Abstract
Background/Objectives: The potential of DNA as an information-dense storage medium has inspired a broad spectrum of creative systems. In particular, hybrid biomolecular systems that integrate new materials and chemistries with DNA could drive novel functions. In this work, we explore the potential for proteins to serve as molecular file addresses. We stored DNA-encoded data in yeast and leveraged yeast surface display to readily produce the protein addresses and make them easy to access on the cell surface. Methods: We generated yeast populations that each displayed a distinct protein on their cell surfaces. These proteins included binding partners for cognate antibodies as well as chromatin-associated proteins that bind post-translationally modified histone peptides. For each specific yeast population, we transformed a library of hundreds of DNA sequences collectively encoding a specific image file. Results: We first demonstrated that the yeast retained file-encoded DNA through multiple cell divisions without a noticeable skew in their distribution or a loss in file integrity. Second, we showed that the physical act of sorting yeast displaying a specific file address was able to recover the desired data without a loss in file fidelity. Finally, we showed that analog addresses can be achieved by using addresses that have overlapping binding specificities for target peptides. Conclusions: These results motivate further exploration into the advantages proteins may confer in molecular information storage.
1. Introduction
The growing interest in DNA as a potential substrate for digital data storage derives predominantly from its high theoretical information density and low resource utility. Yet, it is also intriguing to consider how DNA might confer additional advantages through its unique biophysical properties and through currently unexplored interactions with novel chemistries and other materials. As just a few examples, the physical structure of DNA itself or epigenetic modifications to the DNA could store information or confer new functionalities [1,2,3,4,5]. Hybrid systems incorporating materials like polymers, silica particles, and soft dendritic colloids provide additional functionalities ranging from repeatable multiplexed data access, in-storage computation, and long-term storage [6,7,8,9].
Proteins may present a powerful class of molecular materials to integrate into DNA-based information systems. They could provide diverse molecular recognition and search capabilities, evident in the natural role some proteins hold as antibodies. Their molecular recognition capabilities derive from the high degrees of three-dimensional and chemical freedom conferred by the twenty amino acid building blocks. They do face several potential limitations, including their lower stability, higher cost, and orders of magnitude lower synthesis scalability compared to DNA.
Yeast surface display presents an interesting system by which to leverage the advantages of both DNA and proteins. Yeast can be lyophilized and kept stable for up to 20 years [10]. It can encode proteins in synthesized DNA, bypassing the need for chemical or recombinant protein synthesis and purification. It can display those proteins on its cell surface, providing a potential molecular address by which to find and manipulate specific yeast cells [11]. Recent work also demonstrates that yeast can be transformed to hold large synthetic DNA cassettes and even whole artificial chromosomes [12,13,14,15]. In a related biomolecular system, promising work has shown file-encoded DNA stored and accessed in bacteria [16,17,18,19,20,21].
Here, we probed the potential utility of yeast surface display leveraging protein addresses for DNA-based information storage. We first tracked how the distribution of yeast populations, collectively comprising data files, were maintained over multiple cell divisions. We then implemented file access via sorting based upon protein addresses. Finally, we demonstrated how proteins can confer unique types of addresses that extend beyond simple one-to-one interactions.
2. Materials and Methods
2.1. Yeast Strains
Frozen yeast strains from prior work were used for this study, and all derived from EBY100 (Table S1) [22,23,24]. All frozen yeast were streaked on dropout agar plates except for the parent EBY100, which was streaked on a YPD agar plate. EBY100 is BJ5465 (MATa aga1::gal1-aga1::ura3 ura3-52 trp1 leu2-delta200 his3-delta200 pep4::HIS3 prbd1.6R can1 GAL) and is an auxotroph for Leu and Trp. Single colonies were then picked and placed into a 2 mL culture of their appropriate synthetic dropout or YPD media to grow for 2 days at 30 °C, 250 rpm. From this yeast stock, a fresh SDCAA-Tryptophan (SD-trp) culture was seeded before each experiment. For an experiment, an SD-trp culture of yeast was grown for 24 h before passage into SGCAA-Tryptophan (SG-trp) induction media, where it was grown at 20 °C, 250 rpm for 16–24 h. SG-trp cultures were all induced at an OD600nm of 1. In addition, 1 µM biotin in DMSO was used in inducing the yeast strain with the FLAG tag to aid in its folding. After the file library was electroporated in the yeast strains, they were grown in SDCAA-Tryptophan–Leucine culture (SD-trp-leu) instead, with the corresponding SGCAA -Tryptophan–Leucine (SG-trp-leu) media for induction.
2.2. Labeling Epitope Tag Yeast Before and After File Transformation with Flow Cytometry
Freshly induced yeast cultures were processed for flow cytometry by aliquoting 4 × 105 cells into individual wells of a 96-well plate for antibody labeling. Each sample was labeled with a mixture of primary and secondary antibodies. First, 50 µL of a primary antibody dilution (Chicken anti-MYC for the MYC tag and Rabbit anti-FLAG for the FLAG tag) was added to each well and allowed to incubate at room temperature, shaking at 800 rpm for 30 min. The samples were then washed of unbound primary antibody by resuspension in 200 µL of 0.1% BSA 1× PBS and centrifugation at 3000× g for 2 min and then aspirated. The samples were then labeled with a secondary antibody corresponding to the host animal of the primary antibody (donkey anti-Chicken 647 for the MYC tag and donkey anti-rabbit 647 for the FLAG tag). The secondary antibodies were added in 50 µL aliquots of 1:250 dilutions of their stock concentrations. The secondary antibodies were incubated with the samples for at least 15 min in the dark, at 4 °C. The secondary antibodies were washed in the same manner as the primary antibodies. The final sample pellets were resuspended in 200 µL of 0.1% BSA 1× PBS when the plate was loaded into the flow cytometer (MACSQuant® VYB, Auburn, CA, USA). Flow cytometry data were analyzed with FlowJo software (FlowJo 10.10). All experiments were performed in triplicate with three biological replicates showing the same trend. In total, 50,000 cells were collected for analysis for each sample.
