Genome Editing Against HPV-Driven Cancers: From Bench to Clinic
Abstract
1. Introduction
2. Methods
2.1. Literature Search Strategy
2.2. Inclusion and Exclusion Criteria
- Original in vitro, in vivo, or clinical studies investigating genome editing technologies (CRISPR, TALENs, ZFNs) in HPV infection or HPV-related cancers.
- Studies reporting mechanistic, therapeutic, or translational outcomes.
- Articles published in English.
- Non-HPV related studies.
- Editorials, commentaries, and conference abstracts without primary data.
- Duplicate publications.
2.3. Study Selection and Data Extraction
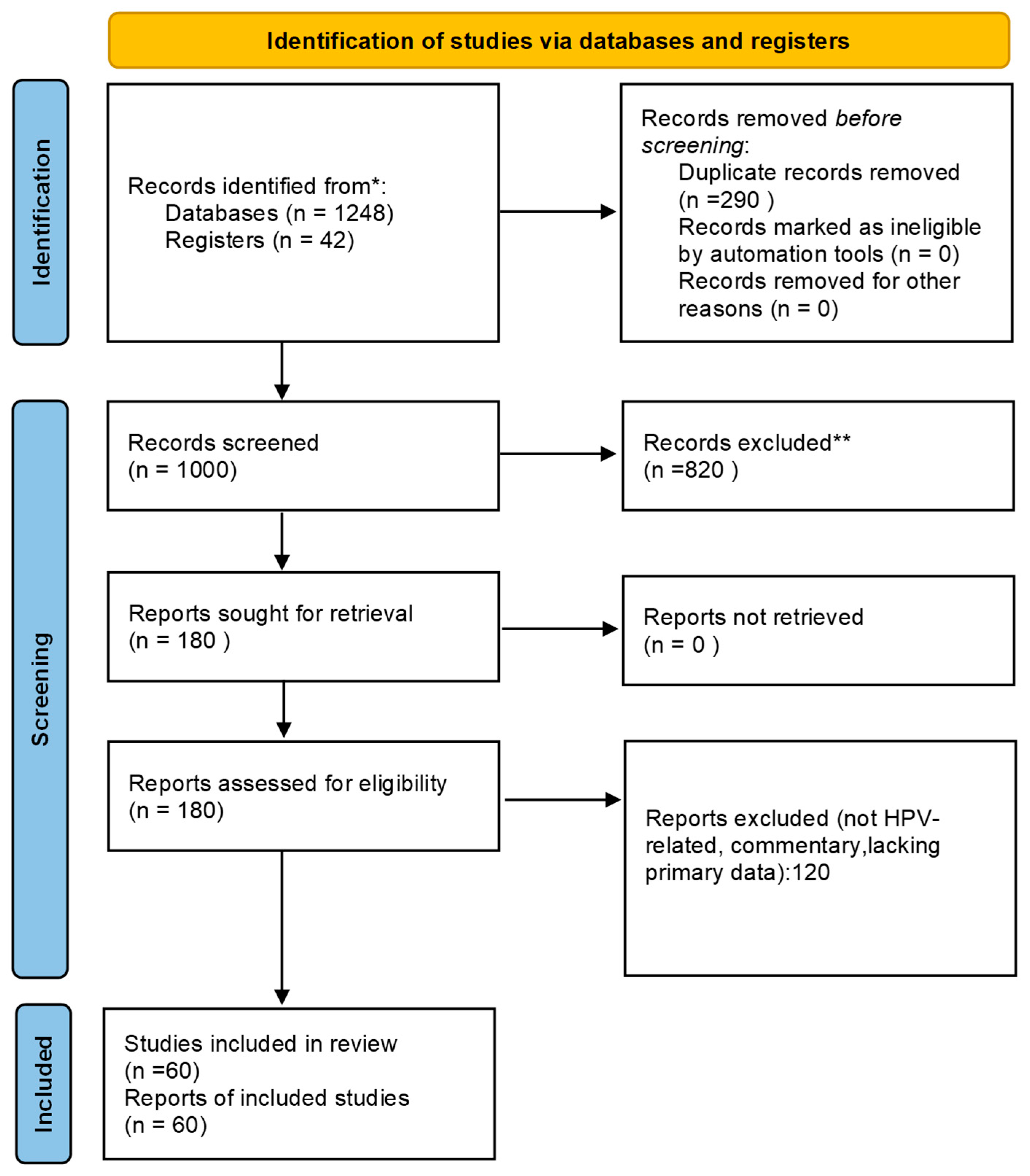
2.4. PRISMA Flow Diagram
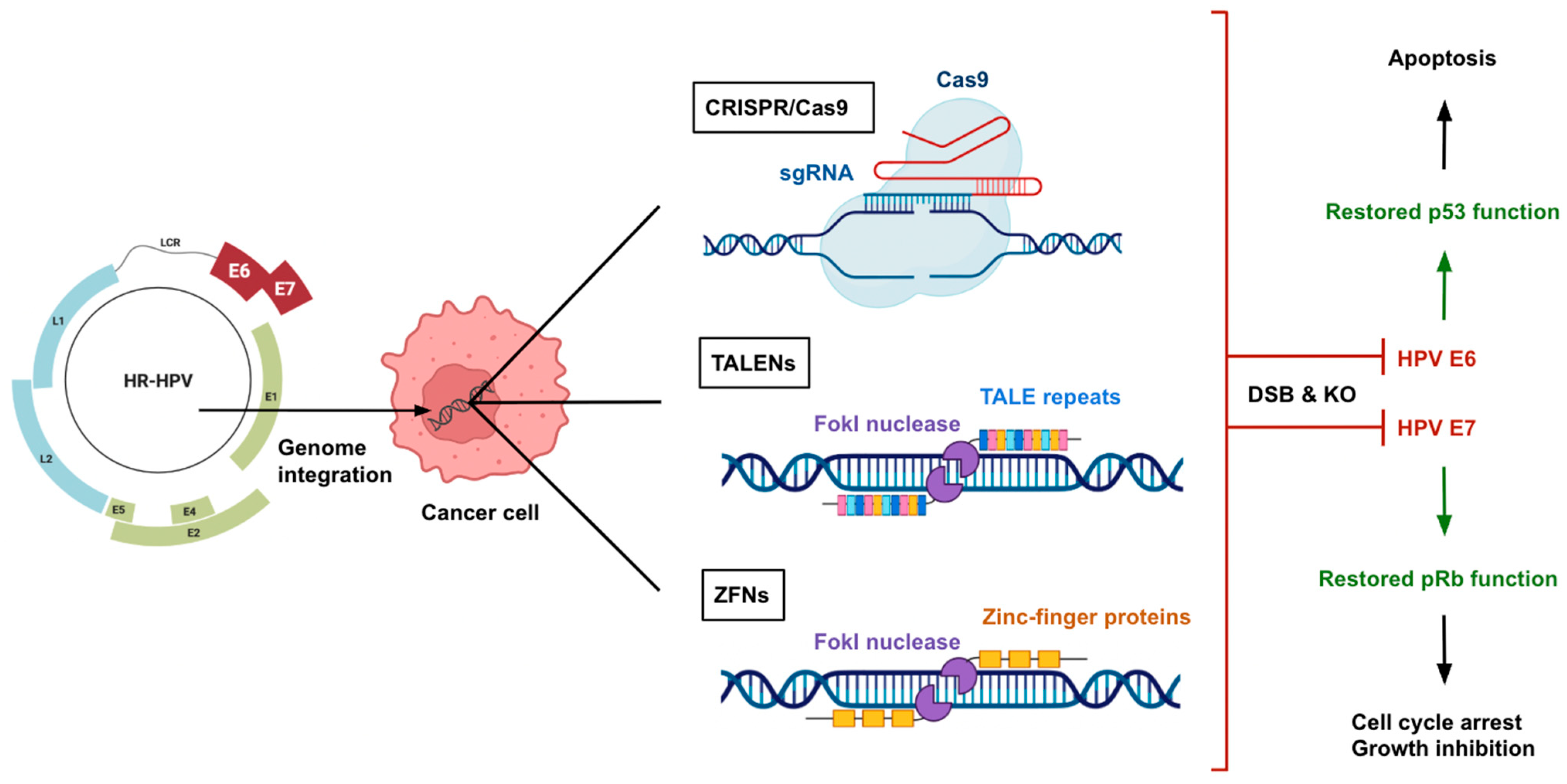
3. Genome Editing Technologies
3.1. CRISPR/Cas9
3.2. TALENs
3.3. ZFNs
4. Therapeutic Applications of Genome Editing for HPV
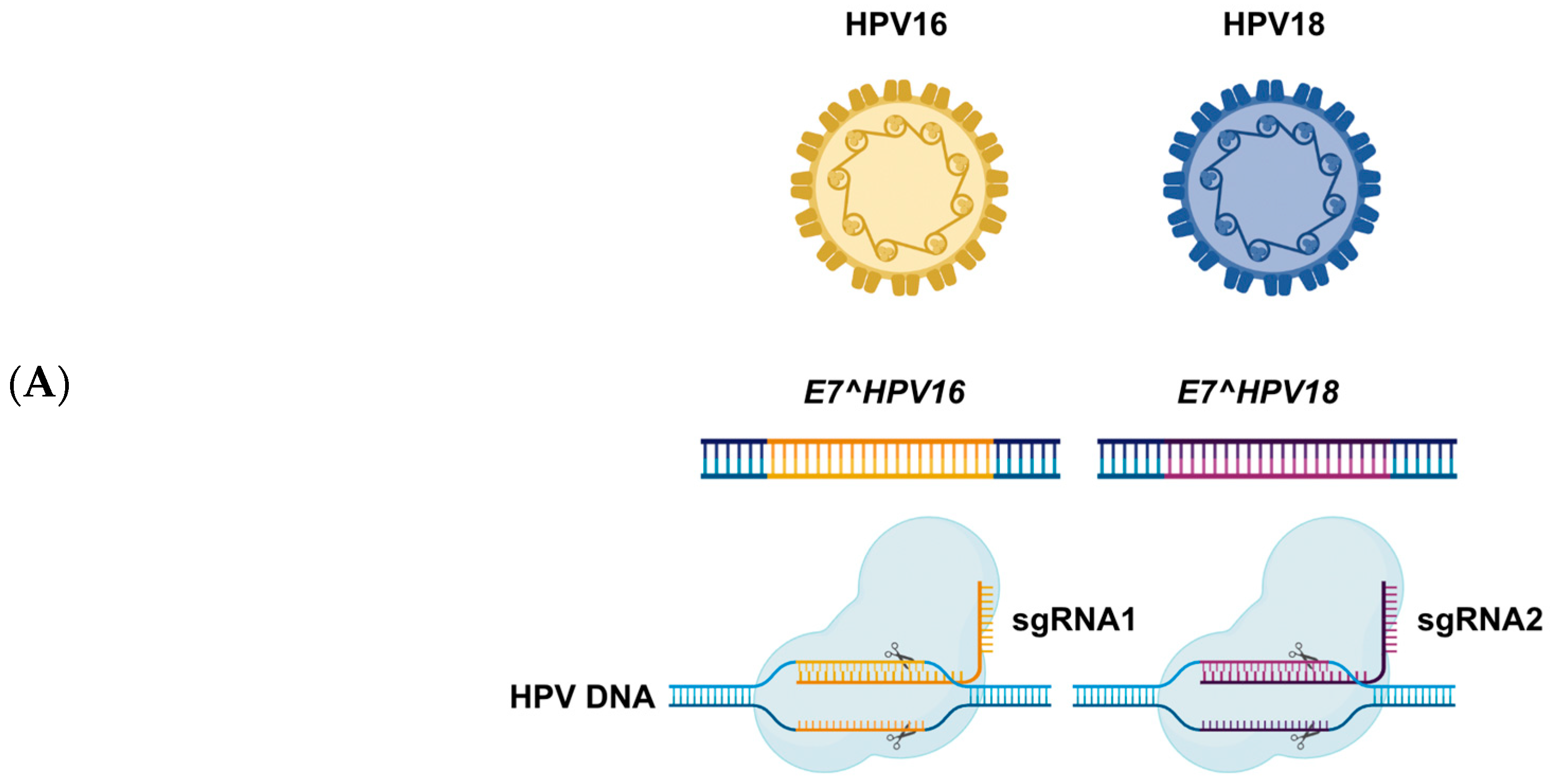
4.1. Editing the HPV to Eliminate Infection
4.2. Targeting HPV Oncogenes (E6 Aand E7)
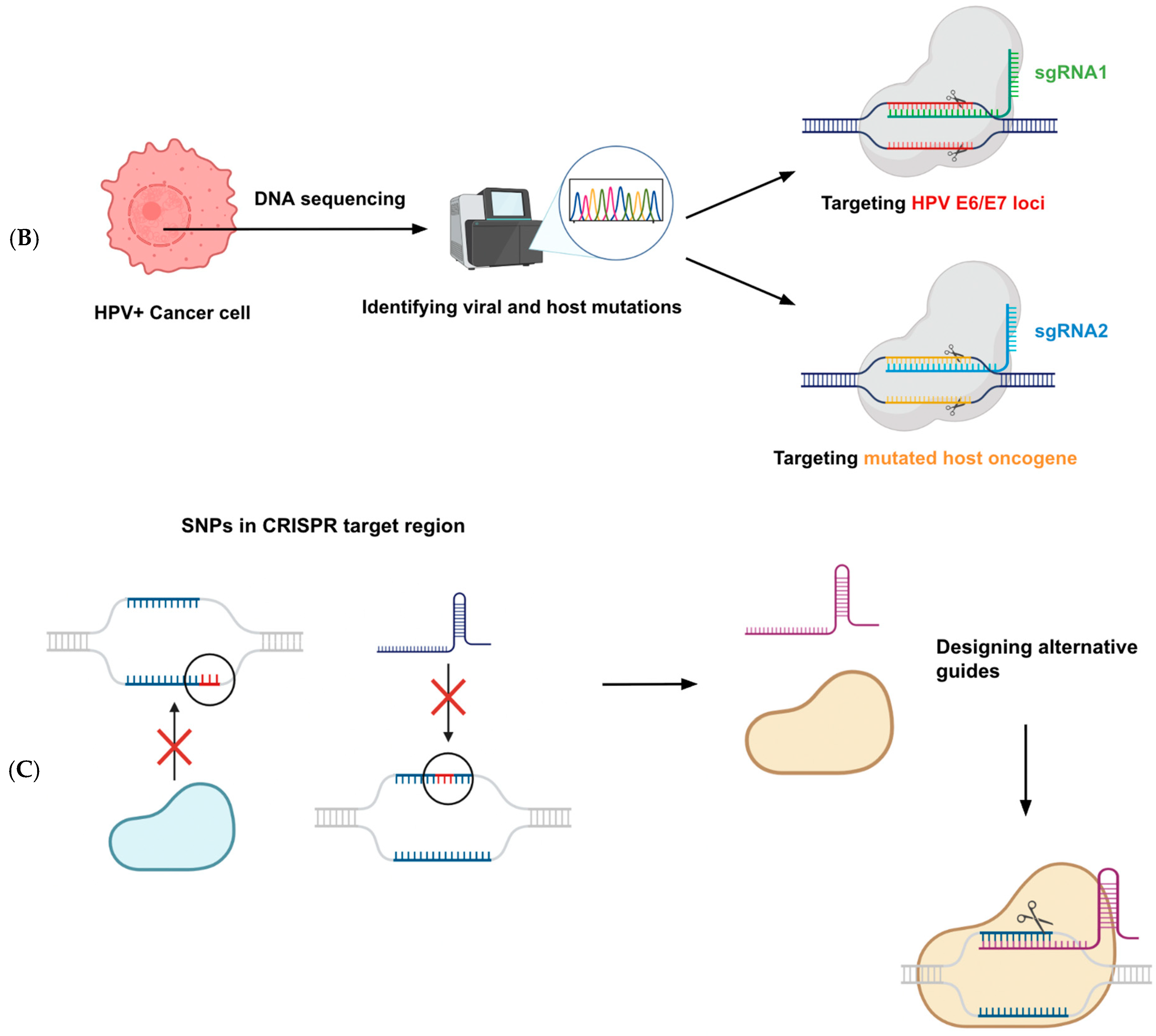
4.3. Potential for Personalized Medicine
5. Preclinical and Clinical Research Progress
5.1. Preclinical Efficacy in Models
5.2. Clinical Trials and Studies
6. Oncological Perspectives and Integration into Treatment
6.1. Complement to Existing Therapies
6.2. Comparison with Immunotherapy
6.3. Versus Chemoradiation
6.4. Unique Benefit—Viral Specificity
6.5. Safety Considerations in Oncology
6.6. Patient Acceptance and Feasibility
7. Challenges and Future Prospects
7.1. Technical Hurdles
7.2. Efficiency of Editing
7.3. Immune Responses and Safety
7.4. Ethical and Regulatory Concerns
7.5. Future Research Directions
7.6. Translational Potential
8. Discussion
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stanley, M. Pathology and epidemiology of HPV infection in females. Gynecol. Oncol. 2010, 117 (Suppl. S2), S5–S10. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Hausen, H. Papillomaviruses and cancer: From basic studies to clinical application. Nat. Rev. Cancer 2002, 2, 342–350. [Google Scholar] [CrossRef]
- Moody, C.A.; Laimins, L.A. Human papillomavirus oncoproteins: Pathways to transformation. Nat. Rev. Cancer 2010, 10, 550–560. [Google Scholar] [CrossRef]
- McLaughlin-Drubin, M.E.; Münger, K. The human papillomavirus E7 oncoprotein. Virology 2009, 384, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Münger, K.; Howley, P.M. Human papillomavirus immortalization and transformation functions. Virus Res. 2002, 89, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 2006, 110, 525–541. [Google Scholar] [CrossRef] [PubMed]
- Koutsky, L. Epidemiology of genital human papillomavirus infection. Am. J. Med. 1997, 102, 3–8. [Google Scholar] [CrossRef]
- Arbyn, M.; Castellsagué, X.; de Sanjosé, S.; Bruni, L.; Saraiya, M.; Bray, F.; Ferlay, J. Worldwide burden of cervical cancer in 2008. Ann. Oncol. 2011, 22, 2675–2686. [Google Scholar] [CrossRef]
