Abstract
Probiotics as antibiotic alternatives for preventing and controlling infectious diseases are a relatively eco-friendly strategy in aquaculture. The bacteriocin-producing probiotic Paenibacillus ehimensis NPUST1 was isolated from tilapia culture pond water in our previous study. The present study demonstrated that P. ehimensis NPUST1 produced hydrolytic enzymes, including protease, amylase, cellulase, xylanase and lipase. The effects of P. ehimensis NPUST1 on zebrafish nutrient metabolism, growth performance and innate immunity were evaluated by measuring the expression of indicator genes in zebrafish after feeding P. ehimensis NPUST1 at doses of 106 and 107 CFU/g for 8 weeks. There was an obvious increase in the hepatic mRNA expression of carbohydrate metabolism-related genes, including glucokinase, hexokinase 1, glucose-6-phosphatase, and pyruvate kinase, and growth-related genes, including the growth hormone receptor and insulin-like growth factor-1. The expression of the innate immune-related genes including interleukin (IL)-1β, IL-6, IL-15, tumor necrosis factor-α, toll-like receptor (TLR)-1, TLR-4, complement component C3b and lysozyme were significantly increased in P. ehimensis NPUST1-supplemented fish. A significant reduction in cumulative mortality was exhibited in P. ehimensis NPUST1-supplemented fish after infection with Aeromonas hydrophila and Streptococcus iniae. In conclusion, our data suggested that P. ehimensis NPUST1 supplementation in feed could potentially improve nutrient metabolism and growth in addition to enhancing innate immunity and disease resistance against A. hydrophila and S. iniae in zebrafish.
1. Introduction
Aquaculture is the fastest growing production sector for animal protein in the world and plays an important role in supporting the demand for food for human consumption [1]. However, infectious diseases can cause massive mortality, and severe economic loss in fish farming is considered one of the major issues challenging the sustainable development of aquaculture. Among infectious diseases, bacterial pathogen-induced diseases exhibit the greatest incidences in aquaculture because bacteria are capable of multiplying in an aquatic environment independent of their host. For instance, Aeromonas hydrophila and Streptococcus iniae are typical pathogens that cause hemorrhagic septicemia and red fin disease in most freshwater fish species [2,3], and vibriosis caused by Vibrio pathogens usually causes massive mortality in animals cultured in marine water [4]. The use of antibiotics or chemicals is an effective way to prevent disease outbreaks or to support therapeutic purposes in fish farming. However, the ban on the use of antibiotics and chemicals in feed additives has become a global consensus due to issues such as the rapid spread of antibiotic-resistant pathogens in microbial ecosystems and the increased risk to food safety by residual antibiotic contamination. Thus, it is urgent and important to develop alternatives to antibiotics for the biocontrol infectious diseases in aquaculture.
Probiotics can confer positive effects on host health and have been considered an eco-friendly alternative to antibiotics for preventing disease outbreaks in aquaculture [5]. Numerous reviews have shown that the use of probiotics in aquaculture provides multiple benefits, such as improving nutrient metabolism, growth performance and feed efficiency, improving the beneficial gut microflora in the gastrointestinal (GI) tract and enhancing immunity and resistance to pathogen infection [6]. In addition to lactic acid bacteria, the Bacillus genus is the most commonly used probiotic in aquaculture due to its ability to produce endospores that are tolerant to the low pH of the GI tract and to produce hydrolytic enzymes that improve nutrient utilization and feed efficiency. The Bacillus genus is also known for its long-term safety in mammalian species. A recent review reported that potential probiotics isolated for use in fish farming should have criteria including nonpathogenicity and nonantibiotic resistance in addition to the ability to produce hydrolytic enzymes and antimicrobial substances [7]. Hydrolytic enzyme-producing probiotics have been shown to possess beneficial effects on improving nutrient utilization, feed efficiency and growth performance in fish. For instance, the dietary administration of hydrolytic enzyme-producing probiotics (enzymes including protease, cellulase, amylase, xylanase and phytase) of the Bacillus genus can enhance nutrient metabolism, feed efficiency and growth in tilapia, Labeo rohita, shrimp and zebrafish [8,9,10,11,12].
Species of Paenibacillus are gram-positive and endospore-forming bacteria that were originally categorized in the Bacillus genus because their morphological and physiological characteristics are quite similar to those of Bacillus subtilis. However, in 1993, Ash et al. [13] reclassified Paenibacillus as a new genus based on a phylogenetic analysis of 16S rRNA sequences and the characterization of its phenotypic characteristics. Recently, a review determined that the Paenibacillus genus can produce a variety of bacteriocins that can potentially be applied as antimicrobial agents in medicine against human pathogens and in agriculture for controlling plant pathogens [14]. However, there are few investigations on the application of bacteriocin-producing Paenibacillus species as biocontrol agents for disease control in aquaculture. In addition to bacteriocin, Paenibacillus species also exhibited the ability to produce hydrolytic enzymes and play roles in pathogen defense. For instance, amylase, cellulase, protease and lipase produced from P. polymyxa exhibited antagonistic activity against fish pathogens [15]. Chitinase and β-1,3-glucanase produced from P. ehimensis IB-X-b can degrade the cell walls of fungal mycelia, which are composed of β-1,3-glucan, chitin, cellulose and protein [16]. Our previous study isolated the bacteriocin-producing probiotic Paenibacillus ehimensis NPUST1 from a water sample of tilapia culture pools and demonstrated that dietary supplementation with P. ehimensis NPUST1 can improve feed efficiency, enhance growth and promote innate immunity against pathogen infection in Nile tilapia [17]. The present study evaluated the ability of the probiotic to produce hydrolytic enzymes and investigated the effect of the probiotic on nutrient metabolism, immunity and disease resistance in zebrafish.
2. Materials and Methods
2.1. Experimental Animals and Bacterial Strains
The adult zebrafish AB strain was obtained from the Taiwan Zebrafish Core Facility at Academia Sinica (Taipei, Taiwan). The fish were domesticated for 1 week in a 90 L aquarium at 28 °C with a controlled light cycle (14 h of light/10 h of dark) in the aquatic laboratory animal facility of the National Pingtung University of Science and Technology, which is accredited by the Association for Assessment and Accreditation of Laboratory Animals Care (AAALAC). The fish were fed commercial pellet feed twice daily at 2% body weight (MeM Prime, BERNAQUA, Olen, Belgium). The protocols for fish experiments were implemented according to local animal welfare regulations and approved (approval No. NPUST-105-067) by the Institutional Animal Care and Use Committee (IACUC) of NPUST. The probiotic P. ehimensis NPUST1 isolated from tilapia culture pools was as described in a previous report [17,18]. The pathogens A. hydrophila and S. iniae isolated from diseased tilapia were described in reports [18,19]. All bacteria were cultured in tryptic soy broth (TSB) medium at 28 °C overnight and stored in 20% glycerol at −20 °C until use.
