Abstract
In the present study, the occurrence of Garra substrictorostris Roni & Vishwanath, 2018, (Cypriniformes: Cyprinidae) was recorded for the first time from the Manas River in the Brahmaputra drainage system, thereby expanding the species’ distributional range. Two specimens were collected and identified to the species level using the existing literature. Mitochondrial Cytochrome C Oxidase subunit I (COI) gene barcoding was conducted to confirm the identity of the specimens collected from the Manas River. Morphological examination revealed characteristics consistent with the original description of G. substrictorostris from the Barak River drainage area. These findings were further corroborated by molecular data, specifically mitochondrial COI gene barcoding. The genetic distance between Garra substrictorostris and G. nasuta was observed to be 4.05%, inferring G. nasuta to be the closest relative and G. lissorhynchus to be the most distant species (15.45%), forming a different clade among the reported species within the genus Garra. A maximum likelihood (ML) and Bayesian inference phylogenetic trees were also constructed, which signify the correct identification of the species.
1. Introduction
The rivers of South Asia are known for their extraordinary biodiversity, which is characterized by high species richness that is disproportionate to their geographical area [1]. These river systems are home to numerous rare, endangered, and threatened species, as well as many endemic taxa, contributing to their significant conservation value [2]. Additionally, the rivers of the region support a variety of charismatic species, many of which are of considerable ecological and socio-economic importance [1]. The northeastern region of India, which forms part of the Indo-Burma biodiversity hotspot, is a critical area within the broader South Asian riverine ecosystems [3]. The rich and diverse ichthyofauna of Northeast India, particularly within the Brahmaputra River basin, presents a significant area for ecological and taxonomic discovery. This region is home to numerous freshwater fish species, including members of the genus Garra [4,5,6,7].
The species belonging to the genus Garra Hamilton, 1822 are small- to medium-sized, bottom-dwelling inhabitants of mountainous rivers and streams, characterized by fast-flowing and rocky bottoms, and are widely distributed from southern China and Borneo in the east, through southern Asia, the Arabian Peninsula, and the Middle East to eastern, central, and west Africa [8,9,10]. The genus Garra is diagnosed from the labeonine genera by having a crescentic anteromedian fold of the lower lip equal to or wider than the callous pad, the lateral end of the anteromedian fold on each side reaching the anterolateral lobe of the mental adhesive disc, and three rows of pharyngeal teeth [11]. Recently, numerous species have been described from the various river systems in the eastern Himalayan foothills [12,13,14,15,16].
Currently, there are 41 valid species of the genus Garra distributed in the rivers and streams of the Brahmaputra, Barak, Kaladan, Karnaphuli, and Chindwin drainages [17]. Twenty species of the genus Garra have been considered as valid in the Brahmaputra drainage, viz., G. lamta (Hamilton, 1822), G. gotyla (Gray, 1830), G. nasuta (McClelland, 1838), G. rupecula (McClelland, 1839), G. lissorhynchus (McClelland, 1842), G. annandalei Hora, 1921, G. jenkinsoniana Hora, 1921, G. kempi Hora, 1921, G. arupi (Nebeshwar et al., 2009), G. kalpangi Nebeshwaret et al., 2012, G. arunachalensis Nebeshwar & Vishwanath, 2013, G. quadratirostris Nebeshwar & Vishwanath, 2013, G. birostris Nebeshwar & Vishwanath, 2013, G. magnidiscus Tamang, 2013, G. tamangi Gurumayum & Kosygin, 2016, G. parastenorhynchus Thoni et al., 2016, G. bimaculacauda Thoni et al., 2016, G. biloborostris Roni & Vishwanath, 2017, G. clavirostris Roni et al., 2017, G. magnacavus Shangningam et al., 2019 [17].
Integrative taxonomy, which combines both morphological and molecular data, is essential for accurate species delimitation, especially in areas where cryptic species or complex taxonomic groups are prevalent [18,19,20,21]. In recent times, this comprehensive approach has led to more robust identification and classification of species [18]. In the present study, a similar approach was adopted for recording the occurrence of G. substrictorostris Roni & Vishwanath, 2018, from the Manas River in the Brahmaputra drainage for the first time.