- The FUTURE II Study Group. Quadrivalent vaccine against HPV to prevent high-grade cervical lesions. N. Engl. J. Med. 2007, 356, 1928–1943. [Google Scholar] [CrossRef]
- Drolet, M.; Bénard, É.; Pérez, N.; Brisson, M.; HPV Vaccination Impact Study Group. Population-level impact and herd effects following the introduction of human papillomavirus vaccination programmes: Updated systematic review and meta-analysis. Lancet 2019, 394, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Pimple, S.A.; Mishra, G.A. Global strategies for cervical cancer prevention and screening. Minerva Ginecol. 2019, 71, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Center, M.M.; DeSantis, C.; Ward, E.M. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol. Biomarkers Prev. 2010, 19, 1893–1907. [Google Scholar] [CrossRef]
- Hu, Z.; Yu, L.; Zhu, D.; Ding, W.; Wang, X.; Zhang, C.; Wang, L.; Jiang, X.; Shen, H.; He, D.; et al. Disruption of HPV16-E7 by CRISPR/Cas System Induces Apoptosis and Growth Inhibition in HPV16 Positive Human Cervical Cancer Cells. BioMed Res. Int. 2014, 1, 612823. [Google Scholar] [CrossRef] [PubMed]
- Ravanlo, Z.Z.; Gholami, S.; Baghi, H.B. HPV-Specific Antivirals: Disarming Viral Entry and Disruption of Replication. Microbe 2025, 9, 100593. [Google Scholar] [CrossRef]
- Egawa, N.; Doorbar, J. The low-risk papillomaviruses. Virus Res. 2017, 231, 119–127. [Google Scholar] [CrossRef]
- Ntuli, L.; Mtshali, A.; Mzobe, G.; Liebenberg, L.J.P.; Ngcapu, S. Role of Immunity and Vaginal Microbiome in Clearance and Persistence of Human Papillomavirus Infection. Front. Cell. Infect. Microbiol. 2022, 12, 927131. [Google Scholar] [CrossRef]
- Cuzick, J.; Adcock, R.; Kinney, W.; Castle, P.E.; Robertson, M.; McDonald, R.M.; Stoler, M.H.; Du, R.; Wheeler, C.M. Impact of HPV testing in Opportunistic Cervical Screening: Support for Primary HPV Screening in the United States. Int. J. Cancer 2023, 153, 83. [Google Scholar] [CrossRef]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for genome editing. Nat. Rev. Mol. Cell. Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef]
- Sander, J.D.; Joung, J.K. CRISPR–Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef]
- Ling, K.; Yang, L.; Yang, N.; Chen, M.; Wang, Y.; Liang, S.; Li, Y.; Jiang, L.; Yan, P.; Liang, Z. Gene Targeting of HPV18 E6 and E7 Synchronously by Nonviral Transfection of CRISPR/Cas9 System in Cervical Cancer. Hum. Gene Ther. 2020, 31, 297–308. [Google Scholar] [CrossRef]
- Khan, S.H. Genome-Editing Technologies: Concept, Pros, and Cons of Various Genome-Editing Techniques and Bioethical Concerns for Clinical Application. Mol. Ther. Nucleic Acids 2019, 16, 326–334. [Google Scholar] [CrossRef]
- Liu, W.; Jiang, Y.; Wang, C.; Wang, M.; Zhang, W.; Ren, H.; Xu, S.; Qin, J.; Liu, P.; Jin, L.; et al. Towards the elimination of infectious HPV: Exploiting CRISPR/Cas innovations. Front. Cell. Infect. Microbiol. 2025, 15, 1627668. [Google Scholar] [CrossRef]