2.3. File DNA Synthesis
Each file DNA library was synthesized from a Twist DNA oligo pool containing all the oligos from the three picture files. Overhang primers were designed to include overlapping regions with the file plasmid (pCT302-SsoFe2-T2A-TOM22) to extend the strand length by 40 bp on each end (Table S2). Multiple PCR reactions were set up to yield a final amount of 12 µg per file library after purification. Each PCR consisted of the following reagents: 1 uL 1 × 1010 st/µL template, 2 µL of each 10 µM primer, 5 µL of the 5× Q5 reaction buffer, 0.5 µL of a 10 mM dNTPs mix, 0.25 µL of the Q5 polymerase (New England Biolabs Cat # M0491L), and 14.25 µL of nanopure water. The amplicons were run on a 1% agarose gel and extracted for gel purification. For the file plasmid, a 500 mL bacteria culture containing the pCT302-SsoFe2-T2A-TOM22 was grown overnight and spun down for plasmid extraction. The purified plasmid was then digested with restriction enzymes EagI and AvrII at 37 °C for 1 h and dephosphorylated for 10 min. The digested product was then run on a 1% agarose gel. The digested 7780 bp band was excised and extracted with a gel purification kit. The purified DNA libraries and digested vector were concentrated to a 30 µL volume each and quantified using nanodrop.
2.4. Inserting Files into Selected Yeast
Each file DNA library was transformed into different yeast strains with different histone binders and epitope tags displayed. Table S3 notes the file DNA library for each protein address used in this manuscript. For each yeast strain, a frozen stock was streaked on an SD-trp agar plate to incubate for 2 days at 30 °C. Then, a single colony was inoculated in 5 mL of SD-trp media and grown at 30 °C to an OD600nm of 3. This culture was passaged into 200 mL of fresh YPD media at an initial OD600nm of 3 and grown to OD600nm of 1.6. The culture was then spun down at 3000 rpm for 3 min. The supernatant was removed and the pellet washed twice with 100 mL of autoclaved water followed by one wash in 100 mL of autoclaved electroporation buffer (1 M sorbitol and 1 mM CaCl2). The pellet was then resuspended in 40 mL of a filter-sterilized LiAC/DTT/HEPES solution (0.1 M LiAc, 50 mM HEPES, 1.5 mg/mL DTT, pH 7.5) and shaken in a 250 mL baffled flask for 30 min at 30 °C. The culture was spun down and washed with 100 mL of electroporation buffer before resuspension in 2 mL of electroporation buffer. Electroporation cuvettes (GenePulser cuvette, 0.2 cm electrode gap, Hercules, CA, USA) were chilled before 400 µL aliquots of the cell mixture were added for each transformation. Two transformations were performed for each yeast strain: one with the file DNA library and one without to serve as the control. 12 µg of the file DNA library and 4 µg of the file plasmid vector were used for each sample, as applicable. After pipetting the sample gently, the mixture was electroporated at 2500 V, 25 F, and 200 ohms. The mixture was then transferred to a separate culture tube, and the cuvette was washed with a 1:1 sorbitol/YPD solution, with each wash transferred to the culture tube. The culture tubes were incubated at 30 °C for 1 h at 250 rpm. The cultures were then transferred to a conical tube, spun down, and washed in SD-trp-leu media. Finally, each pellet was resuspended in 5 mL of SD-trp-leu media and 10-fold serial dilutions of each transformation was plated on SD-trp-leu agar to compare the colony counts of the control and test sample of each transformation. The remaining culture containing the yeast transformations with the file inserts was suspended in 125 mL of SD-trp-leu media and grown overnight. A penicillin-strep stock solution was added to a final concentration of 1× the following morning, and the cultures were used for the sorting experiments after the second passage.
2.5. Analyzing the File3 Yeast Through Multiple Cell Divisions
The transformed yeast strain containing the File3 DNA was used for the serial passaging study. In this experiment, the culture was grown for about 5 growth cycles (from OD600nm 0.05 to OD600nm 3) and aliquoted into 2 mL of fresh SD-trp-leu media to repeat the growth process again for 4 more times. After each growth period, 1 mL of the culture was collected for plasmid extraction. The aliquoted pellet was frozen at −80 °C so that the pellets from each passage could be processed simultaneously. The sequencing primers for File3 (Table S2) were used to amplify the data payload region of the extracted plasmids using the same PCR set-up as previously described, and the amplicons were purified via gel extraction.