2. Materials and Methods
2.1. Study Area and Sample Collection
Specimens of Garra substrictorostris Roni & Vishwanath, 2018, were collected from the Manas River, a transboundary river in the Himalayan foothills between southern Bhutan, India, and China (Figure 1). The river flows through Assam, India, for about 104 km before joining the River Brahmaputra. The specimens were collected on March 16, 2023, from Deosri, Chirang District of Assam (26°47′16.836″ N; 90°28′31.692″ E). Two specimens were collected from the river using a gill net. For further analysis, one pectoral fin clip was cut away from each specimen and preserved in 95% ethanol. Subsequently, specimens were preserved in 10% formalin. The morphometric and meristic characteristics were recorded following Nebeshwar & Vishwanath [22]. For dorsal-fin and anal-fin ray counts, we followed Kottelat [23], who describes that the last two rays articulating on the same pterygiophore are counted as “11/2”. Both specimens were registered (FRMCOFIFM208a and FRMCOFIFM208b) at the Indigenous Fish Museum of the College of Fisheries, Assam Agricultural University.
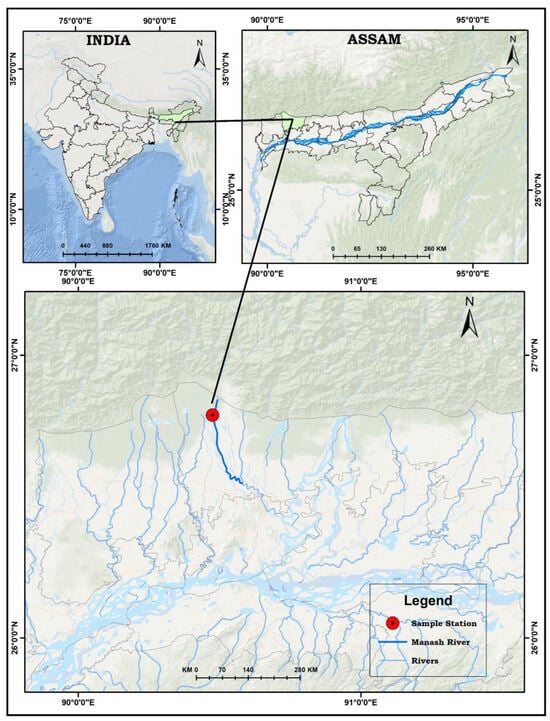
Figure 1.
Map showing the sampling location.
2.2. DNA Extraction and PCR Amplification
For DNA barcoding, total genomic data were extracted from the fin tissues using the phenol–chloroform method [24]. The concentration and purity of the isolated DNA were determined using a NanoDrop spectrophotometer (Nanodrop oneC, ThermoFisher, Waltham, MA, USA). The partial sequence of mitochondrial COI was amplified using Universal Primer FishF1 (5-′TCAACCAACCACAAAGACATTGGCAC-3′) and FishR1 (5′-TAGACTTCTGGGTGGCCAAAGAATCA-3′) (HiMedia, Mumbai, India) [25] in a 25 µL reaction volume containing 12.5 µL Master Mix, 1 µL each of forward and reverse primer, 1.5 µL of the diluted DNA solution, and the rest was adjusted with 9 µL of nuclease free water (NFW). Polymerase Chain Reactions (PCR) were conducted for 35 cycles with the following steps: initial denaturation at 94 °C for 5 min., followed by denaturation at 94 °C for 30 s, annealing at 54 °C for 30 s, extension at 72 °C for 59 s, and a final extension at 72 °C for 10 min. The resultant PCR products were purified using the QIAquick PCR Purification Kit (QIAGEN, Hilden, Germany), and the amplicons were sequenced in both directions using Sanger sequencing.