- Popovitz, J.; Sharma, R.; Hoshyar, R.; Soo Kim, B.; Murthy, N.; Lee, K. Gene editing therapeutics based on mRNA delivery. Adv. Drug Deliv. Rev. 2023, 200, 115026. [Google Scholar] [CrossRef]
- Ren, C.; Li, X.; Mao, L.; Xiong, J.; Gao, C.; Shen, H.; Wang, L.; Zhu, D.; Ding, W.; Wang, H. An effective and biocompatible polyethylenimine based vaginal suppository for gene delivery. Nanomed. Nanotechnol. Biol. Med. 2019, 20, 101994. [Google Scholar] [CrossRef]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and TALE-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef] [PubMed]
- Yee, J.-K. Off-target effects of engineered nucleases. FEBS J. 2016, 283, 3239–3248. [Google Scholar] [CrossRef] [PubMed]
- Sunami, T.; Kono, H. Balance between DNA-binding affinity and specificity enables selective recognition of longer target sequences in vivo. Protein Sci. 2019, 28, 1630–1639. [Google Scholar] [CrossRef]
- Davis, D.; Stokoe, D. Zinc finger nucleases as tools to understand and treat human diseases. BMC Med. 2010, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Cradick, T.J.; Ambrosini, G.; Iseli, C.; Bucher, P.; McCaffrey, A.P. Zfn-site searches genomes for zinc finger nuclease target sites and off-target sites. BMC Bioinform. 2011, 12, 152. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Gupta, A.; Hall, V.L.; Rayla, A.L.; Christensen, R.G.; Dake, B.; Lakshmanan, A.; Kuperwasser, C.; Stormo, G.D.; Wolfe, S.A. Using defined finger–finger interfaces as units of assembly for constructing zinc-finger nucleases. Nucleic Acids Res. 2013, 41, 2455–2465. [Google Scholar] [CrossRef] [PubMed]
- Tebas, P.; Stein, D.; Tang, W.W.; Frank, I.; Wang, S.Q.; Lee, G.; Spratt, S.K.; Surosky, R.T.; Giedlin, M.A.; Nichol, G.; et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014, 370, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Katayama, S.; Watanabe, M.; Kato, Y.; Nomura, W.; Yamamoto, T. Engineering of zinc finger nucleases through structural modeling improves genome editing efficiency in cells. Adv. Sci. 2024, 11, e2310255. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, H.; Zhang, H.; Huang, Z.; Tian, R.; Li, L.; Fan, W.; Chen, Y.; Chen, L.; Zhang, S.; et al. The comparison of ZFNs, TALENs, and SpCas9 by GUIDE-seq in HPV-targeted gene therapy. Mol. Ther. Nucleic Acids 2021, 26, 1466–1478. [Google Scholar] [CrossRef]
- Ding, W.; Hu, Z.; Zhu, D.; Jiang, X.; Yu, L.; Wang, X.; Zhang, C.; Wang, L.; Ji, T.; Li, K.; et al. Zinc Finger Nucleases Targeting the Human Papillomavirus E7 Oncogene Induce E7 Disruption and a Transformed Phenotype in HPV16/18-Positive Cervical Cancer Cells. Clin. Cancer Res. 2014, 20, 6495–6503. [Google Scholar] [CrossRef]