2.6. Yeast Sorting and File Access via Epitope Tag
After vortexing the Protein A-conjugated Dynabeads stock bottle for 30 s, a 950 uL aliquot was transferred to a separate conical tube. Each sample used 1.5 mg (50 µL) Dynabeads, and the Dynabeads were prepped as a master mix. The Dynabeads were washed twice with 1 mL of a buffer containing 0.1% BSA and 0.02% Tween in PBS. This washing buffer was used throughout the experiment. After removing the supernatant by placing the tube against a magnet, the aliquot was resuspended in 1 mL of the washing buffer and divided into two portions for each antibody. The washing buffer was removed, and 50 µL of each antibody stock solution (rabbit anti-FLAG or rabbit anti-MYC) was added to each aliquot and mixed with the washing buffer for a total volume of 2 mL. The tubes were then incubated with rotation for 30 min at room temperature. The antibody-bound beads were then washed twice with 1 mL of the washing buffer and resuspended in 1 mL of the washing buffer. To prep the yeast, 2 × 107 cells of each file-transformed strain were used for each condition. The file-transformed strains were passaged and induced. Additionally, 2 × 108 cells of EBY100 Saccharomyces cerevisiae were added for each condition. The EBY100 Saccharomyces cerevisiae was grown in YPD on the day that the file-transformed yeast strains were induced. Each sample was spun down at 4 G for 2 min. After removing the media, each cell pellet was washed twice with the washing buffer. Each cell pellet was then resuspended in 100 µL of the appropriate antibody-bound bead solution and 900 µL of the washing buffer. The tubes were incubated for 1 h on rotation before each mixture was washed twice with 1 mL of the washing buffer. The supernatant was then removed, and each mixture was resuspended in 2 mL of SD-trp-leu media and placed in a culture block to shake for two days at 250 rpm on 30 °C. Each mixture was then induced at an OD600nm of 1 at 20 °C, and the magnetic bead sorting procedure was repeated upon the sorted cells for refined results. After growing the final sorted mixtures overnight, the plasmids were extracted for a multiplex amplification of the file payload regions using the Zymo Yeast Plasmid Miniprep II kit (Cat #D2004).
Specifically, primers for File2 and File3 were used with Taq polymerase to enrich the extracted DNA, and the amplicons were purified using PCR purification columns before submission to Azenta Amplicon-EZ (Table S2).
2.7. Labeling the Histone Binder Yeast After File Transformation with Flow Cytometry
Freshly induced yeast cultures were processed for flow cytometry by aliquoting 4 × 105 cells into individual wells of a 96-well plate for peptide and antibody labeling. Dilutions of a H3K9me3, H3K27me3, and H3K14ac biotinylated peptide solution were made at a final concentration of 4 uM, 4 uM, and 8 uM, respectively. A dilution of a rabbit anti-MYC solution was also prepped at a final dilution of 1:75. Equivolumes of each diluted peptide and rabbit anti-MYC solution were mixed to prep master mixes for the experiment. Then, 50 µL of the corresponding peptide-myc mixture was added to each well and allowed to incubate at room temperature, shaking at 800 rpm for 30 min. The samples were then washed of unbound primary antibody by resuspension in 200 µL of 0.1% BSA 1× PBS, centrifugation at 3000× g for 2 min, and aspiration. The samples were then stained with a mixture of streptavidin-conjugated PE staining dye and a donkey anti-rabbit 647 antibody. The staining mixture was added in 50 µL aliquots of 1:250 dilutions of their stock concentrations. The staining mixture was allowed to incubate with the samples for at least 15 min in the dark, at 4 °C. The unbound staining mixture was washed in the same manner as the primary labeling step. The final sample pellets were resuspended in 200 µL of 0.1% BSA 1× PBS when the plate was loaded into the flow cytometer (MACSQuant® VYB). Flow cytometry data were analyzed with FlowJo software. All experiments were performed in triplicate with three biological replicates showing the same trend. All fluorescent gates were created based on an unlabeled sample of the same yeast strain (with the same bound peptide when applicable), measured on the same day, as the experimental samples. In total, 50,000 cells were collected for analysis for each sample.
3. Results
3.1. Data Transformed into Yeast Populations Maintain Their Fidelity Through Cell Divisions
An important first consideration for any data storage system is its fidelity over time or over repeated data access. For DNA, the highest risks to data fidelity are mutations or loss of specific sequences or strands of DNA due to chemical or physical manipulation [25]. These risks arise in vitro when DNA strands are copied through enzymatic processes like polymerase chain reaction, and they arise in vivo when cells recombinantly assemble the file-encoded DNA and use DNA polymerase to copy their DNA during cell division. To assess these risks, we transformed yeast with the 507 DNA sequences that altogether encode for File3, an image of a muscle cell (Figure 1). The encoding process is shown in Figure S1. We then cultured the yeast population for 20 cell divisions, harvesting DNA every five cell divisions. Next-generation sequencing (NGS) of the extracted DNA revealed that multiple important parameters of file fidelity were maintained. The normalized strand abundance, a metric describing the spread of the distribution of the DNA library, remained at a log-transformed median of ~0.6 copies per strand through all cell divisions with mutually similar violin plot morphologies (Figure 2a, Equation (S4)). The strand distributions also remarkably matched that of the initial synthesized DNA library prior to transformation into yeast. Each distribution displayed a normal skew (skew = 0.33; 0.28; 0.37; 0.35; 0.32) and mesokurtosis (excess kurtosis = −0.14; −0.28; 0.044; −0.0; −0.18) (Figure 2b).
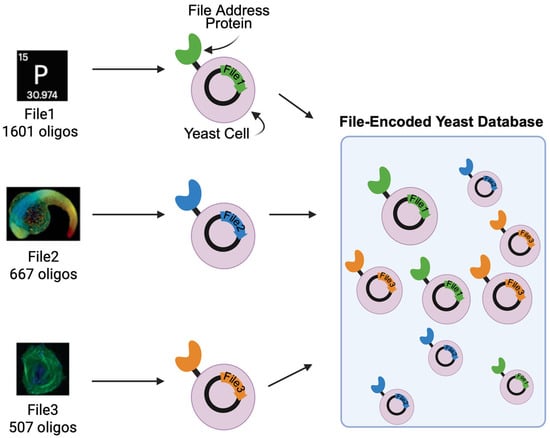
Figure 1.
Overview of hybrid DNA information storage system using file address proteins displayed on the surface of yeast. Yeast strains displaying unique file address proteins are transformed with DNA sequences that collectively encode for distinct files. The File1 DNA library consists of 1601 unique sequences and encodes an image of the phosphorus periodic table icon. The File2 DNA library consists of 667 unique sequences and encodes an image of a zebrafish embryo. The File3 DNA library consists of 507 unique oligos and encodes an image of a muscle cell [8]. Created in BioRender. Lee, M. (2025), https://BioRender.com/p35xx0d (accessed on 7 July 2025).