2.3. Molecular Data Analysis
The quality of the obtained sequence was checked by observing the Phred score of each nucleotide. Subsequently, the resultant sequences were passed through the NCBI ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/ (accessed on 10 March 2025)) to obtain the coding sequences and were searched using the Basic Local Alignment Search Tool (BLAST) nucleotide database (https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 10 March 2025) to confirm the gene and species. The mitochondrial COI DNA sequence was submitted to NCBI GenBank to obtain the gene accession numbers. For the phylogenetic analysis, a total of twenty-three sequences were retrieved from the GenBank database (Table 1). All sequences were aligned using the Clustal W [26] implemented in MEGA 11 (Molecular Evolutionary Genetics Analysis) [27] across computing platforms. The within- and between-species genetic distances were calculated using the Kimura-2 parameter (K2P) [28] distance model implemented in MEGA 11. Subsequently, the phylogeny of the Garra species, with Raiamas bola (Hamilton, 1822) used as an outgroup, was inferred through the maximum likelihood method and general time reversible model [29], and node support was assessed through 1000 bootstrap replicates using MEGA 12 [30]. Furthermore, to complement the maximum likelihood topology, a Bayesian tree was also inferred in the MrBayes v3.2.7 program with Markov chain Monte Carlo (MCMC) methods [31] and the posterior consensus tree was visualized in FigTree v1.4.4 [32].

Table 1.
List of the identified specimens in the present study and the retrieved sequences of the relative species from the GenBank database.
3. Results
3.1. Morphometric Analysis
The present study records a new geographical distribution of Garra substrictorostris from the Manas River, a tributary of the Brahmaputra River drainage (Figure 2 and Figure 3). The specimens of G. substrictorostris were compared using morphological and meristic characteristics, which was originally described as a new species by Roni & Vishwanath [33] from the Leimatak River (Barak River drainage); 24°34′33″ N, 93°40′01″ E; 513 m above sea level. The species was also compared with the congeners recorded from the Brahmaputra drainage. A comparison between the set of key characteristics on the basis of which the holotype was described is presented in Table 2. It was observed that all characters fell within the range originally described for the holotype. The biometric data are presented in Table 3. The global distribution of G. substrictorostris to date (Manipur and Assam, India) is depicted in Figure 4.

Figure 2.
Lateral (a) and dorsal (b) views of Garra substrictorostris collected from the Manas River.
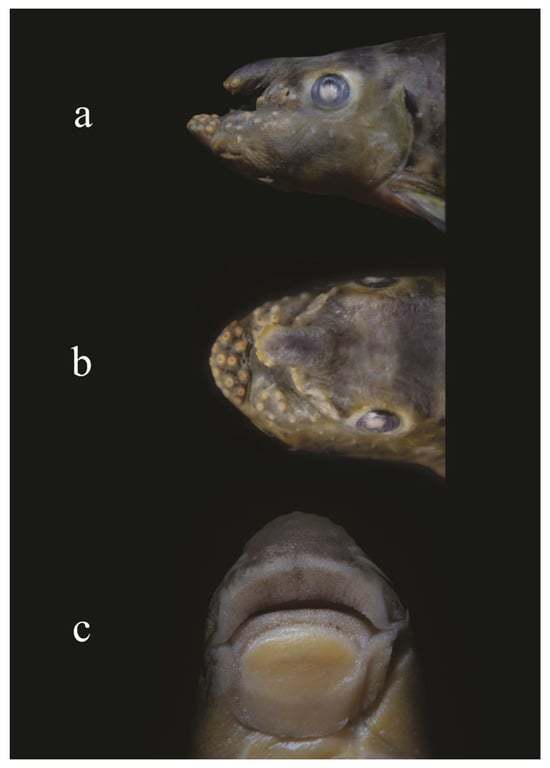
Figure 3.
Lateral (a), dorsal (b) and ventral (c) views of the mouth structure of Garra substrictorostris collected from the Manas River.

Table 2.
Comparison of key characteristics of Garra substrictorostris described from Brahmaputra and Barak River Drainage (original description).

Table 3.
Morpho-meristic data of the specimen Garra substrictorostris.
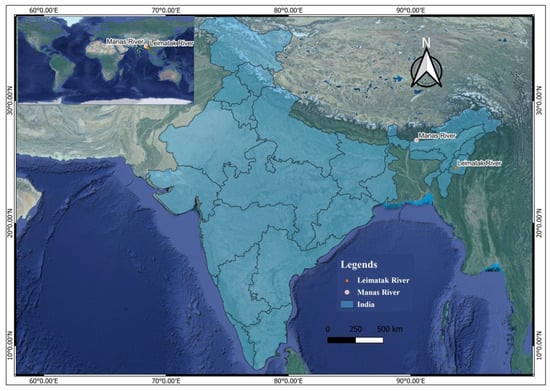
Figure 4.
