Abstract
Since December 2019, the COVID-19 pandemic caused by SARS-CoV-2 has reached approximately 769 million people, leading to more than 7 million deaths worldwide. Faced with the possibility of other respiratory pathogens co-infecting patients and modifying their clinical response to SARS-CoV-2, some researchers have explored this line of investigation. The relationship between these co-infections remains unclear, underscoring the need to deepen our understanding of interactions among pathogens and between pathogens and the host. Thus, the present study employed RT-qPCR to assess the presence of Human Adenovirus (HAdV), Influenza A (Flu A), Influenza B (Flu B), Human Metapneumovirus (HMPV), Respiratory Syncytial Virus (RSV), Human Rhinovirus (HRV), and Parainfluenza Virus (PIV). Nasopharyngeal samples (187) from adult patients exhibiting respiratory symptoms were collected between February 2021 and November 2022 at the University Hospital Polydoro Ernani de São Thiago in Florianópolis, SC, Brazil. The present findings revealed that 25.16% of samples tested positive for non-SARS-CoV-2 respiratory viruses (29.8%—HRV; 5.3%—PIV; 4.3%—RSV; and 1.1%—HMPV). In the 74.84% of SARS-CoV-2-positive patients, co-infection was observed in 9.7% of patients, with 7.5% being HRV, 1.1% HAdV, and 1.1% Influenza A. Since co-infections can potentially alter patient prognoses and impact local epidemiological dynamics, this study highlights the significance of ongoing monitoring and epidemiological assessment through genomic surveillance of other clinically relevant respiratory pathogens.
1. Introduction
In August 2023, the COVID-19 pandemic, caused by SARS-CoV-2, reached approximately 769 million cases and almost 7 million deaths worldwide. Meanwhile, in Brazil, the number of cases approached 37 million, with approximately 700,000 deaths during that period [].
Early reports from China said that co-infections of SARS-CoV-2 with other respiratory viruses are rare; however, studies have shown that the presence of co-infection between SARS-CoV-2 and other respiratory viruses seems to aggravate lung disease in such a way as to increase the need for mechanical ventilation (when co-infected with influenza, for example) [,]. In addition, it also impacts the circulation of seasonal viral infections and patient morbidity in cases where there is co-infection of SARS-CoV-2 with Respiratory Syncytial Virus (RSV), Human Adenovirus (HAdV), Human Rhinovirus (HRV) and human Metapneumovirus (HMPV) [,]. SARS-CoV-2 and Influenza co-infection are the most reported in the literature and given that both viruses share similarities in their respiratory symptoms, it is believed that at the beginning of the pandemic, several cases may have been misdiagnosed, since at that time there were no well-established diagnostic tests for SARS-CoV-2 [].
During the first cases of severe acute respiratory infections, at a time when the pathological agent had not yet been identified, molecular biology techniques for detecting pathogens through respiratory panels were decisive and paved the way for sequencing and classification of SARS-CoV-2. At the beginning of the SARS-CoV-2 pandemic, the number of studies of co-infection with other respiratory pathogens was low, and even today, after the World Health Organization (WHO) declared the end of the pandemic, knowledge about the influences of co-infections of SARS-CoV-2 with other respiratory viruses is unclear in regard to host–pathogen interactions, patterns of infection, transmissibility, and the clinical outcome of patients []. Studies have demonstrated the importance of identifying pathogens that cause acute pneumonia even before COVID-19, given the pulmonary predominance of Influenza A (Flu A) and Influenza B (Flu B), HRV, RSV, HMPV, and Parainfluenza Virus (PIV), which had their epidemiological patterns modified with the insertion of SARS-CoV-2 into population [].
The present study evaluated 187 nasopharyngeal samples from patients with non-serious respiratory symptoms, admitted to a screening unit for the diagnosis of SARS-CoV-2 infection, to determine co-infections by HAdV; Flu A; Flu B; HMPV; RSV; HRV, and PIV. Thus, this study aims to report the profile of co-infections of respiratory viruses and SARS-CoV-2 from February 2021 to November 2022 in the state of Santa Catarina, Brazil.
