Description of Two Fungal Endophytes Isolated from Fragaria chiloensis subsp. chiloensis f. patagonica: Coniochaeta fragariicola sp. nov. and a New Record of Coniochaeta hansenii
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Fungal Isolation
2.2. DNA Sequence Analyses
2.3. Macroscopic and Microscopic Characterization
2.4. Phenotypic Characterization
2.5. Data and Image Processing
3. Results and Discussion
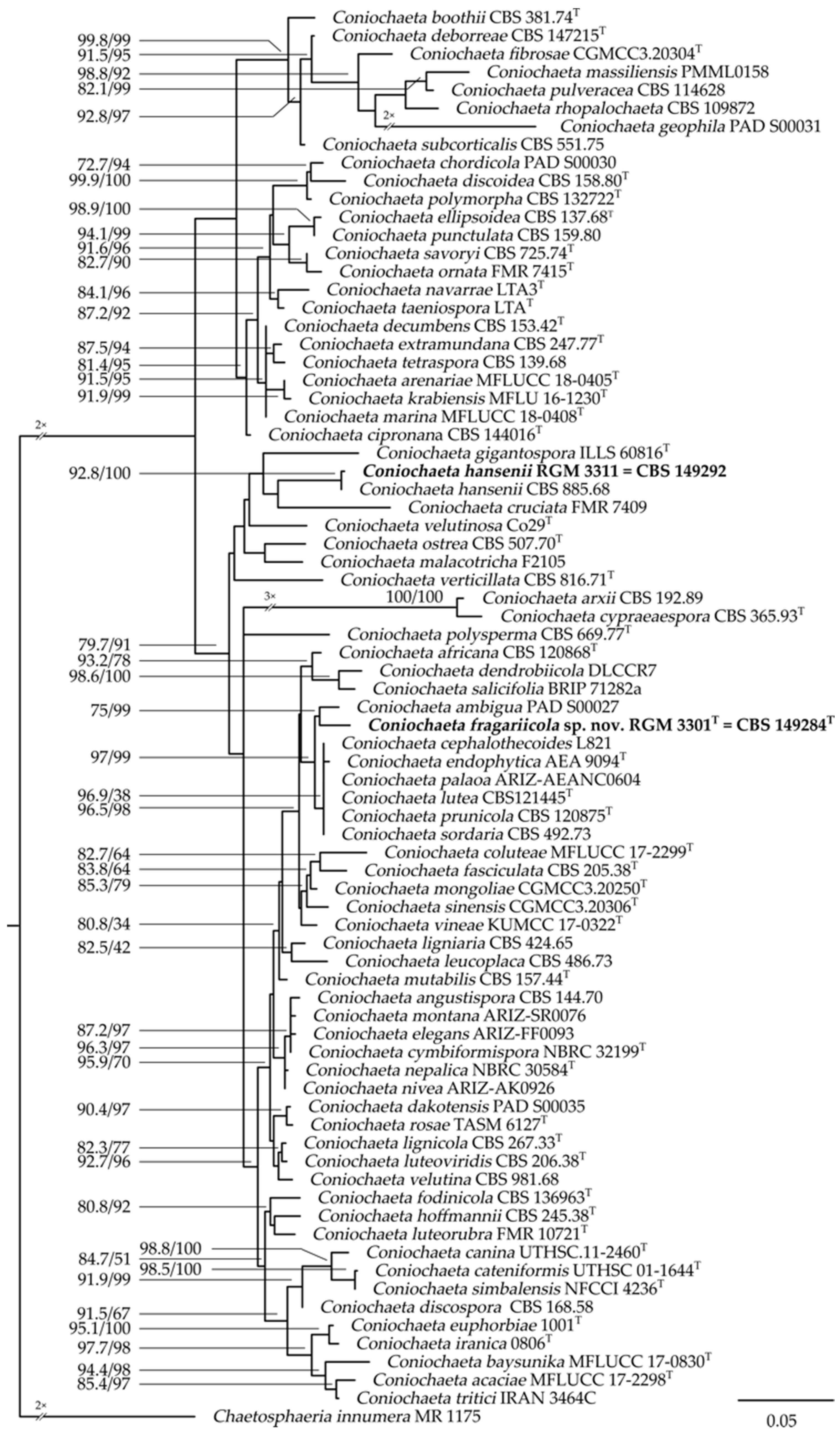
3.1. Taxonomic Position of the Isolates
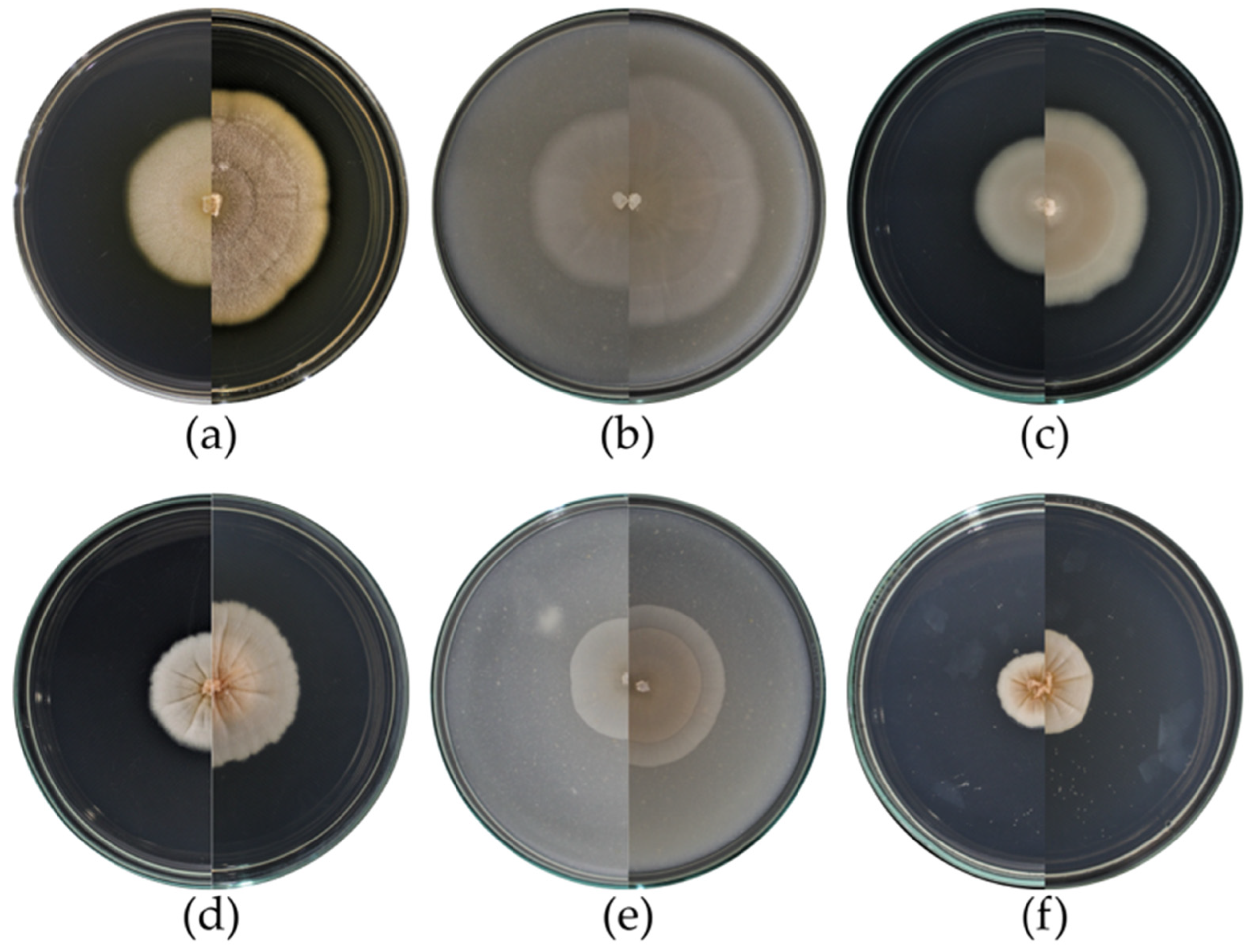
3.2. Macromorphological Characterization of the Isolates
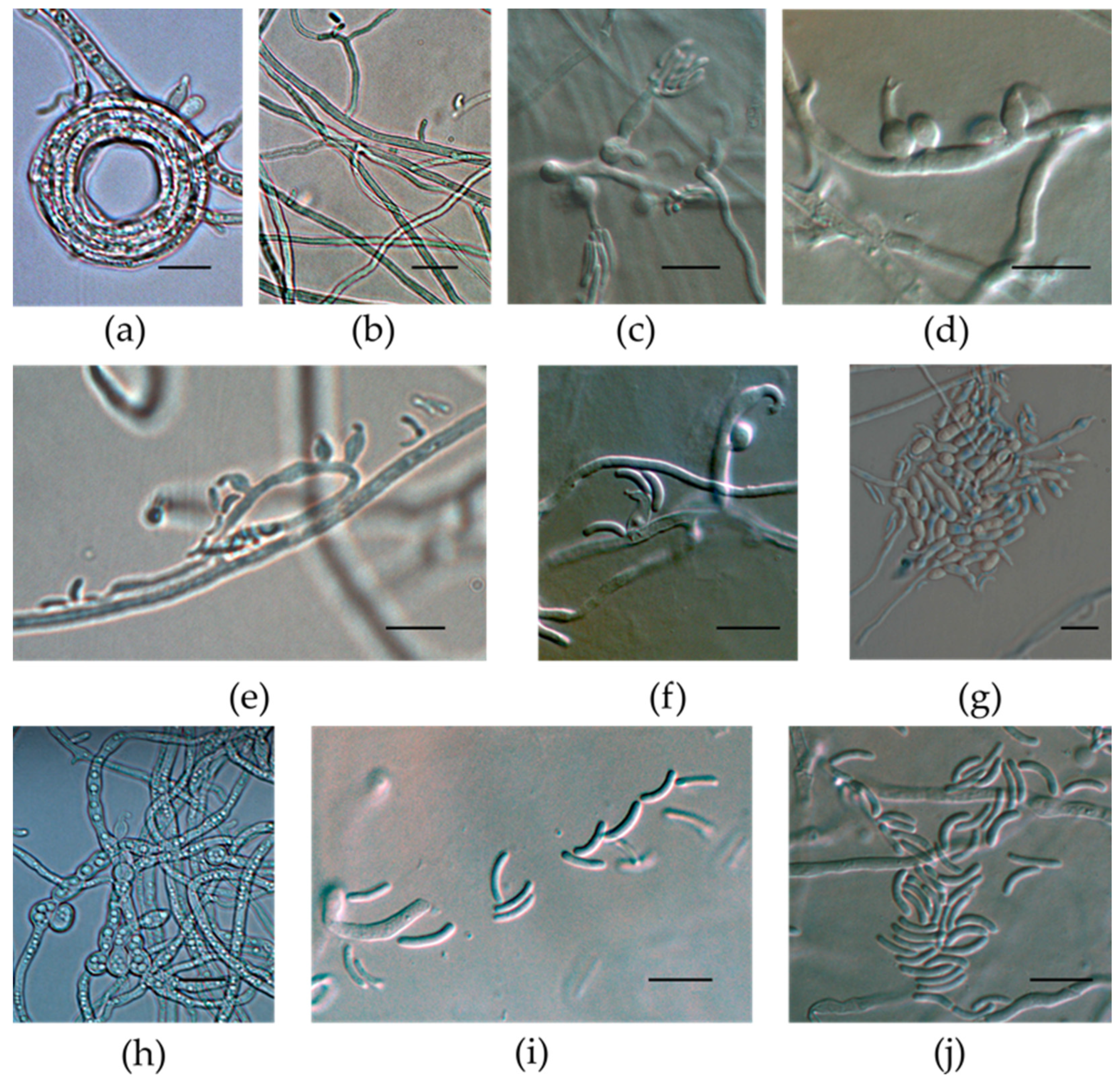
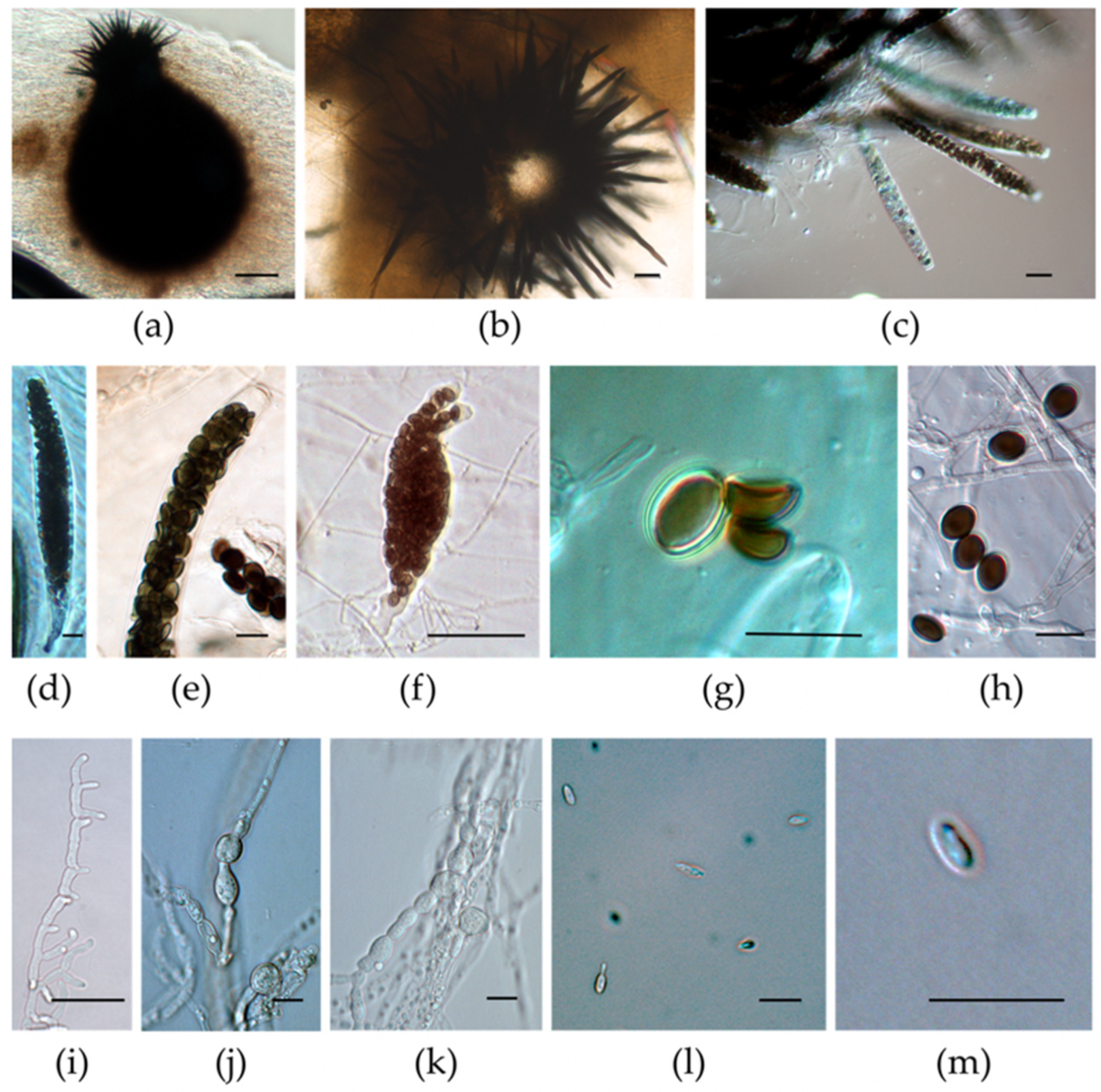
3.3. Micromorphological Characterization of the Isolates
3.4. Biochemical Features of the Isolates
3.5. Taxonomy of the Two Endophytes of Fragaria chiloensis subsp. chiloensis f. patagonica
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cooke, M.C. Coniochaeta, Sacc. Grevillea 1887, 16, 16. [Google Scholar]
- García, D.; Stchigel, A.M.; Cano, J.; Calduch, M.; Hawksworth, D.L.; Guarro, J. Molecular phylogeny of Coniochaetales. Mycol. Res. 2006, 110, 1271–1289. [Google Scholar] [CrossRef] [PubMed]
- Guarro, J.; Gené, J.; Stchigel, A.M.; Figueras, M.J. Atlas of Soil Ascomycetes; CBS-KNAW Fungal Biodiversity Centre: Utrecht, The Netherlands, 2012. [Google Scholar]
- Mahoney, D.P.; LaFavre, J.S. Coniochaeta Extramundana, with a synopsis of other Coniochaeta species. Mycologia 1981, 73, 931–952. [Google Scholar] [CrossRef]
- Dayarathne, M.C.; Jones, E.B.G.; Maharachchikumbura, S.S.N.; Devadatha, B.; Sarma, V.V.; Khongphinitbunjong, K.; Chomnunti, P.; Hyde, K.D. Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 2020, 11, 1–188. [Google Scholar] [CrossRef]
- Si, H.L.; Su, Y.M.; Zheng, X.X.; Ding, M.Y.; Bose, T.; Chang, R.L. Phylogenetic and morphological analyses of Coniochaeta isolates recovered from Inner Mongolia and Yunnan revealed three new endolichenic fungal species. MycoKeys 2021, 83, 105–121. [Google Scholar] [CrossRef]
- Rosa, L.H.; Queiroz, S.C.N.; Moraes, R.M.; Wang, X.; Techen, N.; Pan, Z.; Cantrell, C.L.; Wedge, D.E. Coniochaeta ligniaria: Antifungal activity of the cryptic endophytic fungus associated with autotrophic tissue cultures of the medicinal plant Smallanthus sonchifolius (Asteraceae). Symbiosis 2013, 60, 133–142. [Google Scholar] [CrossRef]
- Xie, J.; Strobel, G.A.; Feng, T.; Ren, H.; Mends, M.T.; Zhou, Z.; Geary, B. An endophytic Coniochaeta velutina producing broad spectrum antimycotics. J. Microbiol. 2015, 53, 390–397. [Google Scholar] [CrossRef]
- Damm, U.; Fourie, P.H.; Crous, P.W. Coniochaeta (Lecythophora), Collophora gen. nov. and Phaeomoniella species associated with wood necroses of Prunus trees. Persoonia 2010, 24, 60–80. [Google Scholar] [CrossRef]
- Checa, J.; Barrasa, J.; Moreno, G.; Fort, F.; Guarro, J. The genus Coniochaeta (Sacc.) Cooke (Coniochaetaceae, Ascomycotina) in Spain. Cryptog. Mycol 1988, 9, 1–34. [Google Scholar]
- Jiménez, D.J.; Hector, R.E.; Riley, R.; Lipzen, A.; Kuo, R.C.; Amirebrahimi, M.; Barry, K.W.; Grigoriev, I.V.; van Elsas, J.D.; Nichols, N.N. Draft genome sequence of Coniochaeta ligniaria NRRL 30616, a lignocellulolytic fungus for bioabatement of inhibitors in plant biomass hydrolysates. Genome Announc. 2017, 5, e01476-16. [Google Scholar] [CrossRef]