Map showing the global distribution of Garra substrictorostris.
3.1.1. Diagnosis
The average body length of the collected G. substrictorostris was recorded as 137.87 mm, showing characteristics similar to the range described originally by Roni & Vishwanath [33]. The transverse lobe is prominent on the snout with 17 multi-cuspid tubercles; a narrow antrorse, unilobed proboscis on the snout, with 5 medium to large multi-cuspid tubercles on its anterior margin and 4–6 multi-cuspid tubercles on its antero-ventral margin; tubercles on the lateral surface of the snout with 7–9 small tubercles; transverse scale rows above the lateral line is 51/2; transverse scale rows between the lateral line and anal-fin origin is 41/2. The distance between the anus and anal-fin origin (in % of pelvic-anal distance) was 23.16 (1) or 24.96 (1).
3.1.2. Coloration
The specimen appears dark gray to yellowish ventrally. The dorsum of the head, snout, and opercles are dark gray with a yellowish tinge. The mouth and abdomen are pale white or light gray. The pectoral fin is darker gray with a yellowish tinge at the base compared with the pelvic and anal fins. Base of branched dorsal-fin rays spotted black. The lower lobe of the caudal fin is darker brown or grayish than the upper lobe. A black spot is present at the upper angle of the gill opening.
3.2. Molecular Analysis
3.2.1. NCBI BLAST Analyses
To confirm the gene and species identification, the barcode sequence generated in the present study was first assessed for quality and subsequently queried using the NCBI Nucleotide BLAST tool. Following sequence validation, the barcode was submitted to GenBank (Accession No. PV336094). BLAST analysis revealed a sequence similarity of 93–96% with members of the Garra species.
3.2.2. Genetic Divergence
The pairwise genetic distance values between Garra substrictorostris and other Garra species were calculated and are presented in Table 4. The overall intraspecific distance was observed to be in the range from 0% to 0.007%, whereas the interspecific distance was in the range from 4.05% to 16.43%. The interspecific distance between G. substrictorostris and the other Garra species was observed to be 11.00%. G. nasuta showed the least genetic divergence from G. substrictorostris (4.05%), whereas G. lissorhynchus exhibited the highest distance (15.45%). The genetic distance between G. substrictorostris and other Garra species found was as follows: G. lamta (9.69%), G. gotyla (10.62%), G. annandalei (10.68%), G. kempi (11.77%), G. quadrirostris (12.48%), and G. arupi (13.50%).

Table 4.
The pair-wise genetic distance value was calculated between the Garra substrictorostris and other Garra species.
3.2.3. Phylogenetic Analyses
A maximum likelihood (ML) phylogenetic tree was generated using the MEGA 12 software to validate the correct identification of the reported species (Figure 5). All species clustered with their respective conspecific sequences, with nodes receiving strong bootstrap values.
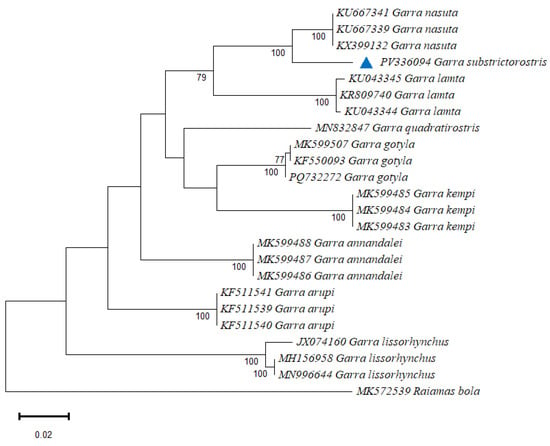
Figure 5.
The maximum likelihood phylogeny of the genus Garra was reconstructed based on 555 bp of the COI 5’ end. The values at the branches represent ML bootstrap probabilities.
Most species formed a single major lineage, whereas G. lissorhynchus, G. arupi, and G. annandalei each formed distinct branches separated from the primary cluster. Within the major clade containing the remaining species, two coherent sub-lineages were recovered: one comprising G. nasuta, G. substrictorostris, and G. lamta, and a second grouping G. quadratirostris, G. gotyla, and G. kempi. From the ML tree, the closest species to G. substrictorostris was observed to be G. nasuta, followed by G. lamta. The Bayesian inference tree (Figure 6) also revealed a similar topology among the Garra species. The clade comprising G. nasuta, G. substrictorostris, and G. lamta was conserved across both methods. However, nodes with moderate support contributed to the difference in the constituents of the remaining clades from those of the ML tree.