- Shankar, S.; Sreekumar, A.; Prasad, D.; Das, A.V.; Pillai, M.R. Genome editing of oncogenes with ZFNs and TALENs: Caveats in nuclease design. Cancer Cell Int. 2018, 18, 169. [Google Scholar] [CrossRef]
- Hong, D.; Liu, J.; Hu, Y.; Lu, X.; Li, B.; Li, Y.; Hu, D.; Lu, W.; Xie, X.; Cheng, X. Viral E6 is overexpressed via high viral load in invasive cervical cancer with episomal HPV16. BMC Cancer 2017, 17, 136. [Google Scholar] [CrossRef]
- Bi, Y.; Sun, L.; Gao, D.; Ding, C.; Li, Z.; Li, Y.; Cun, W.; Li, Q. High-efficiency targeted editing of large viral genomes by RNA-guided nucleases. PLoS Pathog. 2014, 10, e1004090. [Google Scholar] [CrossRef]
- Bhowmik, R.; Chaubey, B. Crispr/cas9: A tool to eradicate HIV-1. AIDS Res. Ther. 2022, 19, 58. [Google Scholar] [CrossRef]
- Zhen, S.; Lu, J.-J.; Wang, L.-J.; Sun, X.-M.; Zhang, J.-Q.; Li, X.; Luo, W.-J.; Zhao, L. In Vitro and In Vivo Synergistic Therapeutic Effect of Cisplatin with Human Papillomavirus16 E6/E7 CRISPR/Cas9 on Cervical Cancer Cell Line. Transl. Oncol. 2016, 9, 498–504. [Google Scholar] [CrossRef]
- Ehrke-Schulz, E.; Heinemann, S.; Schulte, L.; Schiwon, M.; Ehrhardt, A. Adenoviral Vectors Armed with PAPILLOMAVIRUs Oncogene Specific CRISPR/Cas9 Kill Human-Papillomavirus-Induced Cervical Cancer Cells. Cancers 2020, 12, 1934. [Google Scholar] [CrossRef]
- Gao, C.; Wu, P.; Yu, L.; Liu, L.; Liu, H.; Tan, X.; Wang, L.; Huang, X.; Wang, H. The application of CRISPR/Cas9 system in cervical carcinogenesis. Cancer Gene Ther. 2022, 29, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhang, S.; Qin, H.; Yao, K. From bench to bedside: Cutting-edge applications of base editing and prime editing in precision medicine. J. Transl. Med. 2024, 22, 1133. [Google Scholar] [CrossRef]
- Haddad, C.O.; Kalt, I.; Shovman, Y.; Xia, L.; Schlesinger, Y.; Sarid, R.; Parnas, O. Targeting the kaposi’s sarcoma-associated herpesvirus genome with the crispr-cas9 platform in latently infected cells. Virol. J. 2021, 18, 56. [Google Scholar] [CrossRef]
- Tsuchida, C.A.; Wasko, K.M.; Hamilton, J.R.; Doudna, J.A. Targeted nonviral delivery of genome editors in vivo. Proc. Natl. Acad. Sci. USA 2024, 121, e2307796121. [Google Scholar] [CrossRef] [PubMed]
- Sinn, P.L.; Arias, A.C.; Brogden, K.A.; McCray, P.B., Jr. Lentivirus vector can be readministered to nasal epithelia without blocking immune responses. J. Virol. 2008, 82, 10684–10692. [Google Scholar] [CrossRef]
- Zhen, S.; Liu, Y.; Lu, J.; Tuo, X.; Yang, X.; Chen, H.; Chen, W.; Li, X. Human Papillomavirus Oncogene Manipulation Using Clustered Regularly Interspersed Short Palindromic Repeats/Cas9 Delivered by pH-Sensitive Cationic Liposomes. Hum. Gene Ther. 2020, 31, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Excision BioTherapeutics. Clinical Trial EBT-101 for HIV CRISPR Treatment (NCT05144386). ClinicalTrials.gov.; 2022. Available online: https://clinicaltrials.gov/ct2/show/NCT05144386 (accessed on 8 October 2025).
