Description of the Three Complete Mitochondrial Genomes of Click Beetles (Coleoptera, Elateridae) with Phylogenetic Implications
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection and DNA Extraction
2.2. Genome Sequencing, Assembly and Annotation
2.3. Sequence Alignment
2.4. Phylogenetic Reconstruction
3. Results
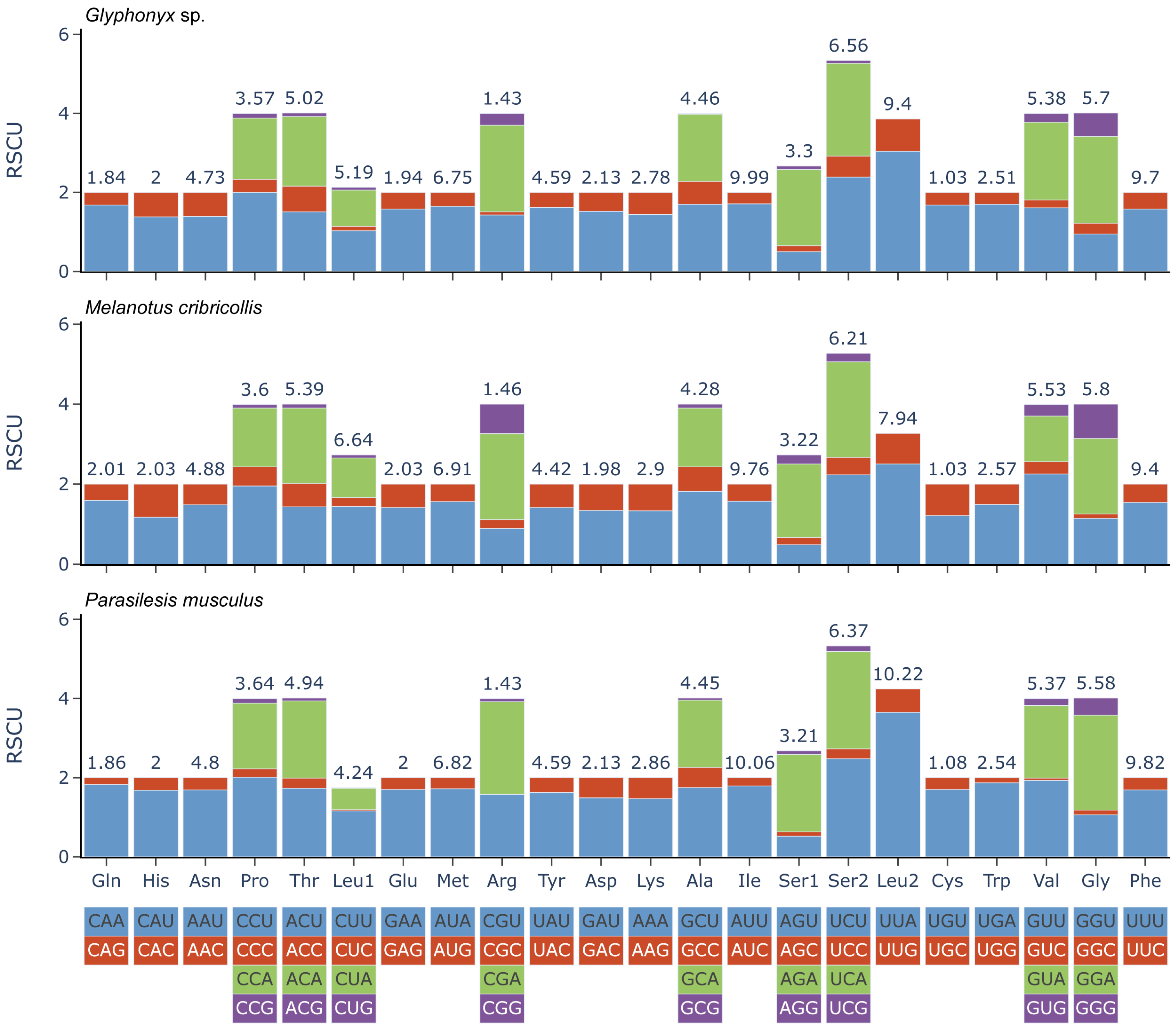
3.1. Characteristics of the New Mitogenomes
3.2. Phylogenetic Inference
4. Discussion
4.1. Monophyly of the Elateridae
4.2. Subfamily Relationships in Elateridae
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Douglas, H.B.; Kundrata, R.; Brunke, A.J.; Escalona, H.E.; Chapados, J.T.; Eyres, J.; Richter, R.; Savard, K.; Ślipiński, A.; McKenna, D. Anchored phylogenomics, evolution and systematics of Elateridae: Are all bioluminescent Elateroidea derived click beetles? Biology 2021, 10, 451. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.; Lawrence, J.F.; Rosa, S.P. 4.7. Elateridae Leach, 1815. In Volume 2 Morphology and Systematics (Elateroidea, Bostrichiformia, Cucujiformia partim); De Gruyter: Berlin, Germany, 2011; pp. 75–103. [Google Scholar]
- Lawrence, J.F.; Newton, A.F., Jr. Evolution and classification of beetles. Annu. Rev. Ecol. Syst. 1982, 13, 261–290. [Google Scholar] [CrossRef]
- Etzler, F.E.; Wanner, K.W.; Morales-Rodriguez, A.; Ivie, M.A. DNA barcoding to improve the species-level management of wireworms (Coleoptera: Elateridae). J. Econ. Entomol. 2014, 107, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Oba, Y.; Ôhira, H.; Murase, Y.; Moriyama, A.; Kumazawa, Y. DNA barcoding of Japanese click beetles (Coleoptera, Elateridae). PLoS ONE 2015, 10, e0116612. [Google Scholar] [CrossRef] [PubMed]
- Stibick, J.N. Classification of the Elateridae (Coleoptera): Relationships and classification of the subfamilies and tribes. Pac. Insects 1979, 20, 145–186. [Google Scholar]
- Muona, J. The phylogeny of Elateroidea (Coleoptera), or which tree is best today? Cladistics 1995, 11, 317–341. [Google Scholar] [CrossRef]
- Kundrata, R.; Bocak, L. The phylogeny and limits of Elateridae (Insecta, Coleoptera): Is there a common tendency of click beetles to soft-bodiedness and neoteny? Zool. Scr. 2011, 40, 364–378. [Google Scholar] [CrossRef]