- Khan, Z.; Gené, J.; Ahmad, S.; Cano, J.; Al-Sweih, N.; Joseph, L.; Chandy, R.; Guarro, J. Coniochaeta polymorpha, a new species from endotracheal aspirate of a preterm neonate, and transfer of Lecythophora species to Coniochaeta. Antonie Leeuwenhoek 2013, 104, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Troy, G.C.; Panciera, D.L.; Pickett, J.P.; Sutton, D.A.; Gene, J.; Cano, J.F.; Guarro, J.; Thompson, E.H.; Wickes, B.L. Mixed infection caused by Lecythophora canina sp. nov. and Plectosphaerella cucumerina in a German shepherd dog. Med. Mycol. 2013, 51, 455–460. [Google Scholar] [CrossRef]
- de Hoog, S.; Guarro, J.; Gené, J.; Ahmed, S.; Al-Hatmi, A.; Figueras, M.; Vitale, R. Atlas of Clinical Fungi, 2nd ed.; Centraalbureau voor Schimmelcultures: Utrecht, The Netherlands; Universitat Rovira i Virgili Reus: Catalonia, Spain, 2020. [Google Scholar]
- Doveri, F. Description of Chaetomium aureum, Corynascus sepedonium and Coniochaeta hansenii newly recorded from Italy, and a key to coprophilous Chaetomiaceae and Coniochaetaceae. Ascomycete Org. 2016, 8, 7–24. [Google Scholar]
- Friebes, G.; Jaklitsch, W.M.; García, S.; Voglmayr, H. Lopadostoma taeniosporum revisited and a new species of Coniochaeta. Sydowia 2016, 68, 87–97. [Google Scholar]
- Forin, N.; Vizzini, A.; Fainelli, F.; Ercole, E.; Baldan, B. Taxonomic re-examination of nine Rosellinia types (Ascomycota, Xylariales) stored in the Saccardo Mycological Collection. Microorganisms 2021, 9, 666. [Google Scholar] [CrossRef]
- Carrasco-Fernández, J.; Guerra, M.; Castro, J.F.; Bustamante, L.; Barra-Bucarei, L.; Ceballos, R.; Fernández, N.; Edgington, S.; France, A. Plant growth promoting rhizobacteria from Juan Fernández archipelago improve germination rate of endangered plant Solanum fernandezianum Phil. Chil. J. Agric. Res. 2020, 80, 41–49. [Google Scholar] [CrossRef]
- Castro, J.F.; Guerra, M.; Carrasco-Fernández, J.; Ortiz-Campos, J.; Cares-Gatica, D.; Campos-Quiroz, C.; Correa, F.; Beltrán, M.F.; Sagredo, B.; Valdés, J.H. Draft genome sequence of Pseudomonas sp. strain RGM 3321, a phyllosphere endophyte from Fragaria chiloensis subsp. chiloensis f. patagonica. Microbiol. Resour. Announc. 2022, 11, e0033522. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J.; Innis, M.; Gelfand, D.; Sninsky, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR—Protocols and Applications—A Laboratory Manual; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; Volume 31, pp. 315–322. [Google Scholar]
- Rehner, S.A.; Samuels, G.J. Molecular systematics of the Hypocreales: A teleomorph gene phylogeny and the status of their anamorphs. Can. J. Bot. 1995, 73, 816–823. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- de Hoog, G.S.; van den Ende, A.H.G.G. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses 1998, 41, 183–189. [Google Scholar] [CrossRef]
- Van den Ende, A.; De Hoog, G. Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana. Stud. Mycol. 1999, 43, 151–162. [Google Scholar]
- Nasr, S.; Bien, S.; Soudi, M.R.; Alimadadi, N.; Shahzadeh Fazeli, S.A.; Damm, U. Novel Collophorina and Coniochaeta species from Euphorbia polycaulis, an endemic plant in Iran. Mycol. Prog. 2018, 17, 755–771. [Google Scholar] [CrossRef]
