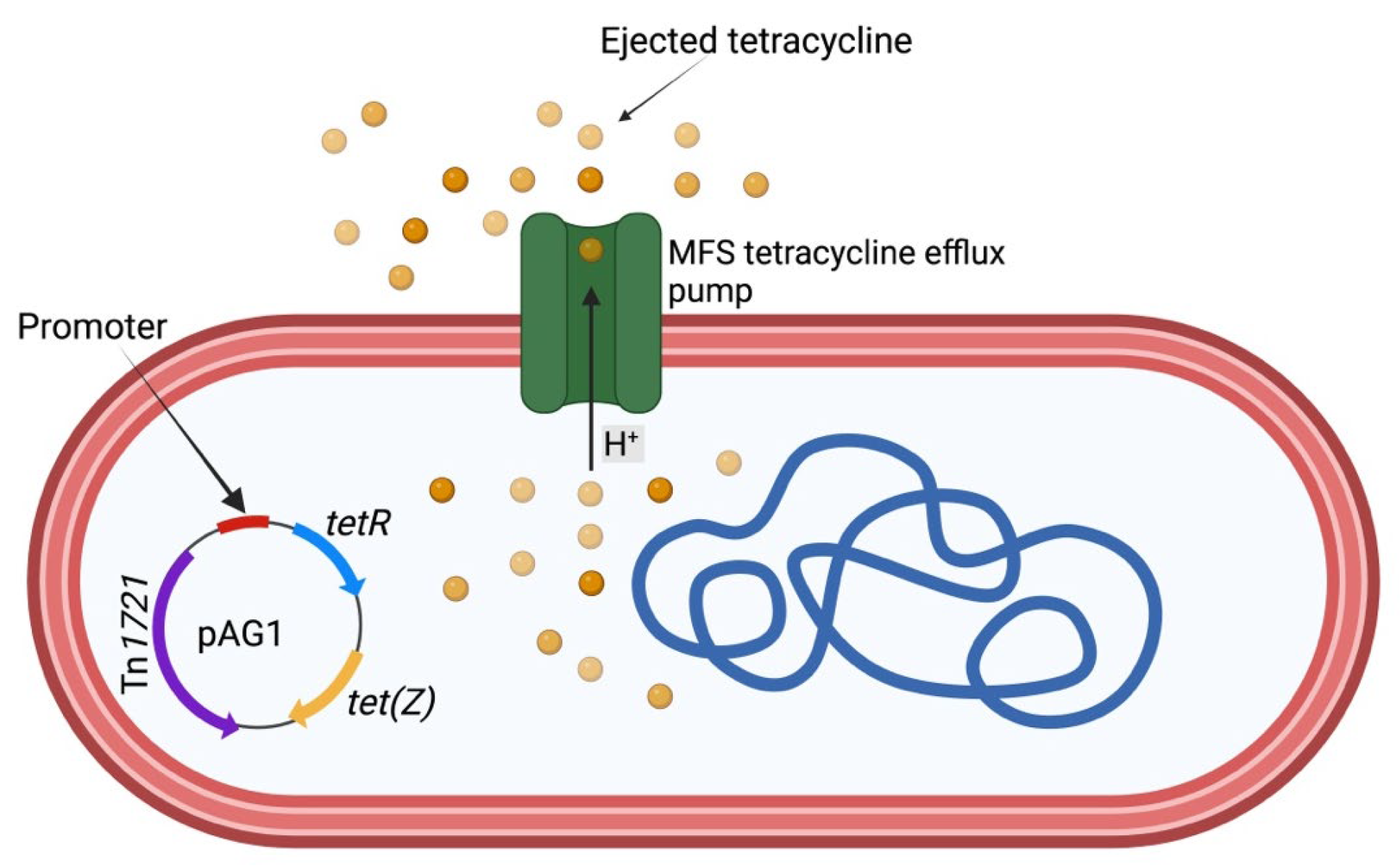
tet(Z) is a rare antibiotic-resistant gene that codes for tetracycline efflux protein
(TetZ). It was first described by [
29]. It is carried by transposon Tn1721, which is encoded on the pAG1 plasmid. The detection of
tet(Z) in
D. congolensis poses a great threat to the treatment of dermatophilosis. TetZ protein uses an efflux pump resistance mechanism to eject tetracycline. Currently, there is no study that has fully investigated this mechanism. However, we propose that the mechanism will not be different from the mechanism used by other Gram-positive bacteria (
Figure 3). More research is needed to ascertain our proposed mechanism. Comparatively, other skin pathogens such as
Staphylococcus aureus and
Nocardia spp. also display efflux pump-mediated resistance [
30], suggesting that stewardship principles developed in these systems may be adapted for
D. congolensis. Implementing routine antibiotic sensitivity testing and selective therapy can reduce the emergence of resistance, while monitoring
tet(Z) prevalence helps tailor regional treatment guidelines.