2.2. Assay of Hydrolytic Enzymes and Antibiotic Susceptibility to Probiotics
The extracellular protease, amylase, cellulase, xylanase and lipase activities of probiotic P. ehimensis NPUST1 were assayed by an agar-well diffusion test as described in a previous report [8]. Bacteria were cultured in a 250 mL flask containing 50 mL of TSB medium at 28 °C for 12 h. Briefly, 10 μL of culture broth was added individually on the surface of differential agar plates and let stand until the added samples were dry. After incubating at 28 °C for 24 h, the plates were washed with sterile water to remove the colonies and then stained with a specific dye. For the protease activity assay, bacterial protease activity was observed as wells surrounded by clear zones that presented on an enriched nutrient agar plate containing 0.5% skimmed milk. For the amylase activity assay, bacterial amylase activity was observed as wells surrounded by clear zones that presented on 0.2% starch-enriched nutrient agar plates after staining with 5% iodine solution. For the cellulase and xylanase activity assays, bacterial cellulase and xylanase activities were observed as wells surrounded by clear zones that presented on 0.5% carboxymethyl cellulose (CMC) and 1% xylan-enriched nutrient agar plates after staining with 0.4% Congo red solution, respectively. For the lipase activity assay, bacterial lipase activity was observed as wells surrounded by a clear zone that presented on the enriched nutrient agar plate containing 1% glyceryl tributyrate and 1% Tween 80. Bacillus amyloliquefaciens R8, which demonstrated protease, amylase, cellulase, xylanase and lipase activities, was used as a positive control. Escherichia coli DH5α, without tested enzyme activities, was used as a negative control. The antibiotic susceptibility of the probiotic was determined by overlaying antibiotic-containing disks on the TSB agar that was previously seeded with approximately 1 × 106 CFU/mL of the probiotic P. ehimensis NPUST1. The tested antibiotics included ampicillin (20 µg), amoxicillin (20 µg), doxycycline (50 µg), erythromycin (50 µg), furazolidone (2 µg), flumequine (20 µg), ormetoprim (50 µg), oxytetracycline (50 µg), chloramphenicol (80 µg) and kanamycin (10 µg). The antibiotic susceptibility phenotypes of P. ehimensis NPUST1 were evaluated by the appearance of inhibition zones on agar plates after incubation at 28 °C for 24 h.
2.3. Feed Preparation and Feeding Experiment
The preparation of experimental diets was carried out according to a protocol described in a previous report [12]. Briefly, the culture broth of P. ehimensis NPUST1b was centrifuged at 6000× g for 15 min at 4 °C to harvest the cell pellet. The pellet was washed three times with phosphate-buffered saline (PBS) and then suspended in sterile water. The number of viable bacterial cells in the suspended solution was determined using a serial dilution and plate counting on TSB agar plates. An appropriate amount of suspended solution was added to the basal diet to obtain a diet containing 106 colony-forming units (CFU)/g and 107 CFU/g of P. ehimensis NPUST1. The ingredients of the basal diet were described in a previous report [12]. The proximate compositions of the basal diets were approximately 39.1% crude protein, 7.6% crude lipid, 11.8% ash and 9.2% moisture, as determined according to the method of the Association of Official Analytical Chemists (AOAC) [20]. The zebrafish (approximately 4.2 ± 0.02 cm in average body length and body weight 0.61 ± 0.04 g) were randomly divided into three groups including one control group and two experimental groups, E1 and E2. The fish in the control group were fed a basal diet. The fish in the E1 and E2 groups were fed a basal diet containing 106 CFU/g and 107 CFU/g of P. ehimensis NPUST1, respectively. Thirty fish per group were cultured in a 10 L tank at 28 °C, and each experiment was conducted in triplicate. The fish in the control, E1 and E2 groups were fed twice daily at 1% of body weight. The tanks were cleaned by siphoning the water daily, and the uneaten food was collected 1 h after feeding. After 8 weeks of feeding, the digestive enzyme activities, the expression levels of indicator genes associated with nutrition metabolism, the innate immunity and challenge tests were evaluated. The viability of probiotic cells in the diet was monitored by performing plate counting of the TSB agar plates every week during the feeding trial.
2.4. Determination of Digestive Enzymes in Intestines
The intestine tissues were sampled from six fish and homogenized in 1 mL of chilled phosphate-buffered saline (PBS; pH 7.4) using a homogenizer (Pro 200, PRO Scientific, Oxford, CT, USA). The buffer was centrifuged at 1000× g for 10 min at 4 °C to remove cell lysates, and then the supernatant was appropriately diluted in the PBS buffer to determine the protein concentration and assess hydrolytic enzyme activities. The protein content was measured with the Bradford method using bovine serum albumin (BSA) as a standard [21]. The protocol for determining protease, amylase, cellulase and xylanase activities was modified from that described in a previous report [22].
2.4.1. Protease Activity Assay
A 200 μL aliquot of supernatant from the fish intestines was mixed with 1.0 mL of a 0.7% (w/v) casein solution in 2.0 mL microcentrifuge tubes and incubated at 37 °C for 15 min. Subsequently, the reaction was terminated by adding 0.5 mL of a 110 mM trichloroacetic acid (TCA) solution and incubating for 20 min at 37 °C. The mixture solution was centrifuged at 12,000× g for 15 min at 4 °C to collect the supernatant. One milliliter of supernatant was mixed with 0.5 mL of Folin, 0.5 mM Ciolcalteau’s reagent and 2.5 mL of 500 mM Na2CO3 in a 10 mL tube and incubated at 37 °C for 30 min. Tyrosine was used as a standard to establish a standard curve by measuring the absorbance at 440 nm with a spectrophotometer (CT-2700, Chrom Tech, Taipei, Taiwan). One protease activity unit was defined as the amount of enzyme that releases 1 μmol of tyrosine per minute.