- Baba, S.K.; Alblooshi, S.S.E.; Yaqoob, R.; Behl, S.; Al Saleem, M.; Rakha, E.A.; Malik, F.; Singh, M.; Macha, M.A.; Akhtar, M.K.; et al. Human papilloma virus (hpv) mediated cancers: An insightful update. J. Transl. Med. 2025, 23, 483. [Google Scholar] [CrossRef]
- Kennedy, E.M.; Kornepati, A.V.R.; Goldstein, M.; Bogerd, H.P.; Poling, B.C.; Whisnant, A.W.; Kastan, M.B.; Cullen, B.R. Inactivation of the Human Papillomavirus E6 or E7 Gene in Cervical Carcinoma Cells by Using a Bacterial CRISPR/Cas RNA-Guided Endonuclease. J. Virol. 2014, 88, 11965–11972. [Google Scholar] [CrossRef]
- Jubair, L.; Fallaha, S.; McMillan, N.A.J. Systemic Delivery of CRISPR/Cas9 Targeting HPV Oncogenes Is Effective at Eliminating Established Tumors. Mol. Ther. 2019, 27, 2091–2099. [Google Scholar] [CrossRef] [PubMed]
- Khairkhah, N.; Bolhassani, A.; Rajaei, F.; Najafipour, R. Systemic delivery of specific and efficient CRISPR/Cas9 system targeting HPV16 oncogenes using LL-37 antimicrobial peptide in C57BL/6 mice. J. Med. Virol. 2023, 95, e28934. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jiang, H.; Wang, T.; He, D.; Tian, R.; Cui, Z.; Tian, X.; Gao, Q.; Ma, X.; Yang, J.; et al. In vitro and in vivo growth inhibition of human cervical cancer cells via human papillomavirus E6/E7 mRNAs’ cleavage by CRISPR/Cas13a system. Antivir. Res. 2020, 178, 104794. [Google Scholar] [CrossRef]
- Hu, Z.; Ding, W.; Zhu, D.; Yu, L.; Jiang, X.; Wang, X.; Zhang, C.; Wang, L.; Ji, T.; Liu, D.; et al. TALEN-mediated targeting of HPV oncogenes ameliorates HPV-related cervical malignancy. J. Clin. Investig. 2015, 125, 425–436. [Google Scholar] [CrossRef]
- Ferreira, D.A.; McMillan, N.A.J.; Idris, A. Genetic deletion of HPV E7 oncogene effectively regresses HPV driven oral squamous carcinoma tumour growth. Biomed. Pharmacother. 2022, 155, 113782. [Google Scholar] [CrossRef]
- McLaughlin-Drubin, M.E.; Münger, K. Oncogenic activities of human papillomaviruses. Virus Res. 2009, 143, 195–208. [Google Scholar] [CrossRef]
- Tony, L.T.; Stabile, A.; Schauer, M.P.; Hudecek, M.; Weber, J. CAR-T Cell Therapy for Solid Tumors. Transfus. Med. Hemother. 2025, 52, 96–108. [Google Scholar] [CrossRef]
- Xinqiao Hospital of Chongqing. HPV-E6-Specific Anti-PD1 TCR-T Cells in the Treatment of HPV-Positive NHSCC or Cervical Cancer (NCT03578406). ClinicalTrials.gov.; 2019. Available online: https://clinicaltrials.gov/study/NCT03578406 (accessed on 8 October 2025).
- Li, X.; Guo, M.; Hou, B.; Zheng, B.; Wang, Z.; Huang, M.; Xu, Y.; Chang, J.; Wang, T. Crispr/cas9 nanoeditor of double knockout large fragments of e6 and e7 oncogenes for reversing drugs resistance in cervical cancer. J. Nanobiotechnol. 2021, 19, 231. [Google Scholar] [CrossRef]
- Ghanaat, M.; Goradel, N.H.; Arashkia, A.; Ebrahimi, N.; Ghorghanlu, S.; Malekshahi, Z.V.; Fattahi, E.; Negahdari, B.; Kaboosi, H. Virus against virus: Strategies for using adenovirus vectors in the treatment of HPV-induced cervical cancer. Acta Pharmacol. Sin. 2021, 42, 1981–1990. [Google Scholar] [CrossRef] [PubMed]
- Hsu, D.S.; Kornepati, A.V.R.; Glover, W.; Kennedy, E.M.; Cullen, B.R. Targeting HPV16 DNA using CRISPR/Cas inhibits anal cancer growth in vivo. Future Virol. 2018, 13, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-C.; Cai, Z.-M.; Zhang, X.-J. Reprogrammed CRISPR-Cas9 Targeting the Conserved Regions of HPV6/11 E7 Genes Inhibits Proliferation and Induces Apoptosis in E7-Transformed Keratinocytes. Asian J. Androl 2016, 18, 475–479. [Google Scholar] [CrossRef]
- Lao, Y.-H.; Li, M.; Gao, M.A.; Shao, D.; Chi, C.-W.; Huang, D.; Chakraborty, S.; Ho, T.-C.; Jiang, W.; Wang, H.-X.; et al. HPV Oncogene Manipulation Using Nonvirally Delivered CRISPR/Cas9 or Natronobacterium gregoryi Argonaute. Adv. Sci 2018, 5, 1700540. [Google Scholar] [CrossRef] [PubMed]
- Yoshiba, T.; Saga, Y.; Urabe, M.; Uchibori, R.; Matsubara, S.; Fujiwara, H.; Mizukami, H. CRISPR/Cas9-mediated cervical cancer treatment targeting human papillomavirus E6. Oncol. Lett. 2018, 17, 2197–2206. [Google Scholar] [CrossRef]
- Li, C.; Guo, L.; Liu, G.; Guo, M.; Wei, H.; Yang, Q.; Wang, J.; Chen, H. Reprogrammed CRISPR-Cas13a Targeting the HPV16/18 E6 Gene Inhibits Proliferation and Induces Apoptosis in E6-Transformed Keratinocytes. Exp. Ther. Med. 2020, 19, 3856–3860. [Google Scholar] [CrossRef]
- Inturi, R.; Jemth, P. CRISPR/Cas9-Based Inactivation of Human Papillomavirus Oncogenes E6 or E7 Induces Senescence in Cer-Vical Cancer Cells. Virology 2021, 562, 92–102. [Google Scholar] [CrossRef]
- Noroozi, Z.; Shamsara, M.; Valipour, E.; Esfandyari, S.; Ehghaghi, A.; Monfaredan, A.; Azizi, Z.; Motevaseli, E.; Modarressi, M.H. Antiproliferative Effects of AAV-Delivered CRISPR/Cas9-Based Degradation of the HPV18-E6 Gene in HeLa Cells. Sci. Rep. 2022, 12, 2224. [Google Scholar] [CrossRef]
- Hu, Z.; Liu, W.; Liu, J.; Zhou, H.; Sun, C.; Tian, C.; Guo, X.; Zhu, C.; Shao, M.; Wang, S. The Anti-Tumor Efficacy of a Recom-Binant Oncolytic Herpes Simplex Virus Mediated CRISPR/Cas9 Delivery Targeting in HPV16-Positive Cervical Cancer. Antivir. Res. 2024, 232, 106035. [Google Scholar] [CrossRef]
- Zhang, A.; Zheng, X.; Chen, S.; Duan, G. In vitro study of HPV18-positive cervical cancer HeLa cells based on CRISPR/Cas13a system. Gene 2024, 921, 148527. [Google Scholar] [CrossRef]
- Shankar, S.; Prasad, D.; Sanawar, R.; Ani, V.; Pillai, M.R. TALEN Based HPV-E7 Editing Triggers Necrotic Cell Death in Cervical Cancer Cells. Sci. Rep 2017, 7, 5500. [Google Scholar] [CrossRef]
- Zhen, S.; Qiang, R.; Lu, J.; Tuo, X.; Yang, X.; Li, X. CRISPR/Cas9-HPV-Liposome Enhances Antitumor Immunity and Treatment of HPV Infection-Associated Cervical Cancer. J. Med. Virol. 2023, 95, 28144. [Google Scholar] [CrossRef]
- First Affiliated Hospital, Sun Yat-Sen University. A Safety and Efficacy Study of TALEN and CRISPR/Cas9 in the Treatment of HPV-Related Cervical Intraepithelial Neoplasia (NCT03057912). ClinicalTrials.gov.; 2017. Available online: https://clinicaltrials.gov/study/NCT03057912 (accessed on 8 October 2025).
- Sichuan University. PD-1 Knockout Engineered T Cells for Metastatic Non-Small Cell Lung Cancer (NCT02793856). ClinicalTrials.gov.; 2021. Available online: https://clinicaltrials.gov/study/NCT02793856 (accessed on 8 October 2025).