2. Materials and Methods
2.1. Sample Processing
A total of 187 nasopharyngeal samples from adults (men and women), with non-serious respiratory symptoms, were collected between February 2021 and November 2022 at Labortaório de Biologia Molecular, Microbiologia e Sorologia (LBMMS/CCS/UFSC)—of which 88 were from healthcare professionals—with 93 positive and 94 negative diagnoses for SARS-CoV-2. The samples were collected using a nasopharyngeal swab in an appropriate transport medium, as recommended by health agencies, for the diagnosis of SARS-CoV-2.
2.2. Genetic Material Extraction and RT-qPCR
The samples were aliquoted and stored at −80 °C until further analyses. Viral genetic material was extracted using the QIAmp Viral RNA Mini Kit (Qiagen, Germantown, MD, USA). The detection of SARS-CoV-2 was performed using the Allplex™ 2019-nCoV Assay Kit or Allplex™ SARS-CoV-2 Assay Kit (Seegene, Seoul, Republic of Korea). Negative and positive samples for SARS-CoV-2 from symptomatic patients were tested using the Allplex™ RV Essential Assay (Seegene, Republic of Korea), a multiplex RT-qPCR that detects seven viruses: HAdV; Flu A; Flu B; HMPV; RSV; HRV, and PIV. Amplification was performed according to the manufacturer’s instructions on the CFX96™ Real-time PCR System thermal cycler (Bio-Rad®, Hercules, CA, USA), and the results were visualized using the Seegene Viewer software v3.33.000.008.
2.3. Statistical Analysis
During this study, 187 nasopharyngeal samples from adult patients with respiratory symptoms were collected between February 2021 and November 2022 at Hospital Universitário Polydoro Ernani de São Thiago in Florianópolis, SC, Brazil. After RT-qPCR testing for the viruses HAdV, Flu A, Flu B, HMPV, RSV, HRV, and PIV, Fisher’s exact test was used to examine the association between SARS-CoV-2 and other viral infections.
3. Results
From February 2021 to November 2022, 187 samples were randomly selected to determine the co-infection of SARS-CoV-2 with HAdV, Flu A, Flu B, HMPV, RSV, HRV, and PIV. None of the 187 samples were positive for Flu B. Between February 2021 and November 2022, one test was positive for HAdV, one for Flu A, one for HMPV, four for RSV, 35 for HRV, and five for PIV.
Absolute and relative frequencies for viral infections are shown in Figure 1. HRV was the most frequent infection for patients with negative RT-qPCR results for SARS-CoV-2 (Figure 2) and showed co-infections with PIV, HMPV, and RSV (Figure 1). SARS-CoV-2-positive patients had more co-infections with HRV than the other tested viruses (Figure 2). Also, Flu A and HAdV were only detected in SARS-CoV-2-positive patients, and these results indicate a possible relationship between SARS-CoV-2 and other viral infections caused by Influenza A and HAdV (Fisher’s exact test p-value = 0.000001327).
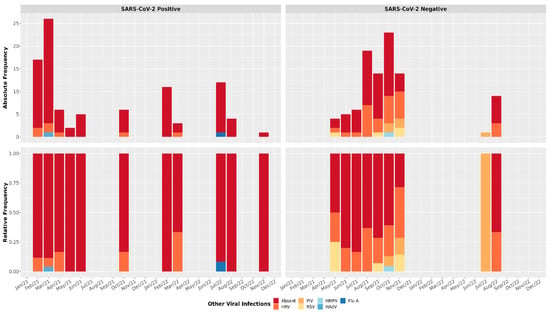
Figure 1.
Viral profile in positive and negative SARS-CoV-2 samples. Absolute and relative frequency for respiratory viral panel tests. Human Adenovirus (HAdV); Influenza A (Flu A); Human Metapneumovirus (HMPV); Respiratory Syncytial Virus (RSV); Human Rhinovirus (HRV); Parainfluenza Virus (PIV).
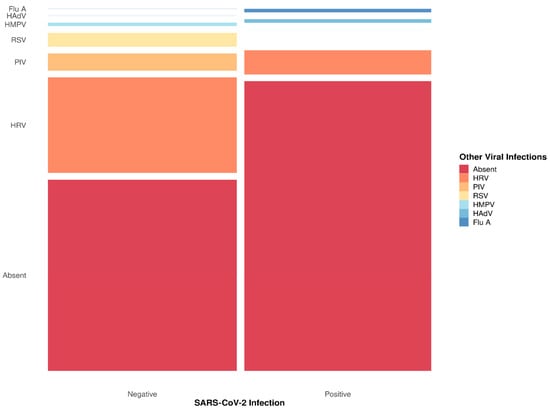
Figure 2.