2.4.2. Amylase Activity Assay
A 200 μL aliquot of enzyme supernatant was mixed with 200 μL of a 1% starch solution and incubated at 37 °C for 30 min, and then 0.5 mL of 3,5-dinitrosalicylic acid (DNS) reagent was added to the mixture in a 10 mL glass tube and incubated in boiling water for 5 min. After cooling to ambient temperature, the absorbance of the solution was determined at 540 nm with a spectrophotometer. Glucose was used as a standard to establish a calibration curve. One amylase activity unit was defined as the amount of enzyme that released 1 μmol of reducing sugar per minute.
2.4.3. Cellulase Activity Assay
A 200 μL aliquot of enzyme supernatant was mixed with 0.2 mL of 0.1 M citrate buffer containing 1% carboxymethyl cellulose (CMC) in a 10 mL glass tube and then incubated at 50 °C for 30 min. Subsequently, 1 mL of the DNS reagent was added and incubated in a boiling water bath for 10 min. After cooling to ambient temperature, 0.5 mL of 40% KNaC4H4O6 was added, and the mixture was allowed to cool to ambient temperature. Glucose generated from the CMC substrate by cellulase activity was measured at 540 nm with a spectrophotometer. One unit of cellulase activity was defined as the amount of enzyme that released 1 μmol of glucose per minute.
2.4.4. Xylanase Activity Assay
A total of 100 μL of enzyme supernatant was mixed with 100 μL of 1% xylan (X-4252, Sigma, St. Louis, MO, USA) substrate solution in a 2 mL microcentrifuge tube and incubated at 55 °C for 10 min. Subsequently, 0.2 mL of the DNS reagent was added and heated to 100 °C for 10 min. After cooling to ambient temperature, 1 mL of deionized water was added to the mixture to terminate the reaction. After centrifugation at 12,000× g for 1 min, the absorbance of the supernatant was measured at 540 nm using a UV-Vis microplate reader (Multiskan Sky, Thermo Fisher Scientific, Waltham, MA, USA). One xylanase activity unit was defined as the amount of enzyme that released 1 mmol of reducing sugars per minute.
2.4.5. Lipase Activity Assay
A 10 μL aliquot of enzyme supernatant was mixed with 190 μL of 25 mM p-nitrophenyl butyrate, which was dissolved in a sodium phosphate buffer (pH 7.0), in a 96-well microplate and incubated at 37 °C for 30 min. The absorbance of the supernatant was measured at 405 nm using a UV-Vis microplate reader. p-Nitrophenol (p-NP) was used as a standard to establish a calibration curve. One unit of lipase activity was defined as the amount of enzyme that released 1 μmol of p-NP per minute.
2.5. Determination of Gene Expression by Real Time Quantitative Polymerase Chain Reaction (Real Time Q-PCR)
The liver tissue and whole-body samples were separately obtained from six zebrafish and used to extract total RNA using the TriPure isolation reagent (Roche, Mannheim, Germany) according to the manufacturer’s protocol. The expression of genes related to glucose metabolism, growth and innate immune responses was determined with real-time q-PCR (Applied Biosystems StepOnePlus, Foster City, CA, USA) using SYBR Green PCR reagents. The specific primers used for detecting genes are listed in Table 1. The expression of the EF-1α gene was used as the internal control. The condition of real-time Q-PCR followed that described in a previous report [12]. The relative expression level of each gene was normalized to that of EF-1α and expressed as the mean ± standard error (SE).

Table 1.
Primer sequences used in this study.
2.6. Challenge Experiment
A. hydrophila and S. iniae were separately cultured in TSB and incubated at 28 °C overnight. Bacterial cells were collected by centrifugation at 6100× g for 15 min at 4 °C and resuspended in the appropriate volumes of PBS to adjust the cell concentration. The 7-day median lethal dose (LD50) was determined by intraperitoneal (IP) injection of serial doses of A. hydrophila and S. iniae separately (105, 106, 107 and 108 CFU/fish) into 10 fish. At the end of the feeding trial, 15 fish from each group were administered 20 μL of diluted A. hydrophila or S. iniae solution by IP injection at a LD50 (1.0 × 106 CFU per fish for A. hydrophi and 1.0 × 105 CFU per fish for S. iniae). The bacterial challenge test was conducted in triplicates. The injected zebrafish were maintained in a tank (15 fish per tank) containing 10 L of fresh water at 28 °C. The fish fed the basal diet and injected with pathogens or with PBS were used as the positive and negative control group, respectively. The cumulative mortality in each group was recorded for 7 days post-infection.
2.7. Statistical Analysis
Significant differences in the relative gene expression levels for each group were analyzed statistically using one-way ANOVA and Tukey’s multiple comparison tests. A probability value of less than 0.05 (p < 0.05) was considered significant. Cumulative mortality in the challenge experiment was analyzed using the Kaplan—Meier method. The data analysis was performed using SAS software (SAS Institute, Cary, NC, USA).
3. Results
3.1. Characterization of Hydrolytic Enzymes and Antibiotic Susceptibility of P. ehimensis NPUST1
The potential probiotic P. ehimensis NPUST1 with bacteriocin activity isolated from a tilapia culture pool was investigated in our previous study [17]. In the present study, the activities of hydrolytic enzymes produced from the probiotic P. ehimensis NPUST1 were evaluated using an agar-well diffusion test. The results revealed that the probiotic P. ehimensis NPUST1 can secrete protease, amylase, cellulase, xylanase and lipase (Figure 1). Moreover, the antibiotic susceptibility of the probiotic P. ehimensis NPUST1 was evaluated using a disc diffusion test. The tested antibiotics showed an inhibition zone against P. ehimensis NPUST1, suggesting that P. ehimensis NPUST1 lacks antibiotic resistance against the tested antibiotics (Figure 2).
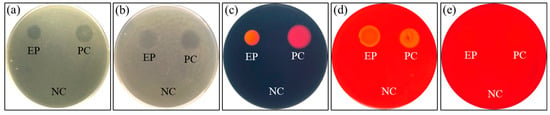
Figure 1.
Analysis of the hydrolytic enzyme activities of P. ehimensis NPUST1 with a selective agar plate after cultivation for 24 h. (a) Protease activity; (b) lipase activity; (c) amylase activity; (d) cellulase activity and (e) xylanase activity. P. ehimensis NPUST1 was used as the experimental group (EP); Escherichia coli DH5α was used as the negative control (NC); Bacillus amyloliquefaciens R8 was used as the positive control (PC).