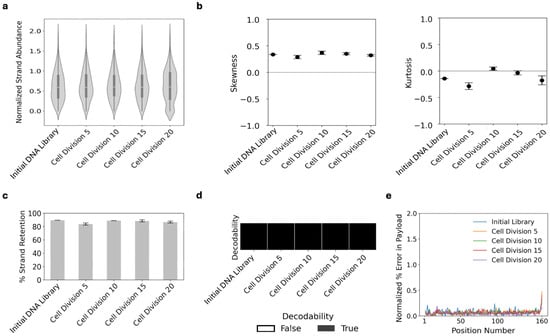
Figure 2.
Data transformed into yeast populations maintain their fidelity through cell divisions. Yeast populations containing File3 DNA were grown for up to 20 cell divisions. DNA was harvested, enriched for the file-encoded regions, sequenced, and normalized to equivalent total reads per sample. Metrics of the NGS data included (a) strand abundance, (b) skewness and kurtosis, (c) percent strand retention, (d) decodability, and (e) percent errors in the payload region of the DNA sequences, including insertions, deletions, and substitutions. There was one replicate measurement of the initial DNA library sample and three replicates from three separate yeast inductions for all other samples. The median normalized strand abundance is the white line in the middle of the thicker dark grey line in each violin plot; the thicker dark grey line depicts the interquartile (IQR) range of the distribution, and the whiskers denote the range of values within 1.5 × IQR from the ends of the box plot. Plotted values of the skewness, excess kurtosis, percent strand retention, and % error are the averages of the three replicate yeast inductions. The % error includes nucleotide substitutions, insertions, and deletions per nucleotide position of the payload sequences. Plotted error bars are the standard errors of the three replicate yeast inductions.
The distribution of yeast also affects the percent of unique DNA sequences recovered. With a limited number of NGS reads, there will often be sequences that are not recovered (drop-outs), with this percentage increasing with fewer NGS reads or with greater skew in the distribution. We observed the percent of unique DNA sequences that were retained remained consistently at ~80% through all cell divisions (Figure 2c). Relatedly, the manner in which the file information is encoded into DNA allows for strand dropouts through some level of redundancy and for some errors in synthesis, sequencing, or mutations through error correction [8,26]. We also analyzed decodability and error rates to determine the efficiency and accuracy of file retrieval. To assess error rates, after filtering the sequencing reads to only include reads with length of 160 nt and normalizing the reads per condition to make sure the same number of reads were used per condition, we ran the normalized subset of sequencing reads through the FrameD sequencing analysis tool (Figure S1) [8,26]. This tool mapped each read to its corresponding encoded strand, and the success of this mapping resulted in error rate statistics. Decodability was defined as “true” if the raw sequencing reads were successfully decoded back to the original image file and “false” if not, without prior knowledge of the file data and DNA sequences. File3 was fully decodable for all samples indicating cell divisions did not catastrophically impact strand retention or error rates (Figure 2d). We also directly measured the error rate and found <0.5% errors per nucleotide position in all samples through 20 cell divisions (Figure 2e and Figure S2). These results indicated that yeast can stably maintain DNA-encoded image files through the transformation process, cell divisions, and DNA extraction.
3.2. Transformation of Yeast with File DNA Partially Impacts Display Effiicency but Not Labeling Specificity
We next asked how file transformation might affect the display of file address proteins. To test this, we transformed yeast that can inducibly display the MYC epitope tag with File2 and yeast that can inducibly display FLAG with File3. Each yeast population was then labeled with a MYC antibody (Figure 3a) or with a FLAG antibody (Figure 3b). The transformed yeast were specifically labeled by their corresponding antibody, but there was a significant decrease in epitope display efficiency upon transformation with file DNA. We attribute this decrease to two potential factors. First, there may be competition for AGA1 partners between the AGA2-epitope fusion protein and an AGA2-TOM22 protein expressed as part of the file plasmid but that was not specifically used in this work (Supplementary Materials Section S2) [24]. Second, the retroactivity of consuming additional resources to copy, transcribe, and translate an additional plasmid could reduce the overall efficiency of surface display of proteins.
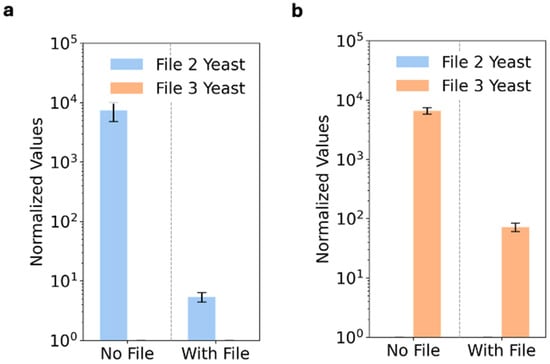
Figure 3.
Transformation of yeast with file DNA partially impacts display efficiency but not labeling specificity. (a) The MYC epitope tag specifically labeled File2 (b) and the FLAG epitope tag specifically labeled File3. Yeast with and without transformed file DNA were compared. The normalized fluorescence values were calculated from the median values of the fluorescence populations using Equations (S1) and (S2), with error bars representing the standard error. Replicates were three different yeast inductions.