Summary of viral infection related to SARS-CoV-2 diagnosis. The respiratory viral panel tests included Human Adenovirus (HAdV), Influenza A (Flu A), Human Metapneumovirus (HMPV), Respiratory Syncytial Virus (RSV), Human Rhinovirus (HRV), and Parainfluenza Virus (PIV). The thinnest lines represent zero counts, and all viruses are ordered in the same sequence in both columns.
4. Discussion
Even after the official WHO declaration putting an end to the public health emergency of international concern related to COVID-19, scientific data still needs to be explored in search of answers that can elucidate the variations in the clinical presentation among patients and the epidemiological aspects of SARS-CoV-2. One of these explorations involves the importance of co-infection with other respiratory pathogens [].
Respiratory viruses such as Influenza (Flu A and Flu B) and RSV were already known in pre-pandemic periods for affecting the population, generating epidemic outbreaks with cardiorespiratory impairment, which could lead to morbidity in the infected population [,]. Early in the pandemic, the diagnosis of other respiratory pathogens helped to identify the virus that later became known as SARS-CoV-2. Some studies have reported the clinical importance of patients co-infected with respiratory viruses combined with SARS-CoV-2, identifying an increased need for mechanical ventilation after co-infection [,].
The epidemiological data shows a clear change in the detection of seasonal viruses such as Influenza, which seems not to have been identified between the years 2020 and 2022, the period with the highest number of cases and deaths caused by COVID-19 []. It is not clear if this lower detection of seasonal respiratory viruses is due to a reduction in the prevalence of pathogens other than SARS-CoV-2, since interactions and competition between pathogens could have occurred, due to a possible underreporting resulting from the overload of health professionals involved in the pandemic, or due to both [].
Of the 187 samples from adult patients (men and women) with non-serious respiratory symptoms, one test was positive for HAdV, one for Flu A, one for HMPV, four for RSV, 35 for HRV, and five for PIV. Thus, the present data corroborates Kim and colleagues who, in 2020, had already demonstrated higher report rates of HRV and RSV infections in patients with respiratory symptoms but undetected for SARS-CoV-2 [,].
It is important to highlight that, during their study, Kim and colleagues [] also evaluated the presence of enterovirus, Chlamydia pneumoniae, and Mycoplasma pneumoniae; this was unlike the present study, in which such infections may have been underreported and may worsen the prognosis of patients infected with SARS-CoV-2. In addition, the number of cases of COVID-19 in this study was higher [,].
Another study [] evaluated, between November 2021 and February 2022, the presence of co-infections among patients detected with SARS-CoV-2 in combination with Flu A (65%), Enterovirus–Rhinovirus (20%), HAdV (2%), RSV (2%), HMPV (2%), Human Parainfluenza virus type 3 (2%), and Human Coronavirus (7%). Unlike the present study, in which patients infected with SARS-CoV-2 had a higher prevalence of co-infection with HRV, Eldesouki and colleagues [] had the highest prevalence of co-infection between SARS-CoV-2 and Flu A. On the other hand, it must be considered that the studies were developed with different sample sizes (41 patients versus 187), in addition to differences in population characteristics.
In Brazil, since 2021, data from the Board of Epidemiological Surveillance of the State of Santa Catarina—DIVE (official website for monitoring infectious diseases) have presented RSV, HRV, HAdV, and Bocavirus, in addition to the unidentified viruses, as the main agents causing acute respiratory syndrome after SARS-CoV-2. These data are partially different from the present results, showing HRV as the main agent detected both in patients detected and undetected for SARS-CoV-2 [].
Severe Acute Respiratory Syndrome (SARS) is a more aggressive infection that can lead to hospitalizations and death. In this study, the largest number of infections reported, except for SARS-CoV-2, was caused by HRV. This discrepancy in the data can be explained in part by the authors’ sampling, which did not cover severe cases, but only classic flu symptoms, a profile different from that monitored by public agencies. Interestingly, although these results demonstrate only one sample co-infected with SARS-CoV-2 and Flu A, epidemiological data from DIVE [] reported 72 deaths in the state of Santa Catarina due to SARS caused by Flu A. Thus, it is hypothesized that this difference is due to the population group of the study, which were symptomatic adults without enough clinical severity to justify hospitalization due to classic signs of SARS.