- Arnold, A.E.; Harrington, A.H.; Huang, Y.-L.; Ren, J.M.; Massimo, N.C.; Knight-Connoni, V.; Inderbitzin, P. Coniochaeta elegans sp. nov., Coniochaeta montana sp. nov. and Coniochaeta nivea sp. nov., three new species of endophytes with distinctive morphology and functional traits. Int. J. Syst. Evol. Microbiol. 2021, 71, 005003. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Pruitt, K.D.; Schoch, C.L.; Sherry, S.T.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2021, 49, D92–D96. [Google Scholar] [CrossRef]
- Priyam, A.; Woodcroft, B.J.; Rai, V.; Moghul, I.; Munagala, A.; Ter, F.; Chowdhary, H.; Pieniak, I.; Maynard, L.J.; Gibbins, M.A.; et al. Sequenceserver: A modern graphical user interface for custom BLAST databases. Mol. Biol. Evol. 2019, 36, 2922–2924. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef]
- Chang, J.M.; Di Tommaso, P.; Lefort, V.; Gascuel, O.; Notredame, C. TCS: A web server for multiple sequence alignment evaluation and phylogenetic reconstruction. Nucleic Acids Res. 2015, 43, W3–W6. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Castro, J.F.C.; Campos-Quiroz, C.; Carrasco-Fernández, J.; Guerra, M.; Santelices, C.; Cares-Gatica, D.; Ortiz-Campos, J.; Ocares, Y.; Barra-Bucarei, L.; Theelen, B. Description of two fungal endophytes isolated from Fragaria chiloensis subsp. chiloensis f. patagonica: Coniochaeta fragariicola sp. nov. and a new record of Coniochaeta hansenii, Dryad, Dataset. 2023. [Google Scholar]
- Samarakoon, M.C.; Gafforov, Y.; Liu, N.; Maharachchikumbura, S.S.; Bhat, J.D.; Liu, J.-K.; Promputtha, I.; Hyde, K.D. Combined multi-gene backbone tree for the genus Coniochaeta with two new species from Uzbekistan. Phytotaxa 2018, 336, 43–58. [Google Scholar] [CrossRef]
- Hawksworth, D.; Yip, H. Coniochaeta angustispora sp. nov. from roots in Australia, with a key to the species known in culture. Aust. J. Bot. 1981, 29, 377–384. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Phukhamsakda, C.; Hyde, K.D.; Jeewon, R.; Lee, H.B.; Gareth Jones, E.B.; Tibpromma, S.; Tennakoon, D.S.; Dissanayake, A.J.; Jayasiri, S.C.; et al. Fungal diversity notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018, 89, 1–236. [Google Scholar] [CrossRef]
- Perdomo, H.; García, D.; Gené, J.; Cano, J.; Sutton, D.A.; Summerbell, R.; Guarro, J. Phialemoniopsis, a new genus of Sordariomycetes, and new species of Phialemonium and Lecythophora. Mycologia 2013, 105, 398–421. [Google Scholar] [CrossRef]
- Han, J.; Liu, C.; Li, L.; Zhou, H.; Liu, L.; Bao, L.; Chen, Q.; Song, F.; Zhang, L.; Li, E.; et al. Decalin-containing tetramic acids and 4-hydroxy-2-pyridones with antimicrobial and cytotoxic activity from the fungus Coniochaeta cephalothecoides collected in Tibetan Plateau (Medog). J. Org. Chem. 2017, 82, 11474–11486. [Google Scholar] [CrossRef]
- Coronado-Ruiz, C.; Avendaño, R.; Escudero-Leyva, E.; Conejo-Barboza, G.; Chaverri, P.; Chavarría, M. Two new cellulolytic fungal species isolated from a 19th-century art collection. Sci. Rep. 2018, 8, 7492. [Google Scholar] [CrossRef]
- Van Der Linde, E.J. Coniochaeta cypraeaspora sp. nov. with a Paecilomyces conidial state. Mycol. Res. 1991, 95, 510–512. [Google Scholar] [CrossRef]
