Medical Specialty Classification: An Interactive Application with Iterative Improvement for Patient Triage †
Abstract
1. Introduction
2. Related Work
3. Model Architecture and Implementation
Model Architecture
- Input Layer: Processes tokenized text with a maximum length of 128 tokens.
- Embedding Layer: Combines token, segment, and position embeddings.
- Transformer Encoder Layers: Comprises 12 layers of self-attention and feed-forward networks.
- Classification Layer: Custom layer added during fine-tuning to map BERT outputs to specialty labels.
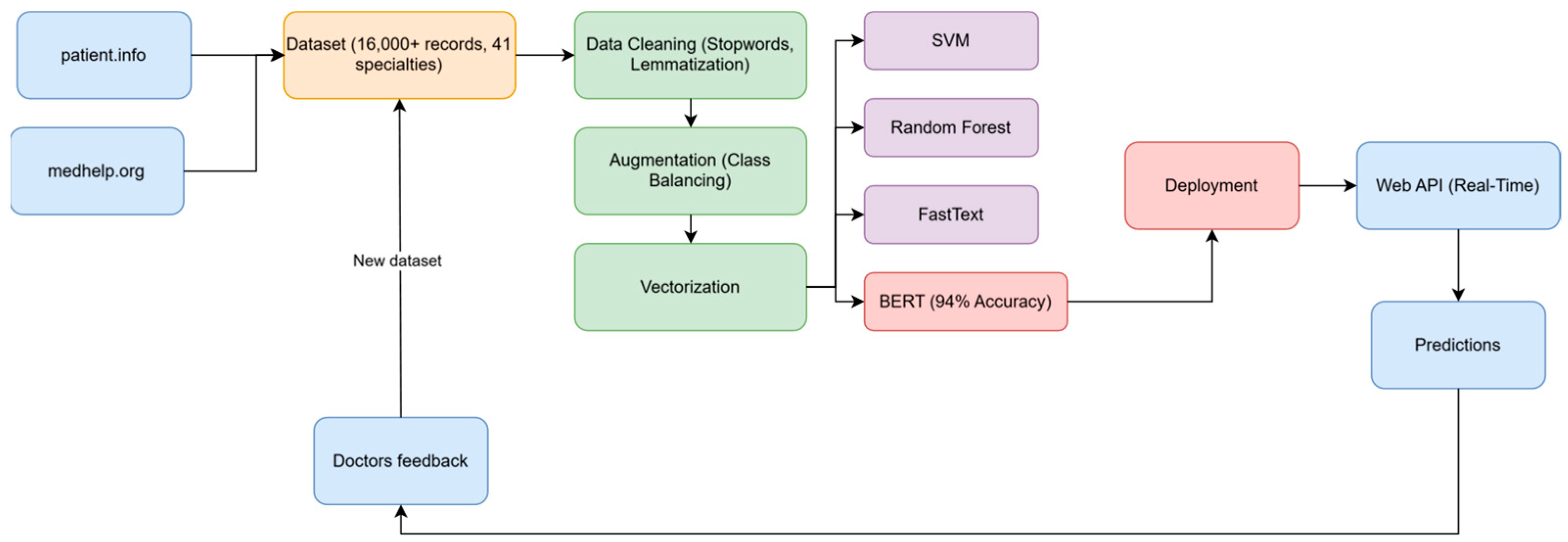
4. Model Enhancement
4.1. Data Collection and Initial Model
4.2. Incorporating User Feedback
4.3. Data Augmentation from Medical Forums
4.4. Model Retraining and Evaluation
4.5. Challenges and Solutions in Model Development