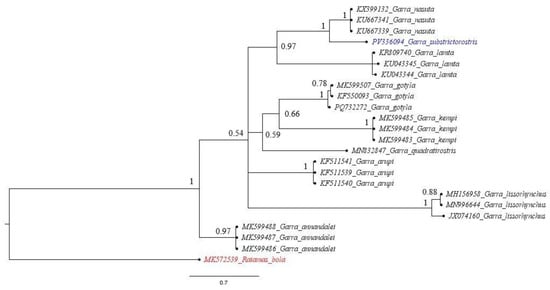
Figure 6.
Bayesian phylogeny of the genus Garra was reconstructed based on 555 bp of the COI 5’ end. The values at the branches represent BI posterior probabilities.
4. Discussion
The genus Garra is highly diversified and known to occur from northern and central Africa to Southeast Asia through the Middle East, southern China, and South Asia [8,34]. A total of nearly 192 species belonging to the genus Garra have been described from different localities worldwide [35], making the genus one of the most diverse genera within the family Cyprinidae. The characteristics of the snout, as well as the size and distribution patterns of the tubercles on the proboscis, are of taxonomic significance in distinguishing species of the genus Garra [19]. Sun et al. [34] hypothesize that the general evolutionary trend of the proboscis in Garra is from no proboscis to a proboscis, from a small proboscis to a large one, and from a simple proboscis to a complex one, i.e., from a unilobed proboscis to a bi-lobed and trilobed proboscis.
Garra substrictorostris has so far been recorded only from the Leimatak River (Barak River drainage) [33]. The present study reveals new information on the type locality and distribution range of G. substrictorostris from the Barak River drainage to the Brahmaputra River drainage. Of the 20 recorded species of the genus Garra from the Brahmaputra River drainage [17], G. substrictorostris distinguished itself with distinctive morphological and meristic characteristics from other species of the genus. One of the most significant morphological features used to distinguish G. substrictorostris from its congeners in the Brahmaputra drainage was the shape and size of the snout and proboscis. Garra substrictorostris is characterized by a narrow, well-defined proboscis with sparsely distributed tubercles, which serves as a key diagnostic feature [33]. The snout is slender and elongated, with a smooth texture, and the body is moderately robust. In contrast, G. clavirostris possesses a broader, club-shaped proboscis with a more robust, conical snout and densely arranged tubercles, making it easily distinguishable [14]. The body of G. clavirostris is more streamlined, and its coloration includes distinct dark lateral stripes, particularly during the breeding season. Garra birostris exhibits a proboscis that is somewhat similar to that of G. substrictorostris but tends to be slightly broader, with tubercles moderately distributed across the snout. Its body is relatively larger, and the caudal fin is more pronounced compared to G. substrictorostris [14]. Morphological differences in snout and body size are useful for distinguishing G. birostris from G. substrictorostris. Garra arunachalensis, another species described from the Brahmaputra drainage with a proboscis, has a notably larger proboscis compared to G. substrictorostris, with a more rounded and broader shape. The tubercles on the proboscis of G. arunachalensis are more densely arranged, and the snout is more flattened, with a slightly broader head than in other species [22]. Furthermore, the distance between the anus and anal fin origin is another diagnostic characteristic of G. substrictorostris, which is reported to be 15–27% of the distance between the pelvic fin and anal fin origin by Roni & Vishwanath [33] in the original description. A similar observation, i.e., 23.16–24.96%, is recorded in the present documentation.