In addition to the
tet(Z) that confers tetracycline resistance in
D. congolensis, twenty-five housekeeping genes have been reported to be associated with its susceptibility against a wide range of antibiotics [
20]. They include: Dfr, PgsA, Iso-tRNA, rpoB, Mtra, EF-G, kasA, rpsL, Ddl, S12p, EF-Tu, folP, gyrA, S10p rpoC, Alr, OxyR, rho, folA, gyrB, inhA, fabl, dxr, MtrB, and gidB. Judicious use of antibiotics in the treatment of dermatophilosis is required in order to prevent the resistance of these genes to their respective antibiotics. Although molecular and resistome studies of
D. congolensis have expanded our understanding of its pathogenicity, much of the literature remains descriptive. Key virulence factors, including proteases (nasp), the agac gene, hyphae, and zoospores, have been characterized; however, their precise interactions with host immune responses are poorly understood
4. Implications for Cattle Health and Disease Control
Diagnosis of
D. congolensis is commonly performed by cytology from lesions or bacterial culture. However, some recent advancements in molecular characterization techniques have allowed researchers to explore the genomic profile of
D. congolensis in greater depth [
16]. The draft genome sequences of multiple
D. congolensis isolates have provided insights into the pathogen’s resistome profile, which is critical for improving disease diagnosis and treatment strategies. Identifying specific genes associated with virulence and antibiotic resistance can aid in developing more accurate diagnostic tools. For instance, a study shows the detection of the
tet(Z) gene, which confers resistance to tetracyclines, in several isolates, highlighting the need for targeted treatments based on the pathogen’s resistance profile [
6]. Molecular characterization, therefore, plays a vital role in enhancing disease diagnosis and guiding veterinarians in selecting appropriate interventions, ultimately improving the overall management of dermatophilosis in cattle.
One of the most pressing concerns in cattle production is the rise in antibiotic-resistant bacterial strains, which complicates the treatment of dermatophilosis. The overuse and misuse of antibiotics in cattle farming have contributed to the development of antibiotic resistance in pathogens such as
D. congolensis. Identifying resistance genes in
D. congolensis isolates underscores the importance of responsible antibiotic stewardship [
6]. By promoting the judicious use of antibiotics and adopting alternative strategies for disease control, such as improving biosecurity measures and the use of vaccines, cattle producers can help mitigate the spread of resistant bacterial strains. Antibiotic stewardship is essential for preserving the efficacy of existing treatments and reducing the overall burden of antibiotic resistance in livestock production systems.