- Crous, P.W.; Hernández-Restrepo, M.; Schumacher, R.K.; Cowan, D.A.; Maggs-Kölling, G.; Marais, E.; Wingfield, M.J.; Yilmaz, N.; Adan, O.C.G.; Akulov, A.; et al. New and interesting fungi. 4. Fungal Syst. Evol. 2021, 7, 255–343. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Carnegie, A.J.; Wingfield, M.J.; Sharma, R.; Mughini, G.; Noordeloos, M.E.; Santini, A.; Shouche, Y.S.; Bezerra, J.D.P.; Dima, B.; et al. Fungal Planet description sheets: 868–950. Persoonia 2019, 42, 291–473. [Google Scholar] [CrossRef] [PubMed]
- Harrington, A.H.; del Olmo-Ruiz, M.; U’Ren, J.M.; Garcia, K.; Pignatta, D.; Wespe, N.; Sandberg, D.C.; Huang, Y.-L.; Hoffman, M.T.; Arnold, A.E. Coniochaeta endophytica sp. nov., a foliar endophyte associated with healthy photosynthetic tissue of Platycladus orientalis (Cupressaceae). Plant Fungal Syst. 2019, 64, 65–79. [Google Scholar] [CrossRef]
- Vázquez-Campos, X.; Kinsela, A.S.; Waite, T.D.; Collins, R.N.; Neilan, B.A. Fodinomyces uranophilus gen. nov. sp. nov. and Coniochaeta fodinicola sp. nov., two uranium mine-inhabiting Ascomycota fungi from northern Australia. Mycologia 2014, 106, 1073–1089. [Google Scholar] [CrossRef]
- Raja, H.A.; Shearer, C.A.; Raja, H.A.; Fournier, J.; Miller, A.N. Freshwater ascomycetes: Coniochaeta gigantospora sp. nov. based on morphological and molecular data. Mycoscience 2012, 53, 373–380. [Google Scholar] [CrossRef]
- Weber, E.; Gorke, C.; Begerow, D. The Lecythophora-Coniochaeta complex. II. Molecular studies based on sequences of the large subunit of ribosomal DNA. Nova Hedwig. 2002, 74, 187–200. [Google Scholar] [CrossRef]
- Weber, E. The Lecythophora-Coniochaeta complex: I. Morphological studies on Lecythophora species isolated from Picea abies. Nova Hedwig. 2002, 74, 159–185. [Google Scholar] [CrossRef]
- Arnold, A.E.; Harrington, A.H.; U’Ren, J.M.; Oita, S.; Inderbitzin, P. Two new endophytic species enrich the Coniochaeta endophytica/C. prunicola clade: Coniochaeta lutea sp. nov. and C. palaoa sp. nov. Plant Fungal Syst. 2021, 66, 66–78. [Google Scholar] [CrossRef]
- Jones, E.B.G.; Devadatha, B.; Abdel-Wahab, M.A.; Dayarathne, M.C.; Zhang, S.-N.; Hyde, K.D.; Liu, J.-K.; Bahkali, A.H.; Sarma, V.V.; Tibell, S.; et al. Phylogeny of new marine Dothideomycetes and Sordariomycetes from mangroves and deep-sea sediments. Bot. Mar. 2020, 63, 155–181. [Google Scholar] [CrossRef]
- Kabtani, J.; Militello, M.; Ranque, S. Coniochaeta massiliensis sp. nov. isolated from a clinical Sampl28. J. Fungi 2022, 8, 999. [Google Scholar] [CrossRef]
- van Heerden, A.; van Zyl, W.H.; Cruywagen, C.W.; Mouton, M.; Botha, A. The lignicolous fungus Coniochaeta pulveracea and its interactions with syntrophic yeasts from the woody phylloplane. Microb. Ecol. 2011, 62, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Romero, A.I.; Carmaran, C.C.; Lorenzo, L.E. A new species of Coniochaeta with a key to the species known in Argentina. Mycol. Res. 1999, 103, 689–695. [Google Scholar] [CrossRef]
- Zare, R.; Asgari, B.; Gams, W. The species of Coniolariella. Mycologia 2010, 102, 1383–1388. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Dong, Y.; Phookamsak, R.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Liu, N.-G.; Abeywickrama, P.D.; Mapook, A.; Wei, D.; et al. Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020, 100, 5–277. [Google Scholar] [CrossRef]
- Réblová, M.; Winka, K. Phylogeny of Chaetosphaeria and its anamorphs based on morphological and molecular data. Mycologia 2000, 92, 939–954. [Google Scholar] [CrossRef]