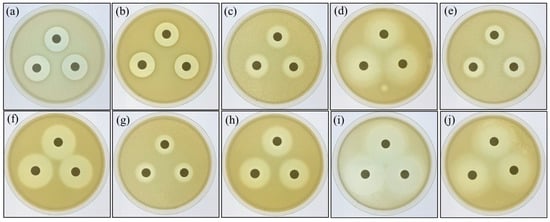
Figure 2.
Antibiotic susceptibility of Paenibacillus ehimensis NPUST1 identified using the agar-well diffusion test after cultivation for 24 h. (a) Amoxicillin; (b) ampicillin; (c) chloramphenicol; (d) doxycycline; (e) erythromycin; (f) flumequine; (g) furazolidone; (h) kanamycin; (i) ormetoprim; (j) oxytetracycline. Experiments were conducted in triplicate on a TSB agar plate.
3.2. Dietary Supplementation with P. ehimensis NPUST1 Enhances Digestive Enzyme Activities and Induces the Expression of Genes Involved in Glucose Metabolism and Growth
To evaluate the efficacy of dietary supplementation with P. ehimensis NPUST1 on nutrient metabolism and growth in fish, digestive enzyme activities, hepatic nutrient metabolism and growth-related genes were determined after feeding the P. ehimensis NPUST1-supplemented diet for 8 weeks. As shown in Table 2, the activity of the digestive enzymes protease, cellulase, xylanase and lipase of the fish in the E1 and E2 groups were significantly increased compared with those of the fish in the control group. The amylase activity in the gut of the fish in the E2 group was significantly higher than that of the fish in the control and E2 groups. The effect of P. ehimensis NPUST1 on nutrient metabolism and growth at the molecular level was also evaluated by determining the hepatic GK, HK1, G6Pase, PK-L, GHR and IGF-1 gene expression. The results showed a significant increase in GK and HK1 mRNA expression for the fish in the E1 and E2 groups compared to the fish in the control group. The results showed that the zebrafish in the E1 and E2 groups produced 1.85- and 2.44-fold and 1.95- and 3.3-fold higher mRNA expression levels of GK and HK1 compared to the zebrafish in the control group, respectively. Moreover, the relative expression level of HK1 exhibited a significant difference between the E1 and E2 groups (Figure 3a,b). The relative mRNA expression level of G6Pase was significantly higher for the fish in the E2 group compared to the fish in the control and E1 groups (Figure 3c). Specifically, zebrafish in the E2 group had a 2.35-fold higher mRNA expression level of G6Pase than the zebrafish in the control group. An obvious increase in hepatic PK-L expression was exhibited in the fish of the E1 group compared to that in the fish of the control group. Moreover, although there was no significant difference in PK-L expression between the E2 and control groups, a higher mRNA expression level of PK-L was detected in the fish of the E2 group than in the fish of the control group (Figure 3d). A significant increase in the GHR and IGF-1 mRNA expression levels were shown in the fish of the E1 and E2 groups compared to the fish of the control. The expression levels of GHR and IGF-1 in the zebrafish of the E1 and E2 groups were 1.71- and 2.48-fold and 2.0- and 2.07-fold higher than those in the zebrafish of the control group, respectively. However, there was no significant difference in GHR and IGF-1 mRNA expression in fish between the E1 and E2 groups (Figure 3e,f).

Table 2.
Intestinal digestive enzyme activities and hepatic metabolic enzyme activities of zebrafish after being fed a diet containing 106 or 107 CFU/g P. ehimensis NPUST1 for 8 weeks.
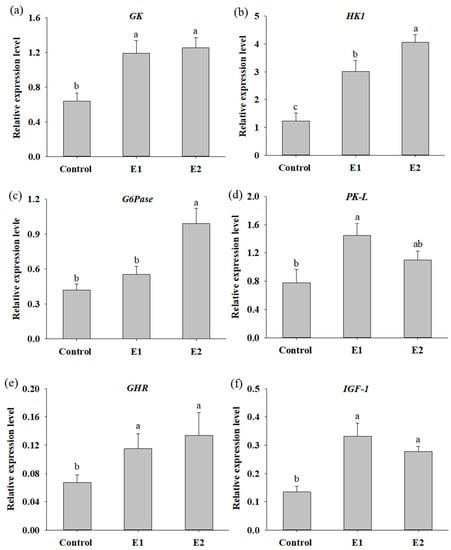
Figure 3.
Relative mRNA expression levels of glucose metabolism genes in the livers of zebrafish fed the basal diet (control), the basal diet containing 106 CFU/g (E1) or the basal diet containing 107 CFU/g (E2) P. ehimensis NPUST1 for 8 weeks. (a) Glucokinase (GK); (b) hexokinase 1 (HK1), (c) gluco-6-phosphatase (G6Pase); (d) liver type pyruvate kinase (PK-L); (e) growth hormone receptor (GHR) and (f) insulin-like growth factor-I (IGF-1). The data are presented as the mean ± S.D. from six individual samples (n = 6). Bars with different superscripts are significantly different (p < 0.05).
3.3. Fish Supplemented with P. ehimensis NPUST1 Modulate the Expression of Innate Immunity Response Genes
The expression of cytokine genes was used as an indicator to evaluate the immunomodulatory function of P. ehimensis NPUST1 in the fish. The mRNA expressions of IL-1β, TNFα, C3b and lysozyme in the whole body were significantly increased in the fish of the E1 and E2 groups compared to that in the fish of the control group. The expression levels of IL-1β, TNFα, C3b and lysozyme in the fish of the E1 and E2 groups were 3.3-, 2.1-, 4.7- and 5.3-fold and 5.71-, 4.0-, 9.43- and 8.05-fold higher than those in the fish of the control group, respectively. Moreover, the expression levels of IL-1β, TNFα, C3b and lysozyme in the fish of the E2 group were significantly higher than that in the fish of the E1 group (Figure 4a,d,g,h). The expression level of TLR-4 was significantly increased in the fish of the E1 and E2 groups compared to that of the fish in the control group; however, there was no significant difference between the fish in the E1 and E2 groups (Figure 4f). The expression levels of TLR-4 in the fish of the E1 and E2 groups were 3.6- and 3.2-fold higher than those in the fish of the control group, respectively. The expression levels of IL-6, IL-15 and TLR-1 were significantly increased in the fish of the E1 group compared to that in the fish of the control and E2 groups. Moreover, the expression of IL-6, IL-15 and TLR-1 in the fish of the E2 group was also significantly higher than that in the fish of the control group (Figure 4b,c,e). In summary, the results suggested that the fish fed a P. ehimensis NPUST1-supplemented diet were subjected to immunomodulatory actions. Moreover, increased immune cytokine gene expression was commonly exhibited in the fish of the E1 and E2 groups, suggesting that a dose of 106 CFU/g P. ehimensis NPUST1 was sufficient to enhance innate immunity.