- Hyslop, J. The phylogeny of the Elateridae based on larval characters. Ann. Entomol. Soc. Am. 1917, 10, 241–263. [Google Scholar] [CrossRef]
- Douglas, H. Phylogenetic relationships of Elateridae inferred from adult morphology, with special reference to the position of Cardiophorinae. Zootaxa 2011, 2900, 1–45. [Google Scholar] [CrossRef]
- Kundrata, R.; Gunter, N.L.; Janosikova, D.; Bocak, L. Molecular evidence for the subfamilial status of Tetralobinae (Coleoptera: Elateridae), with comments on parallel evolution of some phenotypic characters. Arthropod Syst. Phylogeny 2018, 76, 137–145. [Google Scholar]
- Meng, Z.; Lei, C.; Chen, X.; Shi, T.; Chen, Q.; Jiang, S. Phylogenetic analysis of click beetles (Coleoptera: Elateridae) based upon 28S rDNA: Phylogeny and classification. Entomotaxonomia 2018, 40, 231–252. [Google Scholar]
- Kundrata, R.; Gunter, N.L.; Douglas, H.; Bocak, L. Next step toward a molecular phylogeny of click-beetles (Coleoptera: Elateridae): Redefinition of Pityobiinae, with a description of a new subfamily Parablacinae from the Australasian Region. Austral Entomol. 2016, 55, 291–302. [Google Scholar] [CrossRef]
- Kundrata, R.; Bocakova, M.; Bocak, L. The comprehensive phylogeny of the superfamily Elateroidea (Coleoptera: Elateriformia). Mol. Phylogenet. Evol. 2014, 76, 162–171. [Google Scholar] [CrossRef]
- Bocak, L.; Motyka, M.; Bocek, M.; Bocakova, M. Incomplete sclerotization and phylogeny: The phylogenetic classification of Plastocerus (Coleoptera: Elateroidea). PLoS ONE 2018, 13, e0194026. [Google Scholar] [CrossRef] [PubMed]
- Sagegami-Oba, R.; Oba, Y.; Ôhira, H. Phylogenetic relationships of click beetles (Coleoptera: Elateridae) inferred from 28S ribosomal DNA: Insights into the evolution of bioluminescence in Elateridae. Mol. Phylogenet. Evol. 2007, 42, 410–421. [Google Scholar] [CrossRef]
- Bocakova, M.; Bocak, L.; Hunt, T.; Teraväinen, M.; Vogler, A.P. Molecular phylogenetics of Elateriformia (Coleoptera): Evolution of bioluminescence and neoteny. Cladistics 2007, 23, 477–496. [Google Scholar] [CrossRef]
- Karnkowska, A.; Vacek, V.; Zubáčová, Z.; Treitli, S.C.; Petrželková, R.; Eme, L.; Novák, L.; Žárský, V.; Barlow, L.D.; Herman, E.K. A eukaryote without a mitochondrial organelle. Curr. Biol. 2016, 26, 1274–1284. [Google Scholar] [CrossRef]