A study has previously shown that antibiotics such as erythromycin, penicillin, ampicillin, and tetracyclines are effective against
D. congolensis; however, the misuse or overuse of these antibiotics has led to the emergence of resistance in some isolates [
30]. Specific resistance was observed in
D. congolensis against antibiotics like polymyxin B, enrofloxacin, oxacillin, and trimethoprim-sulfamethoxazole. The rise in antibiotic-resistant strains necessitates alternative treatments and management of antibiotic use in both veterinary practice and cattle production. Antibiotic stewardship programs should focus on reducing the dependence on broad-spectrum antibiotics, encouraging targeted therapy based on antibiotic sensitivity testing. In addition to conventional antibiotics, integrated disease management practices, including improved hygiene, pasture rotation, and isolation of affected animals—can reduce transmission. Comparisons with control strategies for Nocardia infections suggest that combining molecular diagnostics with environmental management is particularly effective [
21,
31]. Furthermore, coordinated vaccination programs targeting immunogenic antigens of
D. congolensis could complement these measures, providing a sustainable approach to disease prevention. Proteases likely contribute to immune evasion by degrading host proteins and modulating inflammatory signaling, yet experimental data on cytokine responses or skin barrier disruption are limited. Hyphal structures and motile zoospores facilitate tissue invasion and dissemination, but the host’s innate and adaptive responses to these factors have not been fully characterized. Variation in protease activity among isolates and the limited functional validation of agac indicate further diversity in virulence strategies. In terms of antimicrobial resistance, recent genomic analyses have identified the
tet(Z) gene and other resistance determinants within
D. congolensis populations, highlighting the growing threat of AMR in livestock pathogens. Longitudinal data on the emergence and spread of resistance remain sparse, revealing a critical knowledge gap. Integrating a One Health perspective is essential, as zoonotic transmission has been documented, and environmental factors may facilitate pathogen dissemination.
Going forward, research efforts should focus on further understanding the pathogenic mechanisms of D. congolensis and developing more effective control and management strategies. Scientists should channel more studies into vaccine development targeting specific antigens produced by D. congolensis during infection. Vaccines could offer a long-term solution for preventing dermatophilosis outbreaks, especially in regions where environmental conditions promote the spread of the disease. Additionally, exploring alternative therapies, such as bacteriophage-based treatments, may provide new avenues for controlling D. congolensis infections without relying on traditional antibiotics. There is also a need to understand the environmental and host factors that contribute to disease transmission, which are critical for developing comprehensive management strategies that minimize the impact of dermatophilosis on cattle health and productivity. Therefore, advances in molecular characterization and antibiotic stewardship practices are essential for improving disease management in Dermatophilus congolensis and should receive urgent attention. Future comparative studies with related Actinobacteria could identify conserved virulence pathways and resistance determinants, which may serve as broad-spectrum targets for therapeutics. Additionally, studies assessing host immune responses in different breeds and environmental conditions will refine management practices and inform vaccine development. The government should also work with relevant stakeholders to fund research that explores innovative solutions, such as vaccine development and alternative therapies, to reduce the burden of this pathogen on cattle production systems.
5. Conclusions
In conclusion, the molecular characterization and resistome profiling of Dermatophilus congolensis provide critical insights into its biology, pathogenicity, and antimicrobial resistance. This review highlights key advances, including the identification of virulence factors such as nasp, agac, hyphal structures, and zoospores, as well as the detection of resistance determinants like tet(Z) and associated efflux mechanisms. The presence of mobile genetic elements underscores the potential for horizontal gene transfer and the emergence of multidrug-resistant strains. Collectively, these findings have important translational implications for cattle production, informing the development of rapid and accurate diagnostic tools, evidence-based treatment protocols, and judicious antibiotic stewardship to minimize resistance.
However, several limitations remain. Current genomic datasets are limited, geographically biased, experimental validation of virulence and resistance mechanisms is sparse, and longitudinal epidemiological studies tracking the emergence and spread of resistant strains are lacking. Addressing these gaps will be crucial to translating molecular insights into effective disease management.
Future research should prioritize functional genomics, in vivo and ex vivo host–pathogen interaction studies, expanded AMR surveillance, and the development of vaccines or alternative therapeutics. Such integrated efforts, combining molecular insights with epidemiology and One Health approaches, will be critical to translating research findings into practical strategies for disease control, improved animal welfare, and sustainable cattle production A deeper understanding of D. congolensis at the molecular level will ultimately contribute to more effective disease control strategies, improved animal welfare, and enhanced productivity in cattle production systems.