3.3. Files Accessed with Specificity via Protein Addresses
After demonstrating that the transformed yeast strains preserved their address specificity, we mixed the File2 and File3 yeast populations and attempted to access each file specifically by sorting the yeast using antibodies against each displayed epitope tag. To access each file specifically, an anti-MYC or anti-FLAG antibody-bound Protein A magnetic bead was incubated with the yeast database. Bound yeast were magnetically extracted, grown for 48 h, and then magnetically sorted another time [27]. File DNA was then extracted from the sorted yeast and submitted for NGS. The NGS data indicated that File2 and File3 were successfully enriched from the MYC- and FLAG-sorted populations, respectively (Figure 4a). The strand abundances for the targeted files displayed normal skews (skew values = −0.35; −0.23) and mesokurtosis (excess kurtosis values = −0.77; −0.05) (Figure 4b). Moreover, the strand diversity was retained at a high level with almost 100% of the unique sequences retained for File3 (Figure 4c) and ~80% of the unique sequences retained for File2 (Figure S3). Both File2 and File3 were also decodable (Figure 4d), while the unwanted file was not decodable. In addition, the sorting process did not introduce any additional errors (Figure 4e and Figure S4).
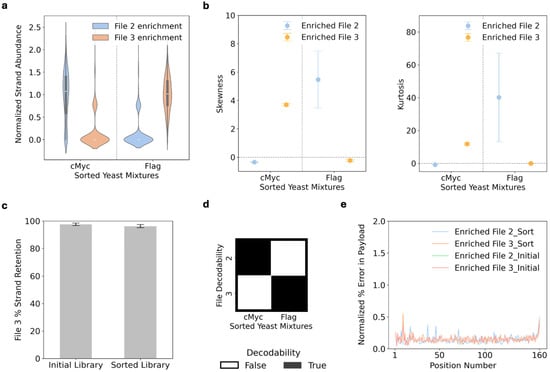
Figure 4.
Files accessed with specificity via protein addresses. Yeast containing File2 and File3 were mixed and magnetically sorted via their respective displayed epitope tags (MYC and FLAG). DNA was extracted from each sorted population and enriched for all file-encoded DNA regions simultaneously. This enriched DNA was then sequenced with the reads normalized to set the same number of reads used per condition. The normalized subsets of reads were analyzed for (a) strand abundance of each file. Three separate populations of induced yeast were used and mixed to generate three replicates of each sorted population. (a) The strand abundance was normalized via log transformation and averaged across the replicates to calculate the plotted strand abundance for each file and sort. The median value of the normalized strand abundance is depicted as the faint white line in the middle of the thicker dark grey line of each violin plot distribution, and the thicker dark grey line depicts the interquartile (IQR) range of the distribution, and the whiskers denote the range of values within 1.5 × IQR from the ends of the box plot. (b) The same samples were analyzed for skewness and kurtosis. The plotted values represent mean skewness and excess kurtosis, and the error bars represent the standard error of the replicates. (c) The strand retention of the initial File3 library and FLAG-sorted library was calculated as the percentage of the unique strands retained, with plotted values representing the mean % strand retention. The initial DNA library contains the synthesized DNA prior to transformation into yeast. The error bars are standard error. The original sequencing reads from the sorted populations were also bulk filtered against the expected sequence length and fed through a decoder to determine the (d) file decodability from the enriched DNA and (e) % error per nucleotide position, which includes nucleotide inserts, deletions, and substitutions. Of the analyzed sequences, only the % errors of the payload sequences were included here. The plotted values represent the averaged % error of the three replicates.
3.4. Combinatorial Peptide Binding Enables Multiplexed File Access
Data can be organized in more complex fashion than simple one-to-one addresses. For example, two emails may share a common label (e.g., inbox) but also retain their own unique labels as well (work vs. personal). Proteins may provide a platform of addresses to implement such overlapping labels. Here, we used histone binding proteins (human BRD2, UHRF1, and MPP8) as file addresses and biotinylated post-translationally modified histone peptides (H3K9me3, H3K27me3, and H3K14ac) to access files in a yeast database [22,23]. We chose histone binders and peptides because of their known binding promiscuity. We created a database of three file-encoded yeast populations (Table S1) and used each biotinylated histone-modified peptide to label yeast populations of interest. The MPP8 histone binding protein displayed on File3-encoded yeast binds to all three peptides, with highest affinity for the H3K9me3 peptide. The UHRF1 histone binding protein displayed on File2-encoded yeast binds to H3K9me3 primarily, with a weaker binding to the H3K14ac peptide. The BRD2 histone binding protein displayed on File1-encoded yeast binds to the H3K27me3 peptide slightly more than with the H3K14ac peptide. We then used streptavidin-PE to label the bound peptide–yeast complexes. The positively labeled yeast cells were analyzed using flow cytometry.
We found that the H3K9me3 peptide was able to label File2 and File3 from the database (Figure 5). The H3K27me3 peptide labeled File1 and File3, and the H3K14ac peptide labeled all three files. These unique combinations therefore enable the use of one histone peptide to extract combinations of multiple files.
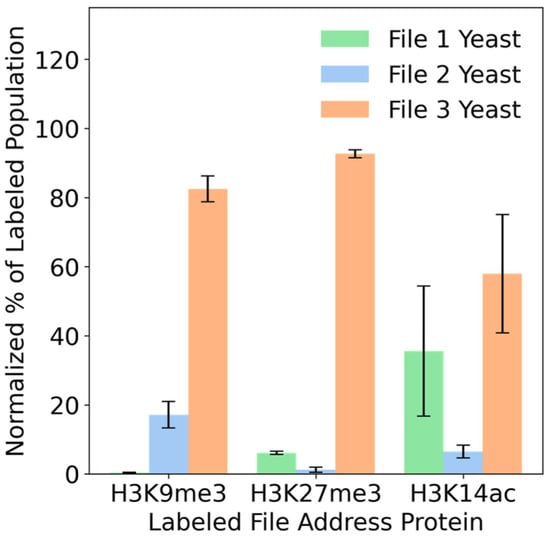
Figure 5.