- Campbell, A.; Mrázek, J.; Karlin, S. Genome signature comparisons among prokaryote, plasmid, and mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1999, 96, 9184–9189. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Liu, Y. The complete mitochondrial genome of click beetle Chiagosnius sulcicollis (Coleoptera: Elateridae) and phylogenetic analysis. Mitochondrial DNA Part B 2019, 4, 2324–2325. [Google Scholar] [CrossRef]
- He, J.; Yao, Y.; Dong, Z.; Ruan, Y.; Chang, Z.; Zhao, R.; Wang, W.; Li, X. Complete mitochondrial genome of Pectocera sp. (Elateridae: Dendrometrinae: Oxynopterini) and its phylogenetic implications. Arch. Insect Biochem. Physiol. 2022, 111, e21957. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Bi, W.; Dong, Z.; Liu, G.; Zhao, R.; Wang, W.; Li, X. The mitochondrial genome of the first luminous click-beetle (Coleoptera: Elateridae) recorded in Asia. Mitochondrial DNA Part B 2019, 4, 565–567. [Google Scholar] [CrossRef]
- Arnoldi, F.G.C.; Ogoh, K.; Ohmiya, Y.; Viviani, V.R. Mitochondrial genome sequence of the Brazilian luminescent click beetle Pyrophorus divergens (Coleoptera: Elateridae): Mitochondrial genes utility to investigate the evolutionary history of Coleoptera and its bioluminescence. Gene 2007, 405, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Candèze, E. Insects recueillis au Japon par Mr. G. LEWIS. Èlatérides Mémoires De La Société R. Des Sci. De Liège 1873, 2, 1–32. [Google Scholar]
- Candèze, E. Monographie des élatérides. Mém. Soc. R. Sci. Liège 1860, 3, 512. [Google Scholar]
- Arimoto, K.; Itô, R.; Noda, R. Elateridae (Insecta, Coleoptera) from Yaku Island (Ryukyu Islands, Japan). Check List 2022, 18, 733–755. [Google Scholar] [CrossRef]
- Arimoto, K.; Itô, R. Elateridae (Insecta, Coleoptera) from Tanegashima Island (Ryukyu Islands, Japan). Check List 2018, 14, 681–692. [Google Scholar] [CrossRef]
- Gillett, C.P.; Crampton-Platt, A.; Timmermans, M.J.; Jordal, B.H.; Emerson, B.C.; Vogler, A.P. Bulk de novo mitogenome assembly from pooled total DNA elucidates the phylogeny of weevils (Coleoptera: Curculionoidea). Mol. Biol. Evol. 2014, 31, 2223–2237. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.; Yiu, S.-M.; Chin, F.Y. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Donath, A.; Jühling, F.; Al-Arab, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Liu, H.-Y.; Yang, X.-J.; Zhao, X.