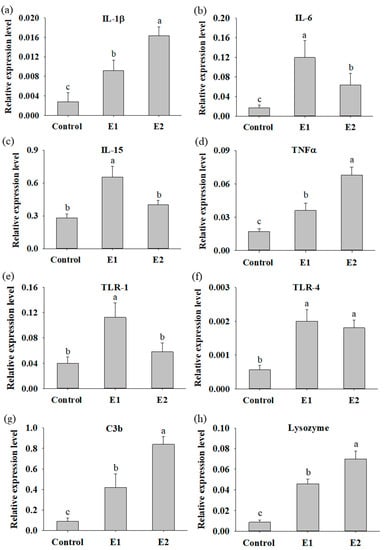
Figure 4.
Quantitative PCR analysis of immune-related expression in zebrafish fed the basal diet (control), the basal diet containing 106 (E1) or the basal diet containing 107 (E2) CFU/g P. ehimensis NPUST1 for 8 weeks. Expression of IL-1β (a), IL-6 (b), IL-15 (c), TNFα (d), TLR-1 (e), TLR-4 (f), C3b (g) and lysozyme (h) in the whole body. The results represent the mean ± S.D. of six fish per group. Bars with different superscripts are significantly different (p < 0.05, one-way ANOVA, Tukey’s multiple comparisons test).
3.4. Dietary Supplementation with P. ehimensis NPUST1 Enhances Disease Resistance against Pathogen Infections
To investigate whether the immunomodulatory function of dietary P. ehimensis NPUST1 could reflect an efficacy in disease resistance, the cumulative mortality of fish was evaluated using a challenge test with A. hydrophila and S. iniae pathogens. As shown in Figure 3, the cumulative mortality was maintained at 0% for the group injected with saline for 7 days post-infection. Conversely, the cumulative mortality in the control group injected with A. hydrophila and S. iniae obviously increased during the first 3 days post-infection and then remained at 78 ± 4.8% and 81 ± 4.8%, respectively, until 7 days post-infection.
Notably, the cumulative mortality was 56 ± 4.8% and 53 ± 5.8% for the fish of the E1 and E2 groups challenged with A. hydrophila, respectively, which was lower than the cumulative mortality for the fish of the control group. Similarly, the cumulative mortality obviously declined in the fish of the E1 and E2 groups after the challenge with S. iniae, which was 61 ± 4.8% and 55 ± 4.8% for the fish of the E1 and E2 groups, respectively. There was no significant difference in cumulative mortality between the E1 and E2 groups (Figure 5). These results suggested that the fish fed a P. ehimensis NPUST1-supplemented diet exhibited disease resistance against A. hydrophila and S. iniae infection, and a dose of 106 CFU/g P. ehimensis NPUST1 was sufficient to achieve protective defense against disease infection.
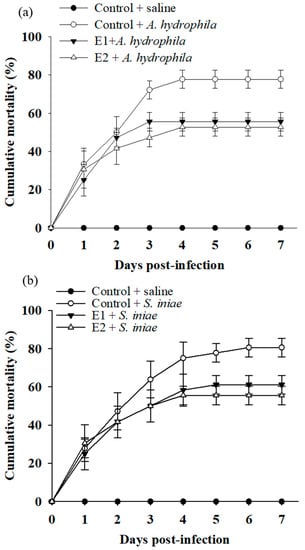
Figure 5.
Cumulative mortality of zebrafish challenged with (a) Aeromonas hydrophila and (b) Streptococcus iniae after being fed the basal diet (control), the basal diet containing 106 (E1) or the basal diet containing 107 (E2) CFU/g P. ehimensis NPUST1 for 8 weeks. Data are presented as the mean ± S.E. from triplicates of each group. The cumulative mortality in the E1 and E2 groups was significantly higher than that in the control group based on the Kaplan–Meier method (p < 0.05).
4. Discussion
Probiotics have been shown to exhibit multiple benefits to fish such as growth promotion, improvement of feed efficiency and gut microbiota, and disease resistance. Thus, probiotics are popular alternatives to antibiotics or chemicals for biocontrol in aquaculture. To date, several studies describing the screening and evaluation of bacteria as potential probiotics for aquaculture have been reported [23,24]; however, the safety assessment of the probiotic candidates is generally limited. Probiotic isolates harboring antibiotic-resistance genes can have negative environmental impacts and potentially cause the spread of multi-drug resistant pathogens by horizontal gene transfer. A recent review of genetic analyses found that multiple probiotics used in the aquaculture industry are antibiotic-resistant bacteria, suggesting that safety assessment for commercial probiotics is needed [25]. The present study showed that the probiotic P. ehimensis NPUST1 was susceptible to the tested antibiotics, suggesting that its use in aquaculture is relatively safe.