5. Conclusions
Between February 2021 and November 2022, patients were tested for Flu B, HMPV, RSV, HRV, and PIV at the University Hospital Polydoro Ernani de São Thiago of the Federal University of Santa Catarina, located in Florianópolis, Santa Catarina, Brazil. During this period, HRV, PIV, RSV, and MPV were detected in patients who were not infected with SARS-CoV-2, in addition to detecting HAdV, Flu A, and HRV co-infecting patients detected with SARS-CoV-2. These data demonstrate the importance of monitoring infectious pathogens that affect the respiratory system, in addition to SARS-CoV-2, aiming to identify the population’s epidemiological pattern to improve prevention and make therapeutic approaches as individualized as possible.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/covid5080133/s1. Table S1: Raw results of molecular screening for Human Adenovirus (HAdV), Influenza A (Flu A), Human Metapneumovirus (HMPV), Respiratory Syncytial Virus (RSV), Human Rhinovirus (HRV), and Parainfluenza Virus (PIV) in nasopharyngeal samples.
Author Contributions
Conceptualization, D.A.P., M.L.B., G.W., and G.F.; methodology, D.A.P., F.H.B., M.A.S., H.B.d.S.G., V.B.F., E.K.K., and D.S.M.S.; formal analysis, D.A.P., G.W., and G.F.; investigation, D.A.P., G.W., and G.F.; resources, M.L.B., G.W., and G.F.; data curation, D.A.P., V.B.F., E.K.K., G.W., and G.F.; writing—original draft preparation, D.A.P., G.W., and G.F.; writing—review and editing, D.A.P., F.H.B., M.A.S., H.B.d.S.G., V.B.F., E.K.K., D.S.M.S., M.L.B., G.W., and G.F.; visualization, D.A.P.; supervision, G.W. and G.F.; project administration, M.L.B., G.W., and G.F.; funding acquisition, M.L.B., G.W., and G.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fundação de Amparo à Pesquisa e Inovação de Santa Catarina (FAPESC—grant number COV2020051000065/COV2020051000099), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), and UFSC (Universidade Federal de Santa Catarina). D.A.P., H.B.S.G., V.B.F., E.K.K., and D.S.M.S. were recipients of FAPESC, CAPES, or CNPq scholarships.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee in Research Humans Beings/UFSC (assent number: 4.035.636, CAAE: 31521920.8.0000.0121, 19 May 2020).
Informed Consent Statement
Patient consent was waived since samples from University Hospital (HU- UFSC/EBSERH) at Federal University of Santa Catarina (Florianópolis, SC, Brazil) were made available anonymously.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Acknowledgments
We express our gratitude to Fundação de Amparo à Pesquisa e Exntensão Universitária (FAPEU/UFSC), FAPESC, CAPES, CNPq, and UFSC for offering financial and structural support for the development of this study.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| COVID-19 | Coronavirus Disease 2019 |
| DIVE | Board of Epidemiological Surveillance of the State of Santa Catarina |
| Flu A | Influenza Virus A |
| Flu B | Influenza Virus B |
| HAdV | Human Adenovirus |
| HMPV | Human Metapneumovirus |
| HRV | Human Rhinovirus |
| PIV | Parainfluenza Virus |
| RSV | Respiratory Syncytial Virus |
| RT-qPCR | Reverse-Transcripted Real-Time Polymerase Chain Reaction |
| SARS-CoV-2 | Severe Acute Respiratory Coronavirus 2 |
| WHO | World Health Organization |
References
- WHO. Corona Virus Dashboard. Available online: https://covid19.who.int/ (accessed on 19 September 2023).