-C.; Lin, A.-L. Complete mitochondrial genome of the darkling beetle Gonocephalum outreyi (Coleoptera: Tenebrionidae) with phylogenetic implications. J. Asia-Pac. Entomol. 2018, 21, 721–730. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kück, P.; Longo, G.C. FASconCAT-G: Extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front. Zool. 2014, 11, 81. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Liu, X.; Long, Y.; Cheng, S.; Zhong, B. The complete mitochondrial genome of click beetle Agrypnus sp. (Coleoptera: Elateridae) and phylogenetic analysis. Mitochondrial DNA Part B 2019, 4, 3354–3355. [Google Scholar] [CrossRef]
- Li, Y.; Wan, Y.; He, H.; Guo, Y.; Jiang, L.; Huang, C.; Zhang, F. The complete mitochondrial genome of Cryptalaus larvatus (Coleoptera: Elateridae). Mitochondrial DNA Part B 2020, 5, 1899–1900. [Google Scholar] [CrossRef]
- Amaral, D.T.; Mitani, Y.; Ohmiya, Y.; Viviani, V.R. Organization and comparative analysis of the mitochondrial genomes of bioluminescent Elateroidea (Coleoptera: Polyphaga). Gene 2016, 586, 254–262. [Google Scholar] [CrossRef]
- Fallon, T.R.; Lower, S.E.; Chang, C.H.; Bessho-Uehara, M.; Martin, G.J.; Bewick, A.J.; Behringer, M.; Debat, H.J.; Wong, I.; Day, J.C.; et al. Firefly genomes illuminate parallel origins of bioluminescence in beetles. eLife 2018, 7, e36495. [Google Scholar] [CrossRef]
- Linard, B.; Arribas, P.; Andújar, C.; Crampton-Platt, A.; Vogler, A.P. Lessons from genome skimming of arthropod-preserving ethanol. Mol. Ecol. Resour. 2016, 16, 1365–1377. [Google Scholar] [CrossRef]
- Gerritsen, A.T.; New, D.D.; Robison, B.D.; Rashed, A.; Hohenlohe, P.; Forney, L.; Rashidi, M.; Wilson, C.M.; Settles, M.L. Full mitochondrial genome sequence of the sugar beet wireworm Limonius californicus (Coleoptera: Elateridae), a common agricultural pest. Genome Announc. 2016, 4, e01628-15. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Zhao, X.; Song, N.; Zhao, T. Analysis of the complete mitochondrial genome of click beetle Agriotes hirayamai (Coleoptera: Elateridae). Mitochondrial DNA Part B 2018, 3, 290–291. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.G.; Choi, S.; Roh, S.J.; Lee, B.W.; Lim, J. A first record of complete mitochondrial genome of Cryptalaus Ohira (Insecta: Coleoptera: Elateridae) based on C. yamato (Nakane). Mitochondrial DNA Part B 2019, 4, 1628–1629. [Google Scholar] [CrossRef]