- Crous, P.W.; Verkley, G.J.M.; Groenewald, J.Z.; Samson, R.A. Fungal Biodiversity, 2nd ed.; CBS-KNAW Fungal Biodiversity Centre: Utrecht, The Netherlands, 2009. [Google Scholar]
- Castro, J.F.; Nouioui, I.; Sangal, V.; Trujillo, M.E.; Montero-Calasanz, M.D.C.; Rahmani, T.; Bull, A.T.; Asenjo, J.A.; Andrews, B.A.; Goodfellow, M. Geodermatophilus chilensis sp. nov., from soil of the Yungay core-region of the Atacama Desert, Chile. Syst. Appl. Microbiol. 2018, 41, 427–436. [Google Scholar] [CrossRef]
- Castro, J.F.; Nouioui, I.; Sangal, V.; Choi, S.; Yang, S.J.; Kim, B.Y.; Trujillo, M.E.; Riesco, R.; Montero-Calasanz, M.D.C.; Rahmani, T.P.D.; et al. Blastococcus atacamensis sp. nov., a novel strain adapted to life in the Yungay core region of the Atacama Desert. Int. J. Syst. Evol. Microbiol. 2018, 68, 2712–2721. [Google Scholar] [CrossRef]
- Kamiya, S.; Uchiyama, S.; Udagawa, S.-I. Two new species of Coniochaeta with a cephaiothecoid peridium wall. Mycoscience 1995, 36, 377–383. [Google Scholar] [CrossRef]
- Petrak, F. Coniochaeta sordaria. Sydowia 1953, 7, 1–4. [Google Scholar]
- Oudemans, C.A.J.A. Sordariae novae. Hedwigia 1882, 21, 123–124. [Google Scholar]
- Phillips, W.; Plowr. Sordaria polyspora. Grevillea 1881, 10, 73. [Google Scholar]
- Cain, R.F. Coniochaeta philocoproides (Griffiths). Univ. Toronto Stud. Biol. Ser. 1934, 38, 65. [Google Scholar]
- Richardson, M. Coniochaeta polymegasperma and Delitschia trichodelitschioides, two new coprophilous ascomycetes. Mycol. Res. 1998, 102, 1038–1040. [Google Scholar] [CrossRef]
- Furuya, K.; Udawaga, S.-i. Coprophilous Pyrenomycetes from Japan IV. Trans. Mycol. Soc. Jpn. 1976, 17, 248–261. [Google Scholar]
- Jiménez, D.J.; Wang, Y.; Chaib de Mares, M.; Cortes-Tolalpa, L.; Mertens, J.A.; Hector, R.E.; Lin, J.; Johnson, J.; Lipzen, A.; Barry, K.; et al. Defining the eco-enzymological role of the fungal strain Coniochaeta sp. 2T2.1 in a tripartite lignocellulolytic microbial consortium. FEMS Microbiol. Ecol. 2020, 96, fiz186. [Google Scholar] [CrossRef]
- Wyatt, T.T.; van Leeuwen, M.R.; Wösten, H.A.B.; Dijksterhuis, J. Mannitol is essential for the development of stress-resistant ascospores in Neosartorya fischeri (Aspergillus fischeri). Fungal Genet. Biol. 2014, 64, 11–24. [Google Scholar] [CrossRef]
| GenBank Accession | ||||||
|---|---|---|---|---|---|---|
| Fungal Species | Collection Number | Host/Source | Country | ITS | LSU | Reference |
| Coniochaeta acaciae | MFLUCC 17-2298T | Acacia sp. | Uzbekistan | MG062735 | MG062737 | [38] |
| Coniochaeta africana | CBS 120868T | Prunus salicina | South Africa | GQ154539 | GQ154601 | [9] |
| Coniochaeta ambigua | PAD S00027T | Sambucus racemosa | Italy | ITS1: MW626895, ITS2: MW626903 | - | [17] |
| Coniochaeta angustispora | CBS 144.70 | Trio compost | Netherlands | MH859528 | MH871308 | [27,39] |
| Coniochaeta arenariae | MFLUCC 18-0405T | Ammophila arenaria | United Kingdom | - | MN017893 | [5] |
| Coniochaeta arxii | CBS 192.89 | Soil | Japan | MH862164 | - | [17,27] |
| Coniochaeta baysunika | MFLUCC 17-0830T | Rosa sp. | Uzbekistan | MG828880 | MG828996 | [40] |
| Coniochaeta boothii | CBS 381.74T | Soil, mud | India | NR_159776 | AJ875226 | [2] |
| Coniochaeta canina | UTHSC 11-2460T R-4810T | Osteolytic lesion in a German shepherd dog | USA | JX481775 | JX481774 | [12,13] |