Classical taxonomy has been used for identification and classification since Linnaeus’s time and is still irreplaceable [36]. However, recently, DNA barcoding has been instrumental in improving the accuracy of species identification, especially for cryptic species that are difficult to distinguish based on morphological characteristics alone [37]. This method has also led to the discovery of many new species, thereby expanding our understanding of biodiversity. Molecular data on Garra species in northeastern Indian waters are scarce and require extensive genetic studies [38]. The present molecular analysis was performed using the sequences available from the NCBI GenBank database, which confirmed that G. substrictorostris shares a relatively close genetic relationship with G. nasuta. The highest interspecific distance was observed between G. lissorhynchus with G. substrictorostris and other Garra species, which is comparable to the findings of Limbu et al. [39], viz., G. lisorhychus with G. lamta (14.3% vs. 16.43%) and G. lissorhynchus with G. gotyla (14.20% vs. 13.80%). The maximum likelihood-based phylogenetic relationship placed all the species into their representative sequences and formed five groups. The observed clade of G. substrictorostris with G. nasuta is the same for both the maximum likelihood and Bayesian inference trees, which highlights the underlying phylogenetic signal of the COI gene. However, inconsistency observed in the arrangement of the clades in the BI tree compared with that of the ML tree may be attributed to the low phylogenetic resolution, arising from limited informative sites or rapid diversification or underlying methods of analysis [40,41,42]. Further studies employing additional genetic markers are likely to improve the understanding of the phylogenetic relationships within the genus.
5. Conclusions
The results of this study provide significant insights into the distributional range of G. substrictorostris in the Brahmaputra drainage, thereby expanding our understanding of its ecological and biogeographical patterns. The integration of morphological and molecular data underscores the ability of an integrative taxonomic approach to accurately characterize species and their distributions in complex and biodiverse ecosystems.
The morphological and meristic diagnosis is further complemented by the results obtained through phylogenetic analysis using both maximum likelihood (ML) and Bayesian inference trees, which validate the distinct taxonomic identity of G. substrictorostris. These resultant topologies highlight the molecular evidence and reinforce the reliability of the inferred lineage relationships. The insights gained here underscore the importance of further research on the study of ecological dynamics, habitat requirements, and population genetics of G. substrictorostris, considering the ongoing environmental changes.
Author Contributions
Sampling, specimen identification, funding acquisition, and original draft preparation, B.P.; Formal analysis and investigation, A.P.K. and M.B.; Manuscript editing, P.D.; Sampling and morphometric analysis, A.T. and R.K.; Supervision and manuscript editing, G.D., K.K.T., and B.K.B.; Supervision, manuscript correction and editing, B.B.N. All authors have read and agreed to the published version of the manuscript.
Funding
The authors are thankful to the project SCoPIF-I under the Government of Assam (Grant Number: FISH/30/2017-FISHERY/27 (eCFNo.43140), India, for the financial support.
Institutional Review Board Statement
All the fish specimens used in the study were collected using traditional gear. The current study followed the Guidelines of the Committee constituted for this purpose (Ethical Approval Committee, EAC) at College of Fisheries, Assam Agricultural University, with vide approval no. AAU/G-9/COF/2022-23/3582 dated 5 January 2023.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
The authors are thankful to the Vice Chancellor of Assam Agricultural University (AAU), Jorhat, Assam, India, for providing the necessary infrastructural facilities for the laboratory analysis. The authors are also thankful to the Director of ICAR-CIFE, Mumbai.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Abbreviations