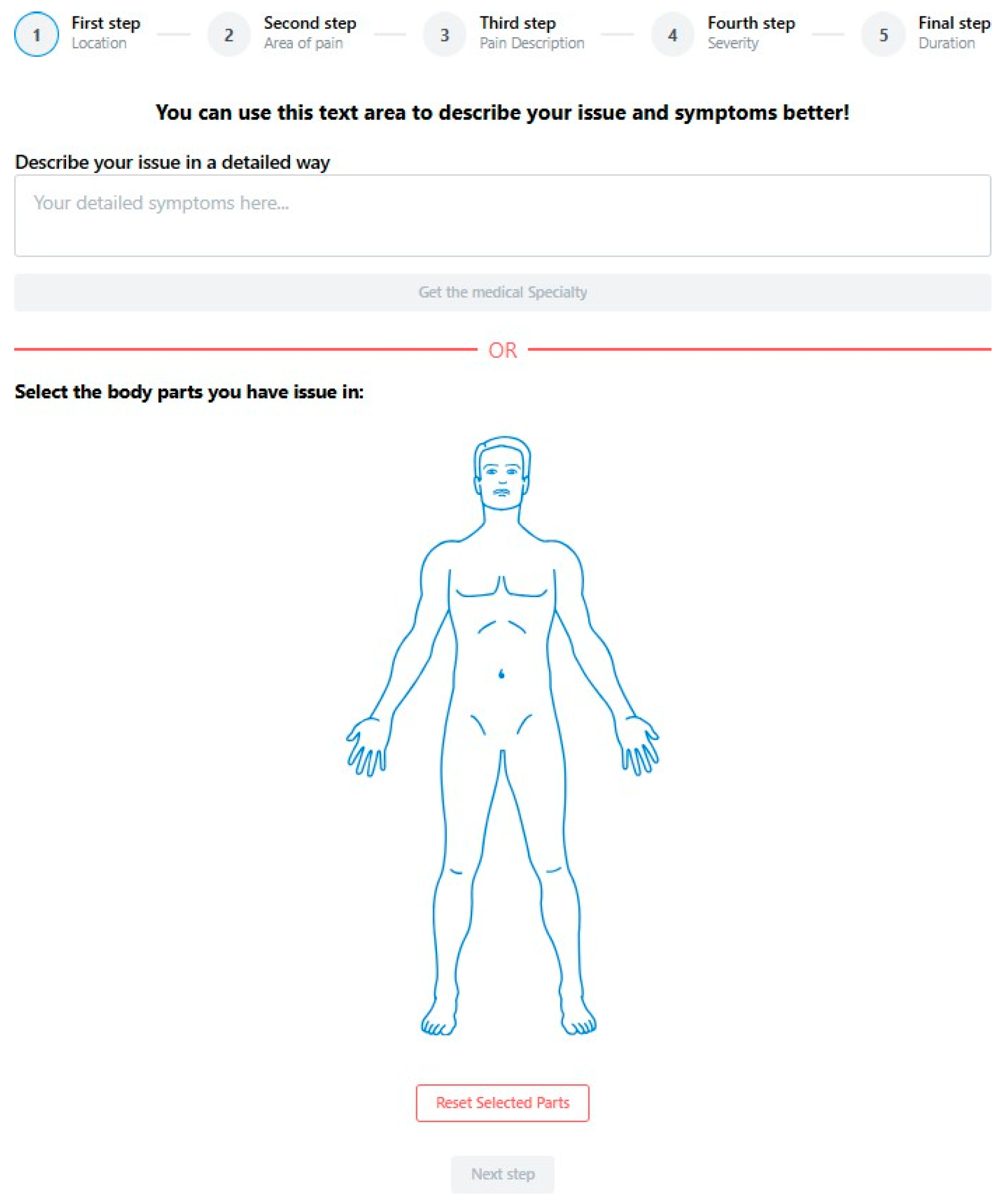
5. The Prediction Application
5.1. Technical Implementation Details
- -
- Frontend: Developed using React.js for responsive design, ensuring compatibility across devices including mobile phones and tablets. The frontend communicates with the backend via RESTful API calls.
- -
- Backend API: Built with Flask, the API handles request processing, text normalization, and model inference. Key features include:
- -
- CPU-based model inference to lower the running cost.
- -
- User feedback collection and storage.
- -
- Authentication for medical professionals.
- -
- Deployment Infrastructure: The application is containerized using Docker and deployed on the Azure App Service with autoscaling capabilities to handle varying loads. This ensures a high availability and consistent performance.