- Crampton-Platt, A.; Timmermans, M.J.; Gimmel, M.L.; Kutty, S.N.; Cockerill, T.D.; Vun Khen, C.; Vogler, A.P. Soup to tree: The phylogeny of beetles inferred by mitochondrial metagenomics of a Bornean rainforest sample. Mol. Biol. Evol. 2015, 32, 2302–2316. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-J.; Wu, Y.-W.; Wang, T.-Y. Characterization of the complete mitochondrial genome of Abscondita cerata (Olivier, 1911) (Coleoptera: Lampyridae) and its phylogenetic implications. Mitochondrial DNA Part B 2021, 6, 2528–2530. [Google Scholar] [CrossRef]
- Chen, X.; Dong, Z.; Liu, G.; He, J.; Zhao, R.; Wang, W.; Peng, Y.; Li, X. Phylogenetic analysis provides insights into the evolution of Asian fireflies and adult bioluminescence. Mol. Phylogenet. Evol. 2019, 140, 106600. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ogoh, K.; Ohba, N.; Liang, X.; Ohmiya, Y. Mitochondrial genomes of two luminous beetles, Rhagophthalmus lufengensis and R. ohbai (Arthropoda, Insecta, Coleoptera). Gene 2007, 392, 196–205. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J. PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Bergsten, J. A review of long-branch attraction. Cladistics 2005, 21, 163–193. [Google Scholar] [CrossRef]
- Felsenstein, J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 1978, 27, 401–410. [Google Scholar] [CrossRef]
- Calder, A.; Lawrence, J.; Trueman, J.H. Austrelater, gen. nov.(Coleoptera: Elateridae), with a description of the larva and comments on elaterid relationships. Invertebr. Syst. 1993, 7, 1349–1394. [Google Scholar] [CrossRef]
- Lawrence, J.F.; Muona, J.; Vahtera, V.; Teräväinen, M.; Lawrence, J. Anischia, Perothops and the phylogeny of Elateroidea (Coleoptera: Elateriformia). Insect Syst. Evol. 2007, 38, 205–239. [Google Scholar]
- Burakowski, B. Development, distribution and habits of Trixagus dermestoides (L.), with notes on the Throscidae and Lissomidae (Coleoptera, Elateroidea). Ann. Zool. 1975, 32, 375–405. [Google Scholar]
- Mertlik, J. New data on the distribution of three species of the family Lissomidae and Melasidae (Coleoptera). Elateridarium 2011, 5, 55–58. [Google Scholar]