In general, the interaction between probiotics and hosts is considered species-specific or strain-specific; thus, in vivo validation of probiotic efficacy is needed. Zebrafish are an excellent animal model for aquaculture research [26]. The present study demonstrated that the probiotic P. ehimensis NPUST1 can produce protease, amylase, lipase, cellulase and xylanase using the agar-well diffusion test. The efficacy of the hydrolytic enzyme-producing (enzymes including protease, amylase, cellulase, xylanase) probiotic P. ehimensis NPUST1 on the nutrition metabolism and healthy status of zebrafish was evaluated. The results showed that intestinal amylase, cellulase, lipase, protease and xylanase were significantly higher in P. ehimensis NPUST1-supplemented fish, potentially suggesting that P. ehimensis NPUST1 could localize to the zebrafish intestine and evoke digestive enzymes. Plant-derived feedstuffs are commonly used as protein sources to partially replace fish meal in aquaculture; however, indigestible non-starch polysaccharides (NPSs) in plant ingredients are considered the main antinutritional factor for feed utilization. Supplementation of hydrolytic enzymes or elevation of digestive enzymes is an efficient approach to solve NPS issues and enhance growth performance and feed efficiency in aquaculture. Reports have shown that dietary supplementation with hydrolytic enzyme-producing probiotics could significantly evoke digestive enzymes or improve growth performance and feed efficiency in Nile tilapia [8,22], zebrafish [12,18], whiteleg shrimp (Litopenaeus vannamei) [27], olive flounder (Paralichthys olivaceus) [28] and golden pompano [29]. It is difficult to evaluate the growth performance and feed efficiency of adult zebrafish because their size almost reaches the maximum limitation (4~5 cm). However, the expression levels of growth- or nutrient metabolism-related genes can be used as indicators of their growth performance and feed efficiency. The liver acts critical roles in growth hormone (GH) regulation of somatic growth and the control of nutrient metabolism. Hepatic IGF-1 secretion and release into the circulatory system, triggered by the GH receptor (GHR) receiving GH ligand, is a major regulatory mechanism that stimulates somatic growth. The increased expression of GHR and IGF-1 in probiotic-supplemented fish indicated that P. ehimensis NPUST1 can stimulate growth performance. GK, HK-1 and PK-L are critical enzymes for regulating the metabolic rate of glycolysis. Hepatic G6Pase is an important enzyme that hydrolyzes glycogen to provide glucose as energy during glycogenolysis. Recently, Nguyen et al. [30] demonstrated that dietary supplementation with probiotics significantly increased growth, nutrient metabolism and glycolysis based on metabolite analysis by mass spectrometry. Significant increases in glycolysis-related gene expression, such as GK, HK-1, PK-L and G6Pase, were observed in Bacillus amyloliquefaciens- and Chromobacterium aquaticum-supplemented fish, which also suggests that dietary supplementation with probiotics can enhance carbohydrate metabolism in fish [12,18]. The present study showed that increased the expression of GK, HK-1, PK-L and G6Pase in zebrafish fed P. ehimensis NPUST1 also supports this conclusion.
The most commonly purported benefit of probiotics is modulating immunity and improving the health status of the host to defend against infections. Few studies have investigated the effect of P. ehimensis NPUST1 on immune modulation. However, some Bacillus species, which are similar probiotics, have shown enhanced immunity against pathogenic infections in many fish. For instance, several reports have demonstrated the beneficial effects of administering probiotics of Bacillus spp. on reducing the mortality of Rhynchocypris lagowskii challenged with A. hydrophila [31], European sea bass (Dicentrarchus labrax) challenged with Vibrio anguillarum [32], Nile tilapia challenged with S. agalcotiae [33], and juvenile olive flounder (Paralichthys olivaceus) challenged with Edwardsiella tarda [34]. In addition, a recent report demonstrated that dietary supplementation with P. ehimensis NPUST1 increased the cumulative survival of Nile tilapia challenged with A. hydropjila and S. iniae [17]. In this study, the cumulative mortality of zebrafish challenged with A. hydrophila and S. iniae was lower for fish in the E1 and E2 groups than in the control group, suggesting that the probiotic P. ehimensis NPUST1 can enhance innate immunity against pathogenic infections. Cytokines, immunomodulating molecules released from immune cells such as macrophages and lymphocytes, mediate local or systemic immune responses in the host to infection and immunological pressure. Probiotics have been shown to augment fish innate immunity by releasing cytokines from immune cells to activate immune responses against infection. The present study showed a significant increase in the gene expression levels of IL-1β, IL-6, TNF-a, TLR-4, C3b and lysozyme in P. ehimensis NPUST1-supplemented fish compared to those of the fish in the control group. Moreover, IL-1β, TNF-α, C3b and lysozyme levels exhibited significant differences between the fish of the E1 and E2 groups. The results of the present study are consistent with those of other studies, which showed that cytokine genes are significantly increased by administration of the probiotic Bacillus spp. in olive flounder (Paralichthys olivaceus) [28], Crucian carp (Carassius auratus) [35], Nile tilapia (O. niloticus) [36] and Labeo rohita [37]. Moreover, a recent study showed that dietary supplementation with P. ehimensis NPUST1 can enhance immunity and increase the expression of IL-1β and TNF-α in the kidney and spleen of Nile tilapia [17]. Serum lysozyme is a cornerstone of host immunity that kills pathogens by splitting the peptidoglycan layers of bacterial cells. Moreover, the degradation and lysis of peptidoglycan by lysozyme action have been shown to activate the complement system and pattern recognition receptors (PPRs) in the host against infections [38]. TLR-1 and TLR-4 are typical PPRs that recognize lipoprotein and lipopolysaccharides, which are components present in the outer membranes of gram-positive and gram-negative bacteria, respectively. Although the expression of TLRs as indicators has been investigated in a wide variety of immunostimulant treatments, there are still few studies on the effects of probiotics on TLR expression in fish. The present study showed a significant increase in TLR-1 and TLR-4 expression in zebrafish in the P. ehimensis NPUST1-supplemented groups, suggesting that dietary supplementation with P. ehimensis NPUST1 can enhance immunity against gram-negative and gram-positive pathogen infections.
5. Conclusions
In conclusion, in the present study, a potential probiotic P. ehimensis NPUST1 with extracellular digestive enzyme activity (protease, amylase, cellulase, and xylanase) was isolated. Zebrafish fed a P. ehimensis NPUST1-supplemented diet exhibited increased expression of indicator genes associated with nutrient metabolism, growth performance and the innate immune response. In addition, the P. ehimensis NPUST1-supplemented diet enhanced resistance to infection with A. hydrophila and S. iniae. The present study showed several beneficial effects of P. ehimensis NPUST1, suggesting that P. ehimensis NPUST1 could be developed as a probiotic for use in aquaculture.
Author Contributions
Conceptualization, S.-Y.H. and Z.-H.W.; Data curation, P.-H.L.; Methodology, P.-H.L. and S.-W.C.; Supervision, S.-Y.H. and Z.-H.W.; Writing-original draft, P.-H.L.; Writing-review and editing, S.-Y.H. and Z.-H.W. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by a grant from the Ministry of Science and Technology (MOST) 106-2313-B-020-007-, Taiwan.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Animal Care and Use Committee (IACUC) of National Pingtung University of Science and Technology (NPUST), Taiwan (approval number: NPUST-105-067).