| Coniochaeta cateniformis | UTHSC 01-1644T | Canine bone marrow | USA | HE610331 | HE610329 | [41] |
| Coniochaeta chordicola | PAD S00030T | On a rope | Italy | ITS1: MW626898, ITS2: MW626905 | - | [17] |
| Coniochaeta cephalothecoides | L821 | Fruiting bodies of Trametes sp. | Tibet, China | KY064029 | KY064030 | [42] |
| Coniochaeta cipronana | CBS 144016T | Art lithograph | Costa Rica | NR_157478 | - | [43] |
| Coniochaeta coluteae | MFLUCC 17-2299T | Colutea paulsenii | Uzbekistan | MG137251 | MG137252 | [38] |
| Coniochaeta cruciata | FMR 7409 | Soil | Nigeria | - | AJ875222 | [2] |
| Coniochaeta cymbiformispora | NBRC 32199T | Swamp soil | Japan | LC146726 | LC146726 | Ban et al., unpublished |
| Coniochaeta cypraeaespora | CBS 365.93T | Dead grass from termite mound | South Africa | MH862420 | MH874073 | [27,44] |
| Coniochaeta dakotensis | PAD S00035T | Crataegus sp. | USA | ITS1: MW626902, ITS2: MW626910 | - | [17] |
| Coniochaeta deborreae | CBS 147215T | Soil | Belgium | MW883413 | MW883808 | [45] |
| Coniochaeta decumbens | CBS 153.42T | Strawberry fruit, Fragaria sp. | The Netherlands | NR_144912 | NG_067257 | [41] |
| Coniochaeta dendrobiicola | MCC 1811T | Roots of Dendriobium lognicornu | Nepal | MK225602 | MK225603 | [46] |
| Coniochaeta discoidea | CBS 158.80T | Paddy soil | Japan | NR_159779 | NG_064120 | [2] |
| Coniochaeta discospora | CBS 168.58T | Horse dung | Canada | MH857740 | MH869278 | [27] |
| Coniochaeta elegans | ARIZ-FF0093T | Juniperus deppeana | USA | MZ262389 | - | [26] |
| Coniochaeta ellipsoidea | CBS 137.68T | Soil | Japan | MH859091 | MH870804 | [27] |
| Coniochaeta endophytica | AEA 9094T | Endophyte of Platycladus orientalis | USA | EF420005 | EF420069 | [47] |
| Coniochaeta euphorbiae | CBS 139768T | Healthy plant of Euphorbia polycaulis | Iran | KP941076 | KP941075 | [25] |
| Coniochaeta extramundana | CBS 247.77T | Soil | USA | MH861057 | MH872828 | [27] |
| Coniochaeta fasciculata | CBS 205.38T | Butter | Switzerland | HE610336 | AF353598 | [41] |
| Coniochaeta fibrosae | CGMCC3.20304T | Medulla of the lichen Candelaria fibrosa | China | MW750760 | MW750758 | [6] |
| Coniochaeta fodinicola | CBS 136963T | Uranium mine raffinate | Australia | JQ904603 | KF857172 | [48] |
| Coniochaeta fragariicola sp. nov. | RGM 3301T = CBS 149284T | Fragaria chiloensis subsp. chiloensis f. patagonica | Chile | OP962067 * | OP962069 * | This work |
| Coniochaeta geophila | PAD S00031T | Sandy ground among mosses | Belgium | ITS2: MW626906 | - | [17] |
| Coniochaeta gigantospora | ILLS 60816T | Submerged wood of Fraxinus excelsior | France | JN684909 | JN684909 | [49] |
| Coniochaeta hansenii | CBS 885.68 | Dung of rabbit | The Netherlands | OP962072 * | AJ875223 | [2] |
| Coniochaeta hansenii | RGM 3311 = CBS 149292 | Fragaria chiloensis subsp. chiloensis f. patagonica | Chile | OP962068 * | OP962070 * | This work |
| Coniochaeta hoffmannii | CBS 245.38T | Butter | Switzerland | HE610332 | AF353599 | [41,50] |
| Coniochaeta iranica | CBS 139767T | Healthy plant of Euphorbia polycaulis | Iran | KP941078 | KP941077 | [25] |
| Coniochaeta krabiensis | MFLU 16-1230T | Submerged marine wood | Thailand | - | MN017892 | [5] |
| Coniochaeta leucoplaca | CBS 486.73 | Agricultural soil | The Netherlands | OP962071 * | MH872465 | [27] |