- Inovio Pharmaceuticals. Phase I of Human Papillomavirus (HPV) DNA Plasmid (VGX-3100) + Electroporation for CIN 2 or 3 (NCT00685412). ClinicalTrials.gov.; 2017. Available online: https://clinicaltrials.gov/study/NCT00685412 (accessed on 8 October 2025).
- Inovio Pharmaceuticals. A Study of VGX-3100 DNA Vaccine with Electroporation in Patients with Cervical Intraepithelial Neoplasia Grade 2/3 or 3 (HPV-003) (NCT01304524). ClinicalTrials.gov.; 2018. Available online: https://clinicaltrials.gov/study/NCT01304524 (accessed on 8 October 2025).
- Inovio Pharmaceuticals. Evaluation of VGX-3100 and Electroporation Alone or in Combination With Imiquimod for the Treatment of HPV-16 and/or HPV-18 Related Vulvar HSIL (Also Referred as: VIN 2 or VIN 3) (NCT03180684). ClinicalTrials.gov.; 2023. Available online: https://clinicaltrials.gov/study/NCT03180684 (accessed on 8 October 2025).
- Inovio Pharmaceuticals. VGX-3100 Delivered Intramuscularly (IM) Followed by Electroporation (EP) for the Treatment of HPV-16 and/or HPV-18 Related Anal or Anal/Peri-Anal, High Grade Squamous Intraepithelial Lesion (HSIL) in Individuals Seronegative for Human Immunodeficiency Virus (HIV)-1/2 (NCT03499795). ClinicalTrials.gov.; 2023. Available online: https://www.clinicaltrials.gov/study/NCT03499795 (accessed on 8 October 2025).
- VGX-3100 and Electroporation in Treating Patients with HIV-Positive High-Grade Anal Lesions (NCT03603808). ClinicalTrials.gov; 2025. Available online: https://clinicaltrials.gov/study/NCT03603808 (accessed on 8 October 2025).
- Kermanshahi, A.Z.; Ebrahimi, F.; Taherpoor, A.; Eslami, N.; Baghi, H.B. HPV-driven cancers: A looming threat and the potential of CRISPR/Cas9 for targeted therapy. Virol. J. 2025, 22, 156. [Google Scholar] [CrossRef]
- Wiik, J.; Kärrberg, C.; Nilsson, S.; Strander, B.; Jacobsson, B.; Sengpiel, V. Associations between cervical intraepithelial neoplasia during pregnancy, previous excisional treatment, cone-length and preterm delivery: A register-based study from western sweden. BMC Med. 2022, 20, 61. [Google Scholar] [CrossRef] [PubMed]
- Zhen, S.; Chen, H.; Lu, J.; Yang, X.; Tuo, X.; Chang, S.; Tian, Y.; Li, X. Intravaginal delivery for CRISPR-Cas9 technology: For example, the treatment of HPV infection. J. Med. Virol. 2023, 95, e28552. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Cai, W.; Wang, H. P53 and pRB induction improves response to radiation therapy in HPV-positive laryngeal squamous cell carcinoma. Clinics 2024, 79, 100415. [Google Scholar] [CrossRef]
- Chung, H.C.; Ros, W.; Delord, J.P.; Perets, R.; Italiano, A.; Shapira-Frommer, R.; Manzuk, L.; Piha-Paul, S.A.; Xu, L.; Zeigenfuss, S.; et al. Efficacy and Safety of Pembrolizumab in Previously Treated Advanced Cervical Cancer: Results From the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2019, 37, 1470–1478. [Google Scholar] [CrossRef]
- Doran, S.L.; Stevanović, S.; Adhikary, S.; Gartner, J.J.; Jia, L.; Kwong, M.L.M.; Faquin, W.C.; Hewitt, S.M.; Sherry, R.M.; Yang, J.C.; et al. T-cell receptor gene therapy for human papillomavirus–associated epithelial cancers: A first-in-human, phase i/ii study. J. Clin. Oncol. 2019, 37, 2759–2768. [Google Scholar] [CrossRef]
- Liu, N.; Wang, X.; Wang, Z.; Kan, Y.; Fang, Y.; Gao, J.; Kong, X.; Wang, J. Nanomaterials-driven in situ vaccination: A novel frontier in tumor immunotherapy. J. Hematol. Oncol. 2025, 18, 45. [Google Scholar] [CrossRef]
- Zheng, S.; Wang, W.; Shen, L.; Yao, Y.; Xia, W.; Ni, C. Tumor battlefield within inflamed, excluded or desert immune phenotypes: The mechanisms and strategies. Exp. Hematol. Oncol. 2024, 13, 80. [Google Scholar] [CrossRef]
- García, E.; Ayoub, N.; Tewari, K.S. Recent breakthroughs in the management of locally advanced and recurrent/metastatic cervical cancer. J. Gynecol. Oncol. 2024, 35, e30. [Google Scholar] [CrossRef]
- Qi, L.; Luo, Q.; Zhang, Y.; Jia, F.; Zhao, Y.; Wang, F. Advances in toxicological research of the anticancer drug cisplatin. Chem. Res. Toxicol. 2019, 32, 1469–1486. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, A.N.; Lee, L.J.; Eswara, J.R.; Horowitz, N.S.; Konstantinopoulos, P.A.; Mirabeau-Beale, K.L.; Rose, B.S.; Von Keudell, A.G.; Wo, J.Y. Complications of pelvic radiation in patients treated for gynecologic malignancies. Cancer 2014, 120, 3870–3883. [Google Scholar] [CrossRef] [PubMed]
- Coventry, B.J. Therapeutic vaccination immunomodulation: Forming the basis of all cancer immunotherapy. Ther. Adv. Vaccines Immunother. 2019, 7, 2515135519862234. [Google Scholar] [CrossRef] [PubMed]
- Zolfi, E.; Khaleghi Mehr, F.; Emtiazi, N.; Moradi, Y. A review of the carcinogenic potential of human papillomavirus (HPV) in urological cancers. Virol. J. 2025, 22, 53. [Google Scholar] [CrossRef]
- Caley, A.; Evans, M.; Powell, N.; Paleri, V.; Tomkinson, A.; Urbano, T.G.; Jay, A.; Robinson, M.; Thavaraj, S. Multicentric human papillomavirus–associated head and neck squamous cell carcinoma. Head Neck 2014, 37, 202–208. [Google Scholar] [CrossRef]
- Khanna, L.; Prasad, S.R.; Yedururi, S.; Parameswaran, A.M.; Marcal, L.P.; Sandrasegaran, K.; Tirumani, S.H.; Menias, C.O.; Katabathina, V.S. Second malignancies after radiation therapy: Update on pathogenesis and cross-sectional imaging findings. Radiat. Oncol. 2021, 41, 876–894. [Google Scholar] [CrossRef]
- Zhou, P.; Liu, W.; Cheng, Y.; Qian, D. Nanoparticle-based applications for cervical cancer treatment in drug delivery, gene editing, and therapeutic cancer vaccines. WIREs Nanomed. Nanobiotechnol. 2021, 13, e1718. [Google Scholar] [CrossRef]
- Calderón, M.; Hedtrich, S. The delivery challenge of genome editing in human epithelia. Adv. Healthc. Mater. 2021, 10, 2100847. [Google Scholar] [CrossRef] [PubMed]