- Mertlik, J.; Platia, G. Catalogue of the family Cebrionidae, Elateridae, Lissomidae, Melasidae and Throscidae (Coleoptera) from Turkey. Elateridarium 2008, 2, 1–40. [Google Scholar]
- Kusy, D.; He, J.W.; Bybee, S.M.; Motyka, M.; Bi, W.X.; Podsiadlowski, L.; Li, X.Y.; Bocak, L. Phylogenomic relationships of bioluminescent elateroids define the ‘lampyroid’clade with clicking Sinopyrophoridae as its earliest member. Syst. Entomol. 2021, 46, 111–123. [Google Scholar] [CrossRef]
- Kusy, D.; Motyka, M.; Bocak, L. Click beetle mitogenomics with the definition of a new subfamily Hapatesinae from Australasia (Coleoptera: Elateridae). Insects 2021, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Ôhira, H. Morphological and Taxonomic Study on the Larvae of Elateridae in Japan (Coleoptera); Okazaki University: Okazaki, Japan, 1962. [Google Scholar]
- Oba, Y. Molecular phylogenetic analysis of click beetles (Coleoptera: Elateridae). Coleopt. News Tokyo 2007, 157, 7–11. [Google Scholar]
- Ôhira, H. The systematics of subfamilies in Elateridae from Japan. In Booklet for a Lecture in the 17th Annual Meeting of the Japanese Society of Coleopterology; The Japanese Society of Coleopterology: Tokyo, Japan, 1999. [Google Scholar]
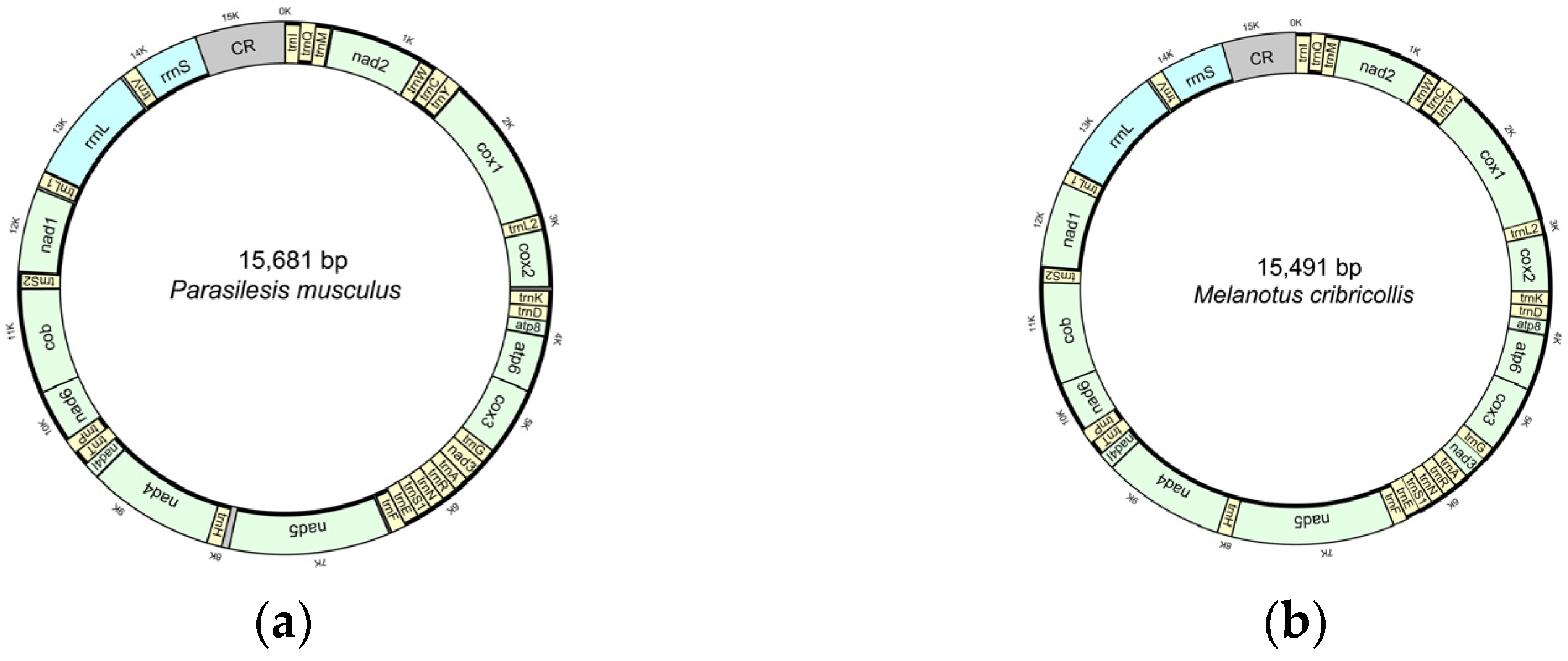







| Family | Subfamily | Species | Accession Number | Reference |
|---|---|---|---|---|
| Elateridae | Agrypninae | Agrypnus sp. YD-2019 | MN370897 | [38] |
| Elateridae | Agrypninae | Cryptalaus larvatus (Candèze, 1874) | NC_047286 | [39] |
| Elateridae | Agrypninae | Cryptalaus Yamato (Nakane, 1957) | NC_046689 | Unpublished |
| Elateridae | Agrypninae | Hapsodrilus ignifer (Germar, 1841) | NC_030058 | [40] |
| Elateridae | Agrypninae | Ignelater luminosus (Illiger, 1807) | MG242621 | [41] |
| Elateridae | Agrypninae | Pyrearinus termitilluminans (Candèze, 1863) | NC_030059 | [40] |
| Elateridae | Agrypninae | Pyrophorus divergens Eschscholtz, 1829 | NC_009964 | [24] |
| Elateridae | Cardiophorinae | Dicronychus cinereus (Herbst, 1784) | KX087283 | Unpublished |
| Elateridae | Cardiophorinae | Dicronychus sp. DIC01 | JX412848 | Unpublished |
| Elateridae | Dendrometrinae | Anostirus castaneus (Linnaeus, 1758) | KX087237 | Unpublished |
| Elateridae | Dendrometrinae | Athous haemorrhoidalis (Fabricius, 1801) | KT876881 | [42] |
| Elateridae | Dendrometrinae | Campsosternus auratus Drury, 1773 | MZ727583 | Unpublished |