Data Availability Statement
The data presented in this study are available in the article.
Acknowledgments
This study was also financially supported by the Research Center for Animal Biologics, from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education and the National Science and Technology Council (MOST 111-2634-F-020-001-), Taiwan, R.O.C.
Conflicts of Interest
The authors declare no conflict of interest.
References
- FAO. The State of Fisheries and Aquaculture 2020. Sustainability in Action; FAO: Rome, Italy, 2020. [Google Scholar]
- Ksepka, S.P.; Bullard, S.A. Morphology, phylogenetics and pathology of “red sore disease” (coinfection by Epistylis cf. wuhanensis and Aeromonas hydrophila) on sportfishes from reservoirs in the South-Eastern United States. J. Fish Dis. 2021, 44, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Ortega, C.; Garcia, I.; Irgang, R.; Fajardo, R.; Tapia-Cammas, D.; Acosta, J.; Avendano-Herrera, R. First identification and characterization of Streptococcus iniae obtained from tilapia (Oreochromis aureus) farmed in Mexico. J. Fish Dis. 2018, 41, 773–782. [Google Scholar] [CrossRef] [PubMed]
- De Souza Valente, C.; Wan, A.H.L. Vibrio and major commercially important vibriosis diseases in decapod crustaceans. J. Invertebr. Pathol. 2021, 181, 107527. [Google Scholar] [CrossRef] [PubMed]
- Yousuf, S.; Tyagi, A.; Singh, R. Probiotic supplementation as an emerging alternative to chemical therapeutics in finfish aquaculture: A review. Probiotics Antimicrob. Proteins 2022, 1–18. [Google Scholar] [CrossRef]
- Ringo, E.; Van Doan, H.; Lee, S.H.; Soltani, M.; Hoseinifar, S.H.; Harikrishnan, R.; Song, S.K. Probiotics, lactic acid bacteria and bacilli: Interesting supplementation for aquaculture. J. Appl. Microbiol. 2020, 129, 116–136. [Google Scholar] [CrossRef]
- Hai, N.V. The use of probiotics in aquaculture. J. Appl. Microbiol. 2015, 119, 917–935. [Google Scholar] [CrossRef]
- Wu, P.S.; Liu, C.H.; Hu, S.Y. Probiotic Bacillus safensis NPUST1 administration improves growth performance, gut microbiota, and innate immunity against Streptococcus iniae in Nile tilapia (Oreochromis niloticus). Microorganisms 2021, 9, 2494. [Google Scholar] [CrossRef]
- Saputra, F.; Shiu, Y.L.; Chen, Y.C.; Puspitasari, A.W.; Danata, R.H.; Liu, C.H.; Hu, S.Y. Dietary supplementation with xylanase-expressing B. amyloliquefaciens R8 improves growth performance and enhances immunity against Aeromonas hydrophila in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2016, 58, 397–405. [Google Scholar] [CrossRef]
- Wang, Y.; Al Farraj, D.A.; Vijayaraghavan, P.; Hatamleh, A.A.; Biji, G.D.; Rady, A.M. Host associated mixed probiotic bacteria induced digestive enzymes in the gut of tiger shrimp Penaeus monodon. Saudi. J. Biol. Sci. 2020, 27, 2479–2484. [Google Scholar] [CrossRef]
- Sumathi, C.; Dillibabu, V.; Madhuri, D.K.; Priya, D.M.; Nagalakshmi, C.; Sekaran, G. Dietary inclusion of protease producing novel Pontibacter spp. and Bacillus megaterium as a probiotic enhances immune responses in Labeo rohita. Pak. J. Biol. Sci. 2014, 17, 451–461. [Google Scholar] [CrossRef][Green Version]
- Lin, Y.S.; Saputra, F.; Chen, Y.C.; Hu, S.Y. Dietary administration of Bacillus amyloliquefaciens R8 reduces hepatic oxidative stress and enhances nutrient metabolism and immunity against Aeromonas hydrophila and Streptococcus agalactiae in zebrafish (Danio rerio). Fish Shellfish Immunol. 2019, 86, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Ash, C.; Priest, F.G.; Collins, M.D. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek 1993, 64, 253–260. [Google Scholar] [CrossRef]
- Olishevska, S.; Nickzad, A.; Deziel, E. Bacillus and Paenibacillus secreted polyketides and peptides involved in controlling human and plant pathogens. Appl. Microbiol. Biotechnol. 2019, 103, 1189–1215. [Google Scholar] [CrossRef] [PubMed]
- Midhun, S.J.; Neethu, S.; Vysakh, A.; Arun, D.; Radhakrishnan, E.K.; Jyothis, M. Antibacterial activity and probiotic characterization of autochthonous Paenibacillus polymyxa isolated from Anabas testudineus (Bloch, 1792). Microb. Pathog. 2017, 113, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Aktuganov, G.; Melentjev, A.; Galimzianova, N.; Khalikova, E.; Korpela, T.; Susi, P. Wide-range antifungal antagonism of Paenibacillus ehimensis IB-X-b and its dependence on chitinase and beta-1,3-glucanase production. Can. J. Microbiol. 2008, 54, 577–587. [Google Scholar] [CrossRef]
- Chen, S.W.; Liu, C.H.; Hu, S.Y. Dietary administration of probiotic Paenibacillus ehimensis NPUST1 with bacteriocin-like activity improves growth performance and immunity against Aeromonas hydrophila and Streptococcus iniae in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2019, 84, 695–703. [Google Scholar] [CrossRef]
- Yi, C.C.; Liu, C.H.; Chuang, K.P.; Chang, Y.T.; Hu, S.Y. A potential probiotic Chromobacterium aquaticum with bacteriocin-like activity enhances the expression of indicator genes associated with nutrient metabolism, growth performance and innate immunity against pathogen infections in zebrafish (Danio rerio). Fish Shellfish Immunol. 2019, 93, 124–134. [Google Scholar] [CrossRef]