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Swets, M.C.; Russell, C.D.; Harrison, E.M.; Docherty, A.B.; Lone, N.; Girvan, M.; E Hardwick, H.; Visser, L.G.; Openshaw, P.J.M.; Groeneveld, G.H.; et al. SARS-CoV-2 co-infection with influenza viruses, respiratory syncytial virus, or adenoviruses. Lancet 2022, 399, 1463–1464. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.J.; Pfotenhauer, B.; Weiner, J.J.; Hilleshiem, J.; Khubbar, M.; Bhattacharyya, S.; Long, S.W. Respiratory Pathogen Coinfections in SARS-CoV-2-Positive Patients in Southeastern Wisconsin: A Retrospective Analysis. Microbiol Spectr. 2021, 9, e0083121. [Google Scholar] [CrossRef] [PubMed]
- Xing, Q.-S.; Li, G.-J.; Xing, Y.-H.; Chen, T.; Li, W.-J.; Ni, W.; Deng, K.; Gao, R.-Q.; Chen, C.-Z.; Gao, Y.; et al. Precautions are needed for COVID-19 patients with coinfection of common respiratory pathogens. medRxiv 2020. [Google Scholar] [CrossRef]
- Burk, M.; El-Kersh, K.; Saad, M.; Wiemken, T.; Ramirez, J.; Cavallazzi, R. Viral infection in community-acquired pneumonia: A systematic review and meta-analysis. Eur. Respir. Rev. 2016, 25, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Denouel, A.; Tietjen, A.K.; Campbell, I.; Moran, E.; Li, X.; Campbell, H.; Demont, C.; O Nyawanda, B.; Chu, H.Y.; et al. Global Disease Burden Estimates of Respiratory Syncytial Virus-Associated Acute Respiratory Infection in Older Adults in 2015: A Systematic Review and Meta-Analysis. J. Infect. Dis. 2020, 222 (Suppl. 7), S577–S583. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, X.; Blau, D.M.; Caballero, M.T.; Feikin, D.R.; Gill, C.J.; A Madhi, S.; Omer, S.B.; Simões, E.A.F.; Campbell, H.; et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: A systematic analysis. Lancet 2022, 399, 2047–2064. [Google Scholar] [CrossRef] [PubMed]
- Maltezou, H.C.; Papanikolopoulou, A.; Vassiliu, S.; Theodoridou, K.; Nikolopoulou, G.; Sipsas, N.V. COVID-19 and Respiratory Virus Co-Infections: A Systematic Review of the Literature. Viruses 2023, 15, 865. [Google Scholar] [CrossRef] [PubMed]
- PAHO—Alerta Epidemiológico—Influenza, Vírus Sincicial Respiratório e SARS-CoV-2. Available online: https://www.paho.org/pt/documentos/alerta-epidemiologico-influenza-virus-sincicial-respiratorio-e-sars-cov-2-6-junho-2023 (accessed on 6 June 2023).
- Dähne, T.; Bauer, W.; Essig, A.; Schaaf, B.; Spinner, C.D.; Pletz, M.W.; Rohde, G.; Rupp, J.; Witzenrath, M.; Panning, M.; et al. The impact of the SARS-CoV-2 pandemic on the prevalence of respiratory tract pathogens in patients with community-acquired pneumonia in Germany. Emerg. Microbes Infect. 2021, 10, 1515–1518. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Quinn, J.; Pinsky, B.; Shah, N.H.; Brown, I. Rates of Co-infection Between SARS-CoV-2 and Other Respiratory Pathogens. JAMA 2020, 323, 2085–2086. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, R.; Goodarzi, P.; Asadi, M.; Soltani, A.; Aljanabi, H.A.A.; Jeda, A.S.; Dashtbin, S.; Jalalifar, S.; Mohammadzadeh, R.; Teimoori, A.; et al. Bacterial co-infections with SARS-CoV-2. IUBMB Life 2020, 72, 2097–2111. [Google Scholar] [CrossRef] [PubMed]
- Eldesouki, R.E.; Uhteg, K.; Mostafa, H.H. The circulation of Non-SARS-CoV-2 respiratory viruses and coinfections with SARS-CoV-2 during the surge of the Omicron variant. J. Clin. Virol. 2022, 153, 105215. [Google Scholar] [CrossRef] [PubMed]
- DIVE. Available online: https://dive.sc.gov.br/phocadownload/Boletins/Boletim%20Epidemiol%C3%B3gico%20SRAG%20N%C2%BA13.pdf (accessed on 20 September 2023).
- DIVE. Available online: https://dive.sc.gov.br/phocadownload/doencas-agravos/Gripe%20(Influenza)/Boletins/boletim-influenza-12-2022.pdf (accessed on 21 September 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).