- Mutombo, A.B.; Simoens, C.; Tozin, R.; Bogers, J.; Van Geertruyden, J.P.; Jacquemyn, Y. Efficacy of commercially available biological agents for the topical treatment of cervical intraepithelial neoplasia: A systematic review. Syst. Rev. 2019, 8, 132. [Google Scholar] [CrossRef]
- Bahrami, K.; Lee, E.; Morse, B.; Lanier, O.L.; Peppas, N.A. Design of nanoparticle-based systems for the systemic delivery of chemotherapeutics: Alternative potential routes via sublingual and buccal administration for systemic drug delivery. Drug Deliv. Transl. Res. 2023, 14, 1173–1188. [Google Scholar] [CrossRef]
- Kotterman, M.A.; Chalberg, T.W.; Schaffer, D.V. Viral vectors for gene therapy: Translational and clinical outlook. Annu. Rev. Biomed. Eng. 2015, 17, 63–89. [Google Scholar] [CrossRef]
- Zhu, D.; Shen, H.; Tan, S.; Hu, Z.; Wang, L.; Yu, L.; Tian, X.; Ding, W.; Ren, C.; Gao, C.; et al. Nanoparticles Based on Poly (β-Amino Ester) and HPV16-Targeting CRISPR/shRNA as Potential Drugs for HPV16-Related Cervical Malignancy. Mol. Ther. 2018, 26, 2443–2455. [Google Scholar] [CrossRef]
- Gao, X.; Jin, Z.; Tan, X.; Zhang, C.; Zou, C.; Zhang, W.; Ding, J.; Das, B.C.; Severinov, K.; Hitzeroth, I.I.; et al. Hyperbranched poly(β-amino ester) based polyplex nanopaticles for delivery of CRISPR/Cas9 system and treatment of HPV infection associated cervical cancer. J. Control. Release 2020, 321, 654–668. [Google Scholar] [CrossRef]
- Xiong, J.; Tan, S.; Yu, L.; Shen, H.; Qu, S.; Zhang, C.; Ren, C.; Zhu, D.; Wang, H. E7-Targeted Nanotherapeutics for Key HPV Afflicted Cervical Lesions by Employing CRISPR/Cas9 and Poly (Beta-Amino Ester). Int. J. Nanomed. 2021, 16, 7609–7622. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, Z.; Lai, L.; Li, Z. Efficient C-to-G Base Editing with Improved Target Compatibility Using Engineered Deaminase-nCas9 Fusions. Cris. J. 2022, 5, 389–396. [Google Scholar] [CrossRef]
- Wilbie, D.; Walther, J.; Mastrobattista, E. Delivery aspects of crispr/cas for in vivo genome editing. Acc. Chem. Res. 2019, 52, 1555–1564. [Google Scholar] [CrossRef] [PubMed]
- Mingozzi, F.; High, K.A. Overcoming the host immune response to adeno-associated virus gene delivery vectors: The race between clearance, tolerance, neutralization, and escape. Annu. Rev. Virol. 2017, 4, 511–534. [Google Scholar] [CrossRef]
- Cencic, R.; Miura, H.; Malina, A.; Robert, F.; Ethier, S.; Schmeing, T.M.; Dostie, J.; Pelletier, J. Protospacer adjacent motif (pam)-distal sequences engage crispr cas9 dna target cleavage. PLoS ONE 2014, 9, e109213. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chao, Z.; Ding, W.; Fang, T.; Gu, X.; Xue, M.; Wang, W.; Han, R.; Sun, W. A Multiplex Rpa-Crispr/Cas12a-Based Poct Technique and Its Application in Human Papillomavirus (HPV) Typing Assay. Cell. Mol. Biol. Lett. 2024, 29, 34. [Google Scholar] [CrossRef]
- Mills, C.; Riching, A.; Keller, A.; Stombaugh, J.; Haupt, A.; Maksimova, E.; Dickerson, S.; Anderson, E.; Hemphill, K.; Ebmeier, C. A novel crispr interference effector enabling functional gene characterization with synthetic guide rnas. Cris. J. 2022, 5, 769–786. [Google Scholar] [CrossRef]
- Wu, Z.; Tang, Y.; Tang, M.; Wu, Z.; Xu, Y. The relationship between the eradication of helicobacter pylori and the occurrence of stomach cancer: An updated meta-analysis and systemic review. BMC Gastroenterol. 2025, 25, 278. [Google Scholar] [CrossRef]
- Aghamiri, S.; Talaei, S.; Roshanzamiri, S.; Zandsalimi, F.; Fazeli, E.; Aliyu, M.; Kheiry Avarvand, O.; Ebrahimi, Z.; Keshavarz-Fathi, M.; Ghanbarian, H. Delivery of genome editing tools: A promising strategy for HPV-related cervical malignancy therapy. Expert Opin. Drug Deliv. 2020, 17, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Badran, A.H.; Liu, D.R. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 2017, 168, 20–36. [Google Scholar] [CrossRef] [PubMed]
| Feature | CRISPR/Cas9 | TALENs | ZFNs |
|---|---|---|---|
| Mechanism | RNA-guided DNA cleavage via Cas9 endonuclease | DNA-binding TALE repeats fused to FokI nuclease | Zinc-finger domains fused to FokI nuclease |
| Target Recognition | 20-nt guide RNA binds complementary DNA adjacent to PAM | TALE repeats recognize individual nucleotides, dimerize FokI | Zinc fingers recognize 3–4 bp each, FokI dimerization |
| Ease of Design | Very high → only guide RNA sequence needs to be changed | Moderate → modular but labor-intensive to assemble | Low → requires protein engineering and optimization |
| Specificity | High with optimized guides, off-targets minimized using high-fidelity variants | Very high → one-to-one nucleotide recognition; no PAM requirement | Variable → some designs highly specific, others very prone to off-targets |
| Delivery | Challenging due to Cas9 size (~4.2 kb), solved via smaller orthologs or mRNA | Large plasmid size (>3 kb per TALEN), amenable to mRNA or protein delivery | Smallest, can fit both ZFNs in one AAV vector |
| Multiplexing | Yes → multiple sgRNAs can be used simultaneously | Difficult → requires multiple TALEN pairs | Very difficult → requires multiple protein constructs |
| Off-target Risk | Moderate → reduced with proper guide design and variants | Low → mismatch discrimination is high | High → may cut hundreds of off-target sites depending on design |
| Gene Editing Tool | Lesion Type | Cell Line | Target Gene | Outcomes/Findings | Reference |
|---|---|---|---|---|---|
| CRISPR/Cas9 | Cervical cancer | SiHa, CaSki | HPV16 E7 |
| Hu et al., 2014 [14] |
| CRISPR/Cas9 | Cervical cancer | Hela, SiHa | HPV18 E6 & E7 |
| Ling et al., 2020 [22] |
| CRISPR/Cas9 | Genital warts | HPV6/11 E7-transformed keratinocytes | HPV6/11 E7 |
| Liu et al., 2016 [63] |
| CRISPR/Cas9 | Cervical cancer | SiHa, C33A | HPV16 E6 & E7 |
| Zhen et al., 2016 [41] |
| CRISPR/Cas9 (micelles) | Cervical cancer | HeLa | HPV18 E7 |
| Lao et al., 2018 [64] |