| Elateridae | Dendrometrinae | Limonius californicus (Mannerheim, 1843) | NC_028541 | [43] |
| Elateridae | Dendrometrinae | Limonius minutus (Mannerheim, 1843) | KX087306 | Unpublished |
| Elateridae | Dendrometrinae | Pectocera sp. | NC_061359 | Unpublished |
| Elateridae | Dendrometrinae | Pheletes quercus (Olivier, 1790) | KX087332 | Unpublished |
| Elateridae | Elaterinae | Adrastus rachifer (Fourcroy, 1785) | KX087232 | Unpublished |
| Elateridae | Elaterinae | Agriotes hirayamai Miwa, 1934 | MG728108 | [44] |
| Elateridae | Elaterinae | Agriotes lineatus (Linnaeus, 1758) | OW618681 | Unpublished |
| Elateridae | Elaterinae | Agriotes obscurus (Linnaeus, 1758) | KT876879 | [42] |
| Elateridae | Elaterinae | Glyphonyx sp. | OQ475943 | This study |
| Elateridae | Elaterinae | Ludioschema sulcicolle (Candèze, 1878) | NC_053929 | [21] |
| Elateridae | Elaterinae | Ludioschema vittiger (Heyden, 1887) | MN306531 | [45] |
| Elateridae | Elaterinae | Melanotus cribricollis (Candèze, 1860) | OQ475941 | This study |
| Elateridae | Elaterinae | Melanotus villosus (Geoffroy, 1785) | KT876904 | [42] |
| Elateridae | Elaterinae | Parasilesis musculus (Candèze, 1873) | OQ475942 | This study |
| Elateridae | Elaterinae | Sericus brunneus (Linnaeus, 1758) | KX087344 | Unpublished |
| Elateridae | N/A | Elateridae sp. 2 ACP-2013 | MH789726 | [46] |
| Elateridae | N/A | Elateridae sp. GENSP01 | JX412817 | Unpublished |
| Elateridae | Negastriinae | Negastrius sabulicola (Boheman, 1852) | KX087320 | Unpublished |
| Elateridae | Physodactylinae | Teslasena femoralis (Lucas, 1857) | KJ938491 | Unpublished |
| Elateridae | Tetralobinae | Sinelater perroti (Fleutiaux, 1940) | NC_065395 | Unpublished |
| Lampyridae | Luciolinae | Abscondita cerata (Olivier, 1911) | MW751423 | [47] |
| Lampyridae | Luciolinae | Curtos fulvocapitalis (Jeng and Sato, 1998) | NC_058281 | Unpublished |
| Omalisidae | N/A | Omalisus fontisbellaquei (Müller, 1764) | JX412744 | Unpublished |
| Rhagophthalmidae | N/A | Rhagophthalmus giganteus (Fairmaire, 1888) | MK292104 | [48] |
| Rhagophthalmidae | N/A | Rhagophthalmus lufengensis Li & Ohba (Li et al., 2008) | DQ888607 | [49] |
| Rhagophthalmidae | N/A | Rhagophthalmus ohbai (Wittmer, 1994) | NC_010964 | [49] |
| Parasilesis musculus | Melanotus cribricollis | Glyphonyx sp. | ||||