Combinatorial peptide binding enables multiplexed file access. Yeast displaying BRD2 (File1), UHRF1 (File2), and MPP8 (File3) were fluorescently labeled using the peptides H3K9me3, H3K27me3, and H3K14ac. The labeled populations were then analyzed, resulting in combinatorial labeling of file sequences. Replicates were three separate yeast inductions, and the mean percentage for file pulled out per peptide was calculated from the median values from the fluorescence populations of each replicate. The means were then normalized using Equation (S3). The error bars are standard error.
4. Discussion
In this work, we demonstrated that yeast-displayed proteins provide a physical address handle for robust storage and access of DNA-based data without requiring recombinant protein production or their sensitive chemical linkages to DNA. Here, we consider some potential limitations for future investigation and engineering.
Protein stability is important to consider. Aside from the obvious need to avoid protein degradation, the structure of the protein also needs to be maintained, as structure determines binding. There are many factors that affect protein stability, including pH, temperature, the existence of other nearby polar groups, and protein chaperones [28]. Conditions favorable for yeast growth will likely place a limit on the types of proteins that can therefore be used with this platform (pH 7.5, 20−30 °C, etc.). However, one advantage of using inducible yeast surface display is that the proteins only need to be expressed and displayed during retrieval. During long-term storage, yeast can be maintained in a lyophilized form [10,25].
Storing DNA within cells has an inherent drawback compared to storage in vitro. The cell contains substantial “overhead” in the form of other cellular machinery that takes up volume. This can be partially addressed by increasing the payload each yeast cell can hold. Synthetic yeast genomes might be one avenue to increase the payload within each yeast, although the throughput of assembling such genomes would need to be improved [13,29]. Each yeast cell could theoretically hold a maximum of 3 MB of information: the Saccharomyces cerevisiae yeast has a total genome size of ~12 megabases from 16 chromosomes and encodes for ~6000 genes, but ~5000 of them are nonessential [30,31]. Therefore, the yeast genome could potentially be re-designed to eliminate dispensable, nonessential genes such that additional payload capacity is available to store information instead [32]. One million yeast cells, which volumetrically fill less than a 6.75 × 10−5 cm3, could therefore hold 3 terabytes of information.
Finally, it is important to consider how a storage system using protein addresses could scale. One of the main potential advantages of proteins is how their diverse chemistry, from at least the twenty canonical amino acids to their three dimensional structure, can confer diverse molecular recognition. The prototypical example is how the human body can synthesize three billion different antibodies to bind diverse antigens [33]. Additionally, there have been recent advances in accurately designing and predicting orthogonal protein–protein interactions de novo using structure guided and machine learning approaches [34,35]. In addition, the protein address space can be further scaled exponentially by displaying combinations of multiple proteins on the yeast surface [2]. The yeast surface display system described here can directly support future investigations into all of these approaches.
This work proposes the concept of using protein–ligand interactions to index and retrieve information from cells. The current implementation as presented here does not prove the proposed advantages of this approach, especially as it did not reach scales, densities, and speeds surpassing other DNA-based information storage systems. However, the theoretical diversity and programmable specificities of proteins and ligands motivates future exploration of this concept, in particular the practical challenges of scaling file organization and access. It is more speculative if densities and speeds could be increased, but there may be design choices that prioritize practical considerations of materials cost, economics, and storage stability and these should be investigated further.
The applications for this system could also extend past storing digital data in DNA. This system might also be used to store and report on information about each cell itself. For example, combinations of proteins could be used to tag and classify different cell types or the activities of different biological networks and mechanisms within the cells. These could facilitate autonomous organization of cells or improve the efficiency and scalability of omics methods including single-cell sequencing and the development of cell atlases [36,37].
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/dna5030034/s1. Section S1: Materials used in this work (Table S1. Main reagents and resources used in this work. Table S2. Oligonucleotide and DNA sequences to assemble file-encoded plasmids and enrich file-encoded DNA regions. Table S3. Breakdown of file diversity and protein addresses correlating to each file.); Section S2: File Plasmid Vector; Section S3: Data analysis; Section S4: Supplementary Figures (Figure S1. File encoding and decoding for image files. Figure S2. File-encoding yeast transformations preserve the sequence integrity without introducing nucleotide errors. Figure S3. The strand diversity of the File2-encoded DNA library is retained in the MYC-sorted yeast population. Figure S4. File-encoding yeast transformations preserve the sequence integrity without intro-ducing nucleotide errors in the header sequences.).
Author Contributions
Conceptualization, M.N.L., B.M.R., J.M.T. and A.J.K.; data curation, M.N.L.; formal analysis, M.N.L. and G.B.; funding acquisition, M.N.L., J.M.T., and A.J.K.; investigation, M.N.L.; methodology, M.N.L., G.B., B.M.R., J.M.T. and A.J.K.; project administration, M.N.L., G.B., J.M.T. and A.J.K.; resources, M.N.L., B.M.R. and A.J.K.; software, M.N.L., G.B. and J.M.T.; supervision, A.J.K.; validation, M.N.L., G.B. and A.J.K.; visualization, M.N.L. and A.J.K.; writing—original draft, M.N.L., B.M.R., J.M.T. and A.J.K.; writing—review and editing, M.N.L. and A.J.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded through the NIH T32 Molecular Biotechnology Training Program Fellowship (5T32GM133366-03), NSF Award 1901324, NSF Award 2027655, NSF Award 2403352, NSF Award 2144539, and NIH R01GM148562.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The research data are available and will be provided upon request.
Acknowledgments
The authors thank K. Volkel, K. Matange, R. Polak, L. Abbott, L. Han, and M. Randall for their scientific support and helpful discussions. The authors also thank Ryan Paerl and the NC State University MicroFACS facility for access to the BD FACSMelody sorting flow cytometer. Some artwork was created with BioRender.