| Coniochaeta ligniaria | CBS 110467 | Picea abies | Germany | - | AF353583 | [41,50] |
| Coniochaeta ligniaria | CBS 424.65 | Decaying wood in herbarium | USA | MH858650 | MH870292 | [51] |
| Coniochaeta lignicola | CBS 267.33T | Unknown | Sweden | NR_111520 | NG_067344 | [41,50] |
| Coniochaeta lutea | CBS 121445T | Necrotic wood of Prunus salicina | South Africa | GQ154541 | GQ154603 | [52] |
| Coniochaeta luteorubra | FMR 10721T | Leg wound | USA | HE610330 | HE610328 | [41] |
| Coniochaeta luteoviridis | CBS 206.38T | Butter | Switzerland | HE610333 | AF353603 | [41,50] |
| Coniochaeta malacotricha | F2105 | Orthotomicus laricis | Germany | - | AF353588 | [50] |
| Coniochaeta marina | MFLUCC 18-0408T | Sweden | Piece of driftwood retrieved from the sea | MK458764 | MK458765 | [53] |
| Coniochaeta massiliensis | PMML0158T | Human body (abscess on hand) | France | OM366153 | OM366268 | [54] |
| Coniochaeta mongoliae | CGMCC3.20250T | Medulla of the lichen Ramalina sinensis | China | MW077645 | MW077646 | [6] |
| Coniochaeta montana | ARIZ-SR0076T | Juniperus deppeana | USA | MZ262414 | - | [26] |
| Coniochaeta mutabilis | CBS 157.44T | River water | Germany | HE610334 | AF353604 | [41,50] |
| Coniochaeta navarrae | CBS 141016T | Bark of Ulmus sp. | Spain | KU762326 | KU762326 | [16] |
| Coniochaeta nepalica | NBRC 30584T | Soil | Nepal | LC146727 | LC146727 | Ban S. et al., unpublished data |
| Coniochaeta nivea | ARIZ-AK0926T | Ophioparma ventosa | USA | MZ262367 | - | [26] |
| Coniochaeta ornata | FMR 7415T | Soil | Russia | - | AJ875228 | [2] |
| Coniochaeta ostrea | CBS 507.70T | Larrea sp. twigs | USA | NR_159772 | NG_064080 | [2,27] |
| Coniochaeta palaoa | ARIZ-AEANC0604T | Endophytic in healthy Hypnum sp. | USA | MZ241149 | - | [52] |
| Coniochaeta polymorpha | CBS 132722T | Endotracheal aspirate of a preterm neonate | Kuwait | HE863327 | HE863327 | [12] |
| Coniochaeta polysperma | CBS 669.77T | Dung of hare | Japan | MH861109 | MH872868 | [27] |
| Coniochaeta prunicola | CBS 120875T | Prunus armeniaca | South Africa | GQ154540 | GQ154602 | [9] |
| Coniochaeta pulveracea | CBS 114628 | Rinsing machine, in soft drink factory | Turkey | MW883414 | GQ351560 | [45,55] |
| Coniochaeta punctulata | CBS 159.80 | River sludge | Japan | MH861254 | MH873024 | [27] |
| Coniochaeta rhopalochaeta | CBS 109872 | Bulnesia retamas, decorticated wood | Argentina | NR_172554 | GQ351561 | [56] |
| Coniochaeta rosae | TASM 6127T | Branches of Rosa hissarica | Uzbekistan | NR_157509 | NG_066204 | [40] |
| Coniochaeta savoryi | CBS 725.74T | Wood of Juniperus scopulorum | United Kingdom | MH860890 | MH872627 | [2] |
| Coniochaeta simbalensis | NFCCI 4236T | Soil | India | NR_164024 | MG917738 | Rana and Singh, unpublished |
| Coniochaeta sinensis | CGMCC3.20306T | Medulla of the lichen Ramalina sinensis | China | MW422269 | MW422265 | [6] |
| Coniochaeta sordaria | CBS 492.73 | - | Germany | - | MH878380 | [6] |
| Coniochaeta subcorticalis | CBS 551.75 | Pinus sylvestris | Norway | MW883416 | AF353593 | [45,50] |
| Coniochaeta taeniospora | CBS 141014T | Quercus petraea | Austria | KU762324 | KU762324 | [16] |
| Coniochaeta tetraspora | CBS 139.68 | Soil | Japan | MH859093 | MH870806 | [27] |
| Coniochaeta velutina | CBS 981.68 | Waste stabilization pond | USA | MH859264 | MH870991 | [27] |