| COI | Cytochrome c oxidase subunit I |
| ML | Maximum likelihood |
| DNA | Deoxyribonucleic acid |
| NFW | Nuclease-Free Water |
| PCR | Polymerase Chain Reaction |
| NCBI | National Center for Biotechnology Information |
| ORF | Open Reading Frames |
| BLAST | Basic Local Alignment Search Tool |
| MEGA | Molecular Evolutionary Genetics Analysis |
References
- Gopal, B. Conserving Riverine Biodiversity—A Himalayan Task? In Rivers for Life: Proceedings of the International Symposium on River Biodiversity: Ganges-Brahmaputra-Meghna River System; Ecosystems for Life, A Bangladesh-India Initiative; Sinha, R.K., Ahmed, B., Eds.; International Union for Conservation of Nature (IUCN): Gland, Switzerland, 2014; pp. 12–15. [Google Scholar]
- Dahanukar, N.; Raut, R.; Bhat, A. Distribution, endemism and threat status of freshwater fishes in the Western Ghats of India. J. Biogeogr. 2004, 31, 123–136. [Google Scholar] [CrossRef]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; Da Fonseca, G.A.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Bhattacharjya, B.K.; Bhaumik, U.; Sharma, A.P. Fish habitat and fisheries of Brahmaputra River in Assam, India. Aquat. Ecosyst. Health Manag. 2017, 20, 102–115. [Google Scholar] [CrossRef]
- Kar, D. Fish and their habitats in North-East India biodiversity hotspot. Oceanogr. Fish. Open Access J. 2021, 13, 24–26. [Google Scholar] [CrossRef]
- Chetry, V.; Das, H.; Saikia, P.K.; Saikia, M.K.; Saikia, B.P.; Sarma, K. Ichthyofaunal diversity in Jia Bharali River of North bank landscape of Assam in Eastern Himalaya, Northeast India. Ecol. Environ. Conserv. 2023, 29. [Google Scholar] [CrossRef]
- Ongh, S.O.; Landge, A.T.; Ramteke, K.; Borah, S.; Barman, J.; Akter, S.; Yadav, A.K.; Das, P.; Majhi, S.K.; Chakraborty, N.; et al. Fish community structure in accordance with environmental signatures in tropical river ecosystem, Eastern Himalayan eco-region. Res. Sq. 2024. [Google Scholar] [CrossRef]
- Zhang, E.; Chen, Y.Y. Garra tengchongensis, a new cyprinid species from the upper Irrawaddy River basin in Yunnan, China (Pisces: Teleostei). Raffles Bull. Zool. 2002, 50, 459–464. [Google Scholar]
- Rath, S.; Shangningam, B.; Kosygin, L. Garra simbalbaraensis, a new species of cyprinid fish (Teleostei: Cyprinidae) from Himachal Pradesh, India. Zootaxa 2019, 4652, 401–410. [Google Scholar] [CrossRef]
- Esmaeili, H.R.; Hamidan, N. Inland fishes of the Arabian Peninsula: Review and a revised checklist. Zootaxa 2023, 5330, 201–226. [Google Scholar] [CrossRef]
- Nebeshwar, K.; Vishwanath, W. On the snout and oromandibular morphology of genus Garra, description of two new species from the Koladyne River basin in Mizoram, India, and redescription of G. manipurensis (Teleostei: Cyprinidae). Ichthyol. Explor. Freshw. 2017, 28, 17–53. [Google Scholar]
- Ezung, S.; Shangningam, B.; Pankaj, P.P. A new fish species of genus Garra (Teleostei: Cyprinidae) from Nagaland, India. J. Threat. Taxa 2021, 13, 18618–18623. [Google Scholar] [CrossRef]
- Catherine, N.; Linthoingambi, I. Garra lungongza, a new species of cyprinid fish (Teleostei: Cyprinidae) from Nagaland, India. Rec. Zool. Surv. India 2023, 123, 1–12. [Google Scholar] [CrossRef]
- Roni, N.; Sarbojit, T.; Vishwanath, W. Garra clavirostris, a new cyprinid fish (Teleostei: Cyprinidae: Labeoninae) from the Brahmaputra drainage, India. Zootaxa 2017, 4244, 367–376. [Google Scholar] [CrossRef]
- Singh, P.; Tudu, A.K.; Gurumayum, S.D. Garra magnarostrum, a new species of cyprinid fish (Teleostei: Cyprinidae) from the Brahmaputra River drainage, Northeastern India. Rec. Zool. Surv. India 2025, 124, 219–227. [Google Scholar] [CrossRef]
- Tenali, D.R.; Chandran, R.; Singh, R.K.; Sarkar, U.K. Garra ngopi sp. n., a new fish species (Cyprinidae: Labeoninae) from the Brahmaputra River basin, Arunachal Pradesh, North–Eastern India. Biologia 2024, 80, 79–88. [Google Scholar] [CrossRef]