Conflicts of Interest
J.M.T. and A.J.K. are co-founders of DNAli Technologies, which holds intellectual property related to DNA-based data storage.
References
- Organick, L.; Chen, Y.-J.; Ang, S.D.; Lopez, R.; Liu, X.; Strauss, K.; Ceze, L. Probing the Physical Limits of Reliable DNA Data Retrieval. Nat. Commun. 2020, 11, 616. [Google Scholar] [CrossRef] [PubMed]
- Tomek, K.J.; Volkel, K.; Simpson, A.; Hauss, A.G.; Indermaur, E.W.; Tuck, J.M.; Keung, A.J. Driving the Scalability of DNA-Based Information Storage Systems. ACS Synth. Biol. 2019, 8, 1241–1248. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Tabatabaei, S.K.; Hossein Tabatabaei Yazdi, S.M.; Hernandez, A.G.; Schroeder, C.M.; Milenkovic, O. Rewritable Two-Dimensional DNA-Based Data Storage with Machine Learning Reconstruction. Nat. Commun. 2022, 13, 2984. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, F.; Chao, J.; Xie, M.; Liu, H.; Pan, M.; Kopperger, E.; Liu, X.; Li, Q.; Shi, J.; et al. DNA Origami Cryptography for Secure Communication. Nat. Commun. 2019, 10, 5469. [Google Scholar] [CrossRef]
- Zhang, C.; Wu, R.; Sun, F.; Lin, Y.; Liang, Y.; Teng, J.; Liu, N.; Ouyang, Q.; Qian, L.; Yan, H. Parallel Molecular Data Storage by Printing Epigenetic Bits on DNA. Nature 2024, 634, 824–832. [Google Scholar] [CrossRef]
- Banal, J.L.; Shepherd, T.R.; Berleant, J.; Huang, H.; Reyes, M.; Ackerman, C.M.; Blainey, P.C.; Bathe, M. Random Access DNA Memory Using Boolean Search in an Archival File Storage System. Nat. Mater. 2021, 20, 1272–1280. [Google Scholar] [CrossRef]
- Bögels, B.W.A.; Nguyen, B.H.; Ward, D.; Gascoigne, L.; Schrijver, D.P.; Makri Pistikou, A.-M.; Joesaar, A.; Yang, S.; Voets, I.K.; Mulder, W.J.M.; et al. DNA Storage in Thermoresponsive Microcapsules for Repeated Random Multiplexed Data Access. Nat. Nanotechnol. 2023, 18, 912–921. [Google Scholar] [CrossRef]
- Lin, K.N.; Volkel, K.; Cao, C.; Hook, P.W.; Polak, R.E.; Clark, A.S.; San Miguel, A.; Timp, W.; Tuck, J.M.; Velev, O.D.; et al. A Primordial DNA Store and Compute Engine. Nat. Nanotechnol. 2024, 19, 1654–1664. [Google Scholar] [CrossRef]
- Zhong, W.; Wang, S.; Geng, C.; Zheng, Y.; Bai, S.; Cao, X.; Liu, K.; Yang, Y.; Lu, C.; Jiang, X. Multiplexed Random Access Approach to DNA Microspheres for High-Capacity Data Storage. Adv. Funct. Mater. 2024, 34, 2408852. [Google Scholar] [CrossRef]
- Ball, N.; Kagawa, H.; Hindupur, A.; Hogan, J.A. Development of Storage Methods for Saccharomyces Strains to Be Utilized for In Situ Nutrient Production in Long-Duration Space Missions. In Proceedings of the 47th International Conference on Environmental Systems, Charleston, SC, USA, 16–20 July 2017. [Google Scholar]
- Shusta, E.V.; Pepper, L.R.; Cho, Y.K.; Boder, E.T. A Decade of Yeast Surface Display Technology: Where Are We Now? Comb. Chem. High Throughput Screen. 2008, 11, 127. [Google Scholar] [CrossRef]
- Chen, W.; Han, M.; Zhou, J.; Ge, Q.; Wang, P.; Zhang, X.; Zhu, S.; Song, L.; Yuan, Y. An Artificial Chromosome for Data Storage. Natl. Sci. Rev. 2021, 8, 2021. [Google Scholar] [CrossRef]
- Annaluru, N.; Muller, H.; Mitchell, L.A.; Ramalingam, S.; Stracquadanio, G.; Richardson, S.M.; Dymond, J.S.; Kuang, Z.; Schifele, L.Z.; Cooper, E.M.; et al. Total Synthesis of a Functional Designer Eukaryotic Chromosome. Science 2014, 344, 55–58. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Ma, Y.; Tian, F.; Zhao, G.; Wu, Y.; Yuan, Y. YLC-Assembly: Large DNA Assembly via Yeast Life Cycle. Nucleic Acids Res. 2023, 51, 8283–8292. [Google Scholar] [CrossRef]
- Lin, Q.; Jia, B.; Luo, J.; Yang, K.; Zeller, K.I.; Zhang, W.; Xu, Z.; Stracquadanio, G.; Bader, J.S.; Boeke, J.; et al. RADOM, an Efficient in Vivo Method for Assembling Designed DNA Fragments up to 10 Kb Long in Saccharomyces Cerevisiae. ACS Synth. Biol. 2015, 4, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Qiao, H.; Gao, Y.; Wang, Z.; Qiao, X.; Chen, X.; Qi, H. A Mixed Culture of Bacterial Cells Enables an Economic DNA Storage on a Large Scale. Nat. Commun. Biol. 2020, 3, 416. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, Y.; Li, J.; Wang, F.; Wang, F.; Ma, C.; Chen, D.; Jiang, X.; Fan, C.; Zhang, H.; et al. In Vivo Processing of Digital Information Molecularly with Targeted Specificity and Robust Reliability. Sci. Adv. 2022, 8, 7415. [Google Scholar] [CrossRef]