|---|---|---|---|---|---|---|
| AA | Count | % | Count | % | Count | % |
| Ala(A) | 165 | 4.45 | 158 | 4.28 | 165 | 4.46 |
| Cys(C) | 40 | 1.08 | 38 | 1.03 | 38 | 1.03 |
| Asp(D) | 79 | 2.13 | 73 | 1.98 | 79 | 2.13 |
| Glu(E) | 74 | 2 | 75 | 2.03 | 72 | 1.94 |
| Phe(F) | 364 | 9.82 | 347 | 9.4 | 359 | 9.7 |
| Gly(G) | 207 | 5.58 | 214 | 5.8 | 211 | 5.7 |
| His(H) | 74 | 2 | 75 | 2.03 | 74 | 2 |
| Ile(I) | 373 | 10.06 | 360 | 9.76 | 370 | 9.99 |
| Lys(K) | 106 | 2.86 | 107 | 2.9 | 103 | 2.78 |
| Leu2(L2) | 379 | 10.22 | 293 | 7.94 | 348 | 9.4 |
| Leu1(L1) | 157 | 4.24 | 245 | 6.64 | 192 | 5.19 |
| Met(M) | 253 | 6.82 | 255 | 6.91 | 250 | 6.75 |
| Asn(N) | 178 | 4.8 | 180 | 4.88 | 175 | 4.73 |
| Pro(P) | 135 | 3.64 | 133 | 3.6 | 132 | 3.57 |
| Gln(Q) | 69 | 1.86 | 74 | 2.01 | 68 | 1.84 |
| Arg(R) | 53 | 1.43 | 54 | 1.46 | 53 | 1.43 |
| Ser2(S2) | 236 | 6.37 | 229 | 6.21 | 243 | 6.56 |
| Ser1(S1) | 119 | 3.21 | 119 | 3.22 | 122 | 3.3 |
| Thr(T) | 183 | 4.94 | 199 | 5.39 | 186 | 5.02 |
| Val(V) | 199 | 5.37 | 204 | 5.53 | 199 | 5.38 |
| Trp(W) | 94 | 2.54 | 95 | 2.57 | 93 | 2.51 |
| Tyr(Y) | 170 | 4.59 | 163 | 4.42 | 170 | 4.59 |
| codon end in A or T | 3263 | 88.02 | 2906 | 78.75 | 3064 | 82.77 |
| codon end in G or T | 1908 | 51.47 | 1892 | 51.27 | 1841 | 49.73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, N.; Lin, X.; Zhao, T. Description of the Three Complete Mitochondrial Genomes of Click Beetles (Coleoptera, Elateridae) with Phylogenetic Implications. Taxonomy 2023, 3, 204-220. https://doi.org/10.3390/taxonomy3020015
Song N, Lin X, Zhao T. Description of the Three Complete Mitochondrial Genomes of Click Beetles (Coleoptera, Elateridae) with Phylogenetic Implications. Taxonomy. 2023; 3(2):204-220. https://doi.org/10.3390/taxonomy3020015
Chicago/Turabian StyleSong, Nan, Xingyu Lin, and Te Zhao. 2023. "Description of the Three Complete Mitochondrial Genomes of Click Beetles (Coleoptera, Elateridae) with Phylogenetic Implications" Taxonomy 3, no. 2: 204-220. https://doi.org/10.3390/taxonomy3020015
APA StyleSong, N., Lin, X., & Zhao, T. (2023). Description of the Three Complete Mitochondrial Genomes of Click Beetles (Coleoptera, Elateridae) with Phylogenetic Implications. Taxonomy, 3(2), 204-220. https://doi.org/10.3390/taxonomy3020015