- Shangningam, B.; Kosygin, L.; Sinha, B. A new species of rheophilic cyprinid fish (Teleostei: Cyprinidae) from the Brahmaputra Basin, northeast India. Zootaxa 2019, 4695, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.A.; Nanda, P.; Pant, B.; Mane, S.S.; Kolipakam, V. Redescription of an endemic mahseer, Tor mahanadicus (David, 1953) from Mahanadi River basin, India based on integrated morphological and molecular techniques. PLoS ONE 2023, 18, e291436. [Google Scholar] [CrossRef]
- Shao, W.; Cheng, J.; Zhang, E. Eight in one: Hidden diversity of the bagrid catfish Tachysurus albomarginatus s.l. (Rendhal, 1928) widespread in lowlands of South China. Front. Genet. 2021, 12, 713793. [Google Scholar] [CrossRef]
- Sheraliev, B.; Peng, Z. Molecular diversity of Uzbekistan’s fishes assessed with DNA barcoding. Sci. Rep. 2021, 11, 16894. [Google Scholar] [CrossRef]
- Vicente, F.; Loeb, M.V.; de Paiva, A.C.G.; Sampaio, C.L.S.; Argôlo, L.A.; Jacobina, U.P. Integrative systematics unveils the controversial identity of Engraulidae fishing stocks in a Neotropical estuary, northeast Brazil. Neotrop. Ichthyol. 2020, 18, e200037. [Google Scholar] [CrossRef]
- Nebeshwar, K.; Vishwanath, W. Three new species of Garra (Pisces: Cyprinidae) from north-eastern India and redescription of G. gotyla. Ichthyol. Explor. Freshw. 2013, 24, 97–120. [Google Scholar]
- Kottelat, M. Freshwater fishes of Northern Vietnam: A preliminary checklist of the fishes known or expected to occur in Northern Vietnam with comments on systematics and nomenclature. In Freshwater Fishes of Northern Vietnam; Kottelat, M., Ed.; World Bank: Washington, DC, USA, 2001; pp. 1–64. [Google Scholar]
- Sambrook, J.; Russell, D.W. The Condensed Protocols from Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 2006. [Google Scholar]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D.N. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 1–22. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MRBAYES 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A. Figtree Ver 1.4.4. Institute of Evolutionary Biology; University of Edinburgh: Edinburgh, UK, 2018. [Google Scholar]
- Roni, N.; Vishwanath, W. A new species of the genus Garra (Teleostei: Cyprinidae) from the Barak River drainage, Manipur, India. Zootaxa 2018, 4374, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Li, X.; Zhou, W.; Li, F. A Review of Garra (Teleostei: Cypriniformes) From Two Rivers in West Yunnan, China with Description of a New Species. Zootaxa 2018, 4378, 49–70. [Google Scholar] [CrossRef]
- Fricke, R.; Eschmeyer, W.N.; van der Laan, R. (Eds.) Eschmeyer’s Catalog of Fishes: Genera, Species, References. 2024. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 22 November 2024).
- Barman, M.; Bhushan, S.; Phukan, B.; Kumar, A.P.; Jaiswar, A.K.; Talukdar, A.; Kalita, R.; Silpa, S. Molecular identification and phylogenetic relationship of fishes belonging to the family Danionidae from Brahmaputra Basin, Assam, Northeast India. Mol. Biol. Rep. 2024, 51, 875. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Zirkunga, M.C. On the definition, status of research, diversity and prospect of exploration of the genus Garra (Cypriniformes: Cyprinidae) from Mizoram, Northeast India. Sci. Vis. 2021, 21, 1–5. [Google Scholar] [CrossRef]
- Limbu, J.H.; Rajbanshi, D.; Khanal, L.; Adhikari, R.C. First record of Garra kempi Hora, 1921 (Cypriniformes: Cyprinidae) from Lohandra River of Nepal. J. Threat. Taxa 2024, 16, 25440–25445. [Google Scholar] [CrossRef]
- Kolaczkowski, B.; Thornton, J.W. Long-branch attraction bias and inconsistency in Bayesian phylogenetics. PLoS ONE 2009, 4, 7891. [Google Scholar] [CrossRef]
- Zhang, R.; Stull, G.W.; Jin, J.J.; Wang, Y.H.; Guo, Y.; Yang, Z.Y.; Li, H.T.; An, K.L.; Charboneau, J.L.; Folk, R.A.; et al. Phylogenetic resolution and conflict in the species-rich flowering plant family leguminosae. Syst. Biol. 2025, 1–23. [Google Scholar] [CrossRef]
- Lemmon, A.R.; Brown, J.M.; Stanger-Hall, K.; Lemmon, E.M. The effect of ambiguous data on phylogenetic estimates obtained by maximum likelihood and Bayesian inference. Syst. Biol. 2009, 58, 130–145. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).