- Bhattarai-Kline, S.; Lear, S.K.; Shipman, S.L. One-Step Data Storage in Cellular DNA. Nat. Chem. Biol. 2021, 17, 232–233. [Google Scholar] [CrossRef]
- Liu, F.; Li, J.; Zhang, T.; Chen, J.; Ho, C.L. Engineered Spore-Forming Bacillus as a Microbial Vessel for Long-Term DNA Data Storage. ACS Synth. Biol. 2022, 11, 3583–3591. [Google Scholar] [CrossRef]
- Hou, Z.; Qiang, W.; Wang, X.; Chen, X.; Hu, X.; Han, X.; Shen, W.; Zhang, B.; Xing, P.; Shi, W.; et al. “Cell Disk” DNA Storage System Capable of Random Reading and Rewriting. Adv. Sci. 2024, 11, 2305921. [Google Scholar] [CrossRef]
- Luo, H.; Huang, W.; He, Z.; Fang, Y.; Tian, Y.; Xiong, Z. Engineered Living Memory Microspheroid-Based Archival File System for Random Accessible In Vivo DNA Storage. Adv. Mater. 2025, 37, 2415358. [Google Scholar] [CrossRef]
- Meanor, J.N.; Keung, A.J.; Rao, B.M. Modified Histone Peptides Linked to Magnetic Beads Reduce Binding Specificity. Int. J. Mol. Sci. 2022, 23, 1691. [Google Scholar] [CrossRef]
- Waldman, A.C.; Rao, B.M.; Keung, A.J. Mapping the Residue Specificities of Epigenome Enzymes by Yeast Surface Display. Cell Chem. Biol. 2021, 28, 1772–1779.e4. [Google Scholar] [CrossRef] [PubMed]
- Bacon, K.; Bowen, J.; Reese, H.; Rao, B.M.; Menegatti, S. Use of Target-Displaying Magnetized Yeast in Screening MRNA-Display Peptide Libraries to Identify Ligands. ACS Comb. Sci. 2020, 22, 738–744. [Google Scholar] [CrossRef]
- Matange, K.; Tuck, J.M.; Keung, A.J. DNA Stability: A Central Design Consideration for DNA Data Storage Systems. Nat. Commun. 2021, 12, 1358. [Google Scholar] [CrossRef] [PubMed]
- Volkel, K.D.; Lin, K.N.; Hook, P.W.; Timp, W.; Keung, A.J.; Tuck, J.M. FrameD: Framework for DNA-Based Data Storage Design, Verification, and Validation. Bioinformatics 2023, 39, 572. [Google Scholar] [CrossRef]
- Cruz-Teran, C.A.; Bacon, K.; McArthur, N.; Rao, B.M. An Engineered Sso7d Variant Enables Efficient Magnetization of Yeast Cells. ACS Comb. Sci. 2018, 20, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.D.; Murphy, K.P. Protein Structure and the Energetics of Protein Stability. Chem. Rev. 1997, 97, 1251–1267. [Google Scholar] [CrossRef]
- Richardson, S.M.; Mitchell, L.A.; Stracquadanio, G.; Yang, K.; Dymond, J.S.; Dicarlo, J.E.; Lee, D.; Huang, C.L.V.; Chandrasegaran, S.; Cai, Y.; et al. Design of a Synthetic Yeast Genome. Science 2017, 355, 1040–1044. [Google Scholar] [CrossRef]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 Genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef]
- Engel, S.R.; Dietrich, F.S.; Fisk, D.G.; Binkley, G.; Balakrishnan, R.; Costanzo, M.C.; Dwight, S.S.; Hitz, B.C.; Karra, K.; Nash, R.S.; et al. The Reference Genome Sequence of Saccharomyces Cerevisiae: Then and Now. G3 Genes Genomes Genet. 2014, 4, 389–398. [Google Scholar] [CrossRef]
- Caspeta, L.; Navarrete, P.C.S. Reduction of the Saccharomyces Cerevisiae Genome: Challenges and Perspectives. Minimal Cells: Des. Constr. Biotechnol. Appl. 2020, 117–139. [Google Scholar] [CrossRef]
- Briney, B.; Inderbitzin, A.; Joyce, C.; Burton, D.R. Commonality despite Exceptional Diversity in the Baseline Human Antibody Repertoire. Nature 2019, 566, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Boldridge, W.C.; Ljubetic, A.; Kim, H.; Lubock, N.; Szilagyi, D.; Lee, J.; Jerala, R.; Kosuri, S. A Multiplexed Bacterial Two-Hybrid for Rapid Characterization of Protein–Protein Interactions and Iterative Protein Design. Nat. Commun. 2023, 14, 1–11. [Google Scholar] [CrossRef]
- Chen, Z.; Boyken, S.E.; Jia, M.; Busch, F.; Flores-Solis, D.; Bick, M.J.; Lu, P.; VanAernum, Z.L.; Sahasrabuddhe, A.; Langan, R.A.; et al. Programmable Design of Orthogonal Protein Heterodimers. Nature 2019, 565, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Luo, Y.; Gao, H.; Li, F.; Li, J.; Chen, Y.; You, R.; Lv, H.; Hua, K.; Jiang, R.; et al. Toward a Unified Information Framework for Cell Atlas Assembly. Natl. Sci. Rev. 2022, 9, 179. [Google Scholar] [CrossRef]
- Estridge, R.C.; Yagci, Z.B.; Sen, D.; Johnson, T.J.; Kelkar, G.R.; Ptacek, T.S.; Simon, J.M.; Keung, A.J. Loss of UBE3A Impacts Both Neuronal and Non-Neuronal Cells in Human Cerebral Organoids. Commun. Biol. 2025, 8, 838. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).