- Gong, H.Y.; Wu, S.H.; Chen, C.Y.; Huang, C.W.; Lu, J.K.; Chou, H.Y. Complete genome sequence of Streptococcus iniae 89353, a virulent strain isolated from diseased tilapia in Taiwan. Genome Announc. 2017, 5. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analyses, 16th ed.; Association of Official Analytical Chemists: Washington, DC, USA, 1997. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Tan, H.Y.; Chen, S.W.; Hu, S.Y. Improvements in the growth performance, immunity, disease resistance, and gut microbiota by the probiotic Rummeliibacillus stabekisii in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2019, 92, 265–275. [Google Scholar] [CrossRef]
- Govindaraj, K.; Samayanpaulraj, V.; Narayanadoss, V.; Uthandakalaipandian, R. Isolation of lactic acid bacteria from intestine of freshwater fishes and elucidation of probiotic potential for aquaculture application. Probiotics Antimicrob. Proteins 2021, 13, 1598–1610. [Google Scholar] [CrossRef] [PubMed]
- Monzon-Atienza, L.; Bravo, J.; Torrecillas, S.; Montero, D.; Canales, A.F.G.; de la Banda, I.G.; Galindo-Villegas, J.; Ramos-Vivas, J.; Acosta, F. Isolation and characterization of a Bacillus velezensis D-18 Strain, as a potential probiotic in European seabass aquaculture. Probiotics Antimicrob. Proteins 2021, 13, 1404–1412. [Google Scholar] [CrossRef] [PubMed]
- Anokyewaa, M.A.; Amoah, K.; Li, Y.; Lu, Y.S.; Kuebutornye, F.K.A.; Asiedu, B.; Seidu, I. Prevalence of virulence genes and antibiotic susceptibility of Bacillus used in commercial aquaculture probiotics in China. Aquac. Rep. 2021, 21, 100784. [Google Scholar] [CrossRef]
- Dahm, R.; Geisler, R. Learning from small fry: The zebrafish as a genetic model organism for aquaculture fish species. Mar. Biotechnol. 2006, 8, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Won, S.; Hamidoghli, A.; Choi, W.; Bae, J.; Jang, W.J.; Lee, S.; Bai, S.C. Evaluation of potential probiotics Bacillus subtilis WB60, Pediococcus pentosaceus, and lactococcus lactis on growth performance, immune response, gut histology and immune-related genes in whiteleg shrimp, Litopenaeus vannamei. Microorganisms 2020, 8, 281. [Google Scholar] [CrossRef]
- Jang, W.J.; Lee, J.M.; Hasan, M.T.; Lee, B.J.; Lim, S.G.; Kong, I.S. Effects of probiotic supplementation of a plant-based protein diet on intestinal microbial diversity, digestive enzyme activity, intestinal structure, and immunity in olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2019, 92, 719–727. [Google Scholar] [CrossRef]
- Liu, S.B.; Wang, S.F.; Cai, Y.; Li, E.C.; Ren, Z.L.; Wu, Y.; Guo, W.L.; Sun, Y.; Zhou, Y.C. Beneficial effects of a host gut-derived probiotic, Bacillus pumilus, on the growth, non-specific immune response and disease resistance of juvenile golden pompano, Trachinotus ovatus. Aquaculture 2020, 514, 734446. [Google Scholar] [CrossRef]
- Nguyen, T.L.; Chun, W.K.; Kim, A.; Kim, N.; Roh, H.J.; Lee, Y.; Yi, M.; Kim, S.; Park, C.I.; Kim, D.H. Dietary probiotic effect of Lactococcus lactis WFLU12 on low-molecular-weight metabolites and growth of olive flounder (Paralichythys olivaceus). Front. Microbiol. 2018, 9, 2059. [Google Scholar] [CrossRef]
- Yu, M.; Zhang, Y.; Zhang, D.; Wang, Q.; Wang, G.; Elsadek, M.; Yao, Q.; Chen, Y.; Guo, Z. The effect of adding Bacillus amyloliquefaciens LSG2-8 in diets on the growth, immune function, antioxidant capacity, and disease resistance of Rhynchocypris lagowskii. Fish Shellfish Immunol. 2022, 125, 258–265. [Google Scholar] [CrossRef]
- Monzon-Atienza, L.; Bravo, J.; Fernandez-Montero, A.; Charlie-Silva, I.; Montero, D.; Ramos-Vivas, J.; Galindo-Villegas, J.; Acosta, F. Dietary supplementation of Bacillus velezensis improves Vibrio anguillarum clearance in European sea bass by activating essential innate immune mechanisms. Fish Shellfish Immunol. 2022, 124, 244–253. [Google Scholar] [CrossRef]
- Van Doan, H.; Wangkahart, E.; Thaimuangphol, W.; Panase, P.; Sutthi, N. Effects of Bacillus spp. Mixture on growth, immune responses, expression of immune-related genes, and resistance of nile tilapia against Streptococcus agalactiae infection. Probiotics Antimicrob. Proteins 2021, 1–16. [Google Scholar] [CrossRef]
- Choi, W.; Moniruzzaman, M.; Bae, J.; Hamidoghli, A.; Lee, S.; Choi, Y.H.; Min, T.; Bai, S.C. Evaluation of dietary probiotic bacteria and processed yeast (Gropro-aqua) as the alternative of antibiotics in juvenile olive flounder Paralichthys olivaceus. Antibiotics 2022, 11, 129. [Google Scholar] [CrossRef]
- Zhang, D.X.; Kang, Y.H.; Zhan, S.; Zhao, Z.L.; Jin, S.N.; Chen, C.; Zhang, L.; Shen, J.Y.; Wang, C.F.; Wang, G.Q.; et al. Effect of Bacillus velezensis on Aeromonas veronii-induced intestinal mucosal barrier function damage and inflammation in crucian carp (Carassius auratus). Front. Microbiol. 2019, 10, 2663. [Google Scholar] [CrossRef]
- Selim, K.M.; Reda, R.M. Improvement of immunity and disease resistance in the Nile tilapia, Oreochromis niloticus, by dietary supplementation with Bacillus amyloliquefaciens. Fish Shellfish Immunol. 2015, 44, 496–503. [Google Scholar] [CrossRef]
- Giri, S.S.; Sen, S.S.; Chi, C.; Kim, H.J.; Yun, S.; Park, S.C.; Sukumaran, V. Effect of cellular products of potential probiotic bacteria on the immune response of Labeo rohita and susceptibility to Aeromonas hydrophila infection. Fish Shellfish Immunol. 2015, 46, 716–722. [Google Scholar] [CrossRef]
- Ragland, S.A.; Criss, A.K. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 2017, 13, e1006512. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).