| CRISPR/Cas9 (AAV) | Cervical cancer | HeLa, HCS-2, SKG-I, HEK293 | HPV18 E6 |
| Yoshiba et al., 2018 [65] |
| CRISPR/Cas9, FokI-dCas9 | Cervical cancer | CaSki, HeLa | HPV16/18 E6 & E7 |
| Jubair et al., 2019 [52] |
| CRISPR/Cas9 (HCAdV) | Cervical cancer | SiHa, CaSki, HeLa | HPV16/18 E6 |
| Ehrke-Schulz et al., 2020 [42] |
| CRISPR/Cas13a | Cervical cancer | SiHa, HeLa, C33A | HPV16/18 E6 & E7 |
| Chen et al., 2020 [54] |
| CRISPR/Cas13a | Genital warts | HPV16/18 E6-transformed keratinocytes | HPV16/18 E6 |
| Li et al., 2020 [66] |
| CRISPR/Cas9 | Cervical cancer | HeLa | HPV18 E6 & E7 |
| Inturi et al., 2021 [67] |
| CRISPR/Cas9 (inducible) | Oral SCC | UDSCC2 (SCC2) | HPV16 E7 |
| Ferreira et al., 2022 [56] |
| CRISPR/Cas9, TALEN, ZFN | Cervical cancer | SiHa, S12, HeLa, C33A | HPV16 E7 |
| Gao et al., 2022 [43] |
| CRISPR/Cas9 (AAV) | Cervical cancer | HeLa | HPV18 E6 |
| Noroozi et al., 2022 [68] |
| CRISPR/Cas9 | Cervical cancer | C3, TC1, HeLa | HPV16 E5, E6, E7 |
| Khairkhah et al., 2023 [53] |
| CRISPR/Cas9 (SONC103) | Cervical cancer | CaSki | HPV16 E6, E7, E2, E5 |
| Hu et al., 2024 [69] |
| CRISPR/Cas13a | Cervical cancer | HeLa | HPV18 E6 |
| Zhang et al., 2024 [70] |
| TALEN | Cervical cancer | HeLa, SiHa, C33A, HEK293 | HPV16/18 E6 & E7 |
| Hu et al., 2015 [55] |
| TALEN | Cervical cancer | SiHa | HPV16 E7 |
| Shankar et al., 2017 [71] |
| ZFN | Cervical cancer | SiHa, HeLa | HPV16/18 E7 |
| Ding et al., 2014 [36] |
| Gene Editing Tool | Lesion Type | Cell Line | Target Gene | Outcomes/Findings | Reference |
|---|---|---|---|---|---|
| CRISPR/Cas9 | Cervical cancer | Nude mice (HeLa xenograft) | HPV18 E6 & E7 |
| Ling et al., 2020 [22] |
| CRISPR/Cas9 + CDDP | Cervical cancer | BALB/c nude mice (hydrodynamic tail vein) | HPV16 E6 & E7 |
| Zhen et al., 2016 [41] |
| CRISPR/Cas9 (micelles) | Cervical cancer | Nude mice (HeLa xenograft) | HPV18 E7 |
| Lao et al., 2018 [64] |
| CRISPR/Cas9 (AAV) | Anal cancer | Immunodeficient mice (ANA001 PDX) | HPV16 E6 & E7 |
| Hsu et al., 2018 [62] |
| CRISPR/Cas9 (AAV) | Cervical cancer | BALB/c nude mice (SKG-I xenograft + AAV-sgE6) | HPV18 E6 |
| Yoshiba et al., 2018 [65] |
| CRISPR/Cas9 | Cervical cancer | Rag1 mice (CaSki, HeLa xenograft, lipoplex Cas9) | HPV16/18 E7 |
| Jubair et al., 2019 [52] |
| CRISPR/Cas9 + anti-PD1 immunotherapy | Cervical cancer | hu-PBL-SCID (SiHa-Luc xenograft) | HPV16 E6 & E7 + PD1 |
| Zhen et al., 2020 [48] |
| CRISPR/Cas13a | Cervical cancer | BALB/c nude (SiHa/HeLa CRISPR knockdown) | HPV16 E6 |
| Chen et al., 2020 [54] |
| Inducible CRISPR/Cas9 | Oral SCC | Nude mice (SCC2 xenograft) | HPV16 E7 |
| Ferreira et al., 2022 [56] |
| CRISPR/Cas9 | Cervical cancer | BALB/c nude (S12 xenograft) | HPV16 E7 |
| Gao et al., 2022 [43] |
| CRISPR/Cas9 | Cervical cancer | K14-HPV16 transgenic mice | HPV16 E7 |
| Gao et al., 2022 [43] |
| CRISPR/Cas9 + liposomes | Cervical cancer | hu-PBL-SCID (SiHa xenograft) | HPV16 E6 & E7 |
| Zhen et al., 2023 [72] |
| CRISPR/Cas9 + cisplatin | Cervical cancer | C57BL/6 (C3 xenograft) | HPV16 E5, E6, E7 |
| Khairkhah et al., 2023 [53] |
| CRISPR/Cas9 (SONC103 virus) | Cervical cancer | BALB/c (CaSki xenograft) | HPV16 E6, E7, E2, E5 |
| Hu et al., 2024 [69] |
| TALEN | Cervical cancer | BALB/c nude + K14-HPV16 topical T512 | HPV16/18 E6 & E7 |
| Hu et al., 2015 [55] |
| ZFN | Cervical cancer | BALB/c nude (SiHa/HeLa xenografts) | HPV16/18 E7 |
| Ding et al., 2014 [36] |
| Trial | Target/Strategy | Indication | Approach |
|---|---|---|---|
| NCT03057912 | CRISPR/Cas9 or TALENs targeting HPV16/18 E6/E7 | CIN I with persistent HPV16/18 infection | Intravaginal plasmid delivery, open-label Phase I trial |
| NCT02793856 | PD-1 knockout via CRISPR in autologous T cells | HPV+ metastatic non-small cell lung cancer | T cell gene editing and reinfusion |
| NCT03578406 | TCR-engineered T cells recognizing HPV16 E6 | Metastatic cervical cancer | TCR-transduced T cells (not gene-edited yet) |
| NCT00685412 NCT01304524 NCT03185013 (REVEAL1) NCT03721978 (REVEAL2) | VGX-3100 therapeutic vaccine targeting HPV16/18 E6/E7 DNA | Cervical intraepithelial neoplasia (CIN) 2/3 | DNA vaccine + Electroporation Phase I, II and III trials |
| NCT03603808 | VGX-3100 therapeutic vaccine targeting HPV16/18 E6/E7 DNA | Anal high-grade squamous intraepithelial neoplasia (HSIL) or AIN 2/3 in HIV+ individuals | DNA vaccine + Electroporation Phase II trial |
| NCT03499795 | VGX-3100 therapeutic vaccine targeting HPV16/18 E6/E7 DNA | AIN2, AIN3, PAIN2, PAIN3 in HIV– individuals | DNA vaccine + Electroporation Phase II trial |
| NCT03180684 | VGX-3100 therapeutic vaccine targeting HPV16/18 E6/E7 DNA | Vulvar high-grade squamous intraepithelial lesion (HSIL) or VIN 2/3 | DNA vaccine + Electroporation Phase II trial |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Hellenic Society for Microbiology. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cakir, M.O.; Selek, M.; Yilmaz, B.; Ozdogan, M.; Ashrafi, G.H. Genome Editing Against HPV-Driven Cancers: From Bench to Clinic. Acta Microbiol. Hell. 2025, 70, 41. https://doi.org/10.3390/amh70040041
Cakir MO, Selek M, Yilmaz B, Ozdogan M, Ashrafi GH. Genome Editing Against HPV-Driven Cancers: From Bench to Clinic. Acta Microbiologica Hellenica. 2025; 70(4):41. https://doi.org/10.3390/amh70040041
Chicago/Turabian StyleCakir, Muharrem Okan, Melis Selek, Betul Yilmaz, Mustafa Ozdogan, and Gholam Hossein Ashrafi. 2025. "Genome Editing Against HPV-Driven Cancers: From Bench to Clinic" Acta Microbiologica Hellenica 70, no. 4: 41. https://doi.org/10.3390/amh70040041
APA StyleCakir, M. O., Selek, M., Yilmaz, B., Ozdogan, M., & Ashrafi, G. H. (2025). Genome Editing Against HPV-Driven Cancers: From Bench to Clinic. Acta Microbiologica Hellenica, 70(4), 41. https://doi.org/10.3390/amh70040041