| Coniochaeta velutinosa | Co29T | Leaf of Hordeum vulgare | Iran | GU553327 | GU553330 | [57] |
| Coniochaeta verticillata | CBS 816.71T | Agricultural soil | The Netherlands | NR_159774 | AJ875232 | [2] |
| Coniochaeta vineae | KUMCC 17-0322T | Dead vine | China | NR_168225 | [58] | |
| Chaetosphaeria innumera | MR 1175 | Fagus sylvatica | Czech Republic | AF178551 | - | [59] |
| Culture Medium 1 | |||
|---|---|---|---|
| Strain | PDA | OA | MEA |
| Coniochaeta fragariicola RGM 3301 | Orange-brown in the center and faded-yellow towards the margin, flat colony with entire margin; from the bottom, the same characteristics. A growth diameter of 50 mm. Grows between 15 and 35 °C, optimal between 25 and 30 °C. | Pale orange-pink colony in the center and grayish-white towards the margin, flat with entire margin; from the bottom, the same characteristics. A growth diameter of 86 mm. | Bright pale orange-pink in the center and yellowish-gray towards the margin, flat colony with entire margin; from the bottom, the same color. A growth diameter of 45 mm. |
| Coniochaeta hansenii RGM 3311 | Orange with a slightly irregular margin, radial grooves from the center to the edge of the colony; from the bottom, the same coloration but paler. A growth diameter of 37 mm. Grows between 10 and 25 °C, optimal 25 °C. | Pale orange, flat with entire margin; from the bottom, the same characteristic. A growth diameter of 37 mm. | Pale orange, flat colony with entire margin, underside of the same coloration; from the bottom the same characteristics. A growth diameter of 23 mm. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos-Quiroz, C.; Castro, J.F.; Santelices, C.; Carrasco-Fernández, J.; Guerra, M.; Cares-Gatica, D.; Ortiz-Campos, J.; Ocares, Y.; Barra-Bucarei, L.; Theelen, B. Description of Two Fungal Endophytes Isolated from Fragaria chiloensis subsp. chiloensis f. patagonica: Coniochaeta fragariicola sp. nov. and a New Record of Coniochaeta hansenii. Taxonomy 2023, 3, 183-203. https://doi.org/10.3390/taxonomy3020014
Campos-Quiroz C, Castro JF, Santelices C, Carrasco-Fernández J, Guerra M, Cares-Gatica D, Ortiz-Campos J, Ocares Y, Barra-Bucarei L, Theelen B. Description of Two Fungal Endophytes Isolated from Fragaria chiloensis subsp. chiloensis f. patagonica: Coniochaeta fragariicola sp. nov. and a New Record of Coniochaeta hansenii. Taxonomy. 2023; 3(2):183-203. https://doi.org/10.3390/taxonomy3020014
Chicago/Turabian StyleCampos-Quiroz, Carolina, Jean Franco Castro, Cecilia Santelices, Jorge Carrasco-Fernández, Matías Guerra, Diego Cares-Gatica, Javiera Ortiz-Campos, Yocelyn Ocares, Lorena Barra-Bucarei, and Bart Theelen. 2023. "Description of Two Fungal Endophytes Isolated from Fragaria chiloensis subsp. chiloensis f. patagonica: Coniochaeta fragariicola sp. nov. and a New Record of Coniochaeta hansenii" Taxonomy 3, no. 2: 183-203. https://doi.org/10.3390/taxonomy3020014
APA StyleCampos-Quiroz, C., Castro, J. F., Santelices, C., Carrasco-Fernández, J., Guerra, M., Cares-Gatica, D., Ortiz-Campos, J., Ocares, Y., Barra-Bucarei, L., & Theelen, B. (2023). Description of Two Fungal Endophytes Isolated from Fragaria chiloensis subsp. chiloensis f. patagonica: Coniochaeta fragariicola sp. nov. and a New Record of Coniochaeta hansenii. Taxonomy, 3(2), 183-203. https://doi.org/10.3390/taxonomy3020014