5.2. Performance Metrics Pages
5.2.1. Home Page
5.2.2. Result Page
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, Y.; Kim, J.-H.; Kim, Y.-M.; Song, S.; Joo, H.J. Predicting medical specialty from text based on a domain-specific pre-trained BERT. Int. J. Med. Inform. 2022, 170, 104956. [Google Scholar] [CrossRef] [PubMed]
- Faris, H.; Habib, M.; Faris, M.; Alomari, M.; Alomari, A. Medical speciality classification system based on binary particle swarms and ensemble of one vs. rest support vector machines. J. Biomed. Inform. 2020, 109, 103525. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Zhu, Q.; Chen, R.; Su, W. Automatic medical specialty classification based on patients’ description of their symp-toms. BMC Med. Inform. Decis. Mak/ 2023, 23, 15. [Google Scholar]
- Weng, W.-H.; Wagholikar, K.B.; McCray, A.T.; Szolovits, P.; Chueh, H.C. Medical subdomain classification of clinical notes using a machine learning-based natural language processing approach. BMC Med. Inform. Decis. Mak. 2017, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mosa, A.S.M.; Yoo, I.; Sheets, L. A Systematic Review of Healthcare Applications for Smartphones. BMC Med. Inform. Decis. Mak. 2012, 12, 67. [Google Scholar] [CrossRef] [PubMed]
- Madanian, S.; Nakarada-Kordic, I.; Reay, S.; Chetty, T. Patients’ perspectives on digital health tools. PEC Innov. 2023, 2, 100171. [Google Scholar] [CrossRef] [PubMed]
- Semigran, H.L.; Linder, J.A.; Gidengil, C.; Mehrotra, A. Evaluation of symptom checkers for self diagnosis and triage: Audit study. BMJ 2015, 351, h3480. [Google Scholar] [CrossRef] [PubMed]
- Tu, T.; Palepu, A.; Schaekermann, M.; Saab, K.; Freyberg, J.; Tanno, R.; Wang, A.; Li, B.; Amin, M.; Tomasev, N.; et al. Towards Conversational Diagnostic AI. arXiv 2024, arXiv:2401.05654. [Google Scholar] [CrossRef]
- Jutel, A.; Lupton, D. Digitizing diagnosis: A review of mobile applications in the diagnostic process. Diagnosis 2015, 2, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Anas, C.; Mohamed, E.; Ghaouth, B.M.; Wafae, M.; Yassine, C. Medical Specialty Triage Using Patients’ Questions and Symptoms. SSN 2023. [Google Scholar] [CrossRef]
- MedHelp-Health Community, Health Information, Medical Questions, and Medical Apps. Available online: https://www.medhelp.org/ (accessed on 11 November 2023).
- Symptom Checker, Health Information and Medicines Guide, Patient. Available online: https://patient.info/ (accessed on 11 November 2023).


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chahid, A.; Chahid, I.; Emharraf, M.; Belkasmi, M.G. Medical Specialty Classification: An Interactive Application with Iterative Improvement for Patient Triage. Eng. Proc. 2025, 112, 64. https://doi.org/10.3390/engproc2025112064
Chahid A, Chahid I, Emharraf M, Belkasmi MG. Medical Specialty Classification: An Interactive Application with Iterative Improvement for Patient Triage. Engineering Proceedings. 2025; 112(1):64. https://doi.org/10.3390/engproc2025112064
Chicago/Turabian StyleChahid, Anas, Ismail Chahid, Mohamed Emharraf, and Mohammed Ghaouth Belkasmi. 2025. "Medical Specialty Classification: An Interactive Application with Iterative Improvement for Patient Triage" Engineering Proceedings 112, no. 1: 64. https://doi.org/10.3390/engproc2025112064
APA StyleChahid, A., Chahid, I., Emharraf, M., & Belkasmi, M. G. (2025). Medical Specialty Classification: An Interactive Application with Iterative Improvement for Patient Triage. Engineering Proceedings, 112(1), 64. https://doi.org/10.3390/engproc2025112064





