A Method for Estimating the Injection Position of Turbot (Scophthalmus maximus) Using Semantic Segmentation
Abstract
1. Introduction
- In order to accurately recognize the fish body, pectoral fin, and caudal fin of turbot, the classic Deeplabv3+ network was improved by using attention modules. Moreover, the proposed Atten-Deeplabv3+ was successfully executed to calculate the BL and BW;
- Using semantic segmentation, a method for estimating the injection position of the turbot was proposed. The experiments compared the errors of the injection position to prove the efficacy of the proposed approach, which would benefit the development of turbot vaccination machines.
2. Materials and Methods
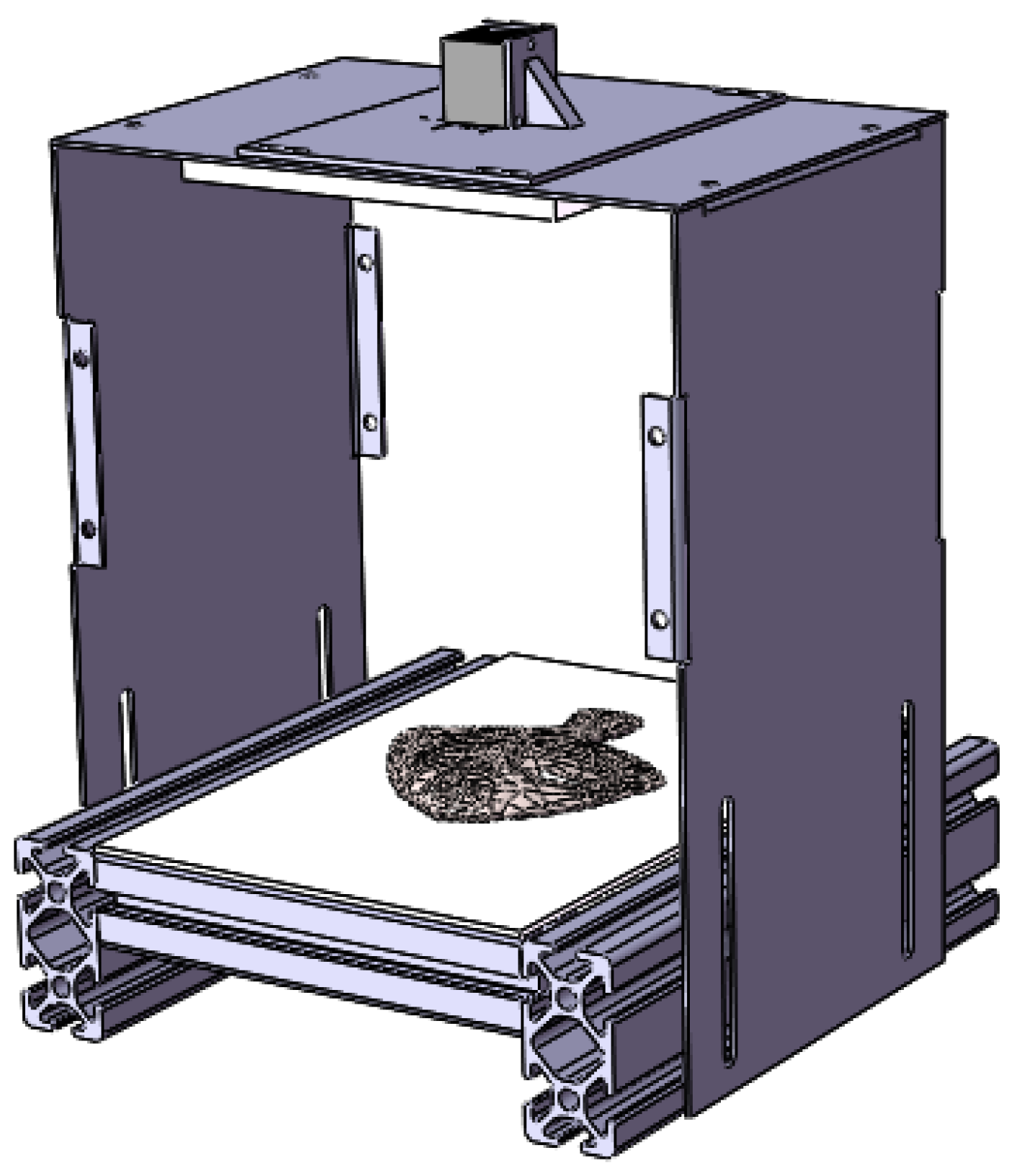
2.1. Image Acquisition and Datasets
2.2. Semantic Segmentation Model Architecture
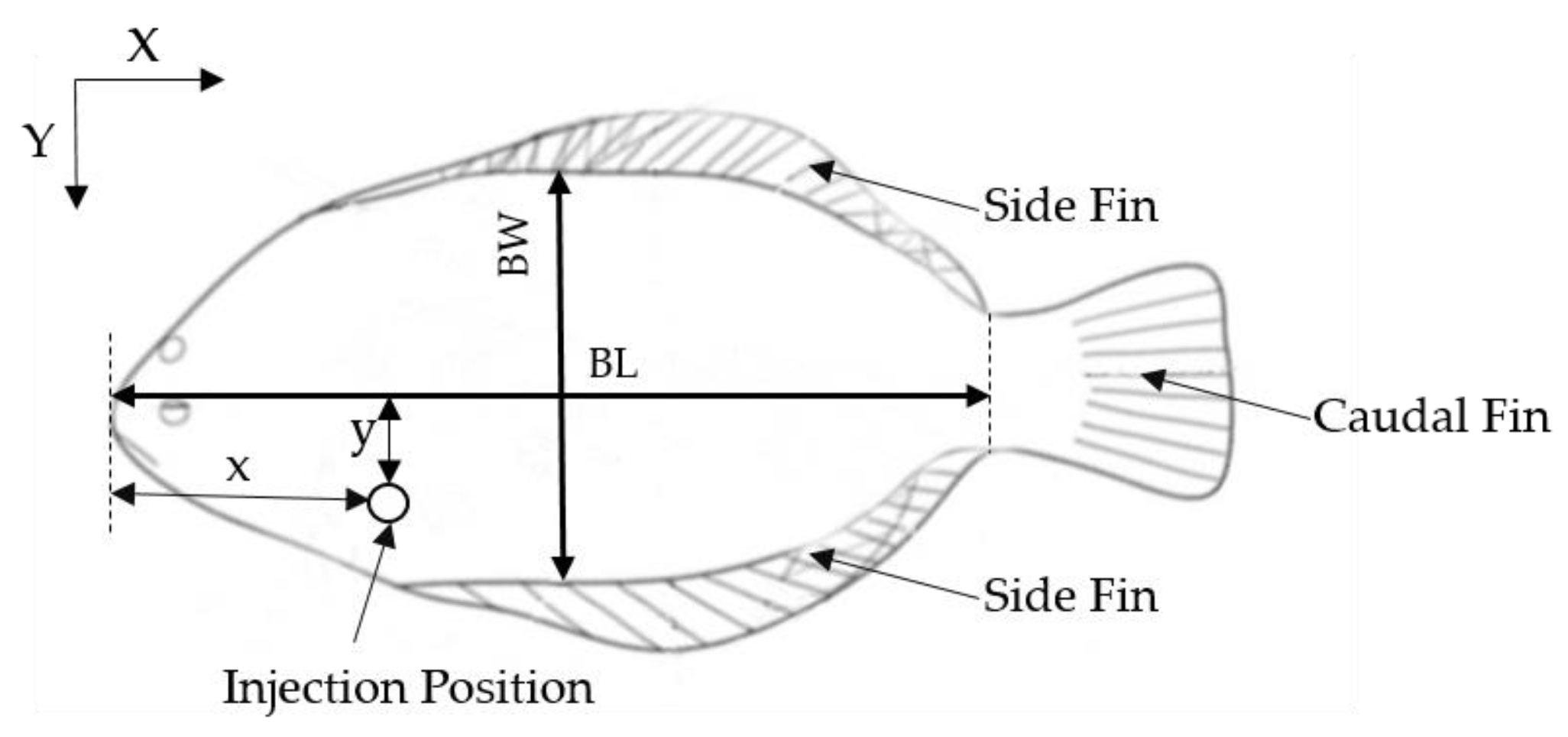
2.3. BL and BW Estimation Algorithm
- Calculate the center of gravity of the fish body and caudal fin
- Calculate the distance of gravity center of the caudal fin and the point of the fish body contour, and determine the point with maximum distance to the tip of the fish mouth as the coordinate origin
- The line connecting the center of gravity of the fish body and the center of gravity of the caudal fin is used as the x-axis, with the positive direction of the x-axis from the center of gravity of the fish body to the center of gravity of the caudal fin. Rotate the x-axis 90 degrees around the origin to obtain the y-axis. The slope of the x-axis (kx) and y-axis (ky) can be calculated by Equation (3) below
- Traverse the contours of the fish body above and below the x-axis, respectively, and determine the longest distance between the point and the x-axis. The body width of the turbot can be found by adding the two results. Then, traverse the contours of the caudal fin to find the nearest point to the coordinate origin, and the distance between the nearest point and the coordinate origin is the body length.
2.4. The Injection Position Estimation Model
2.5. Experimental Setup
2.6. Performance Evaluation
3. Results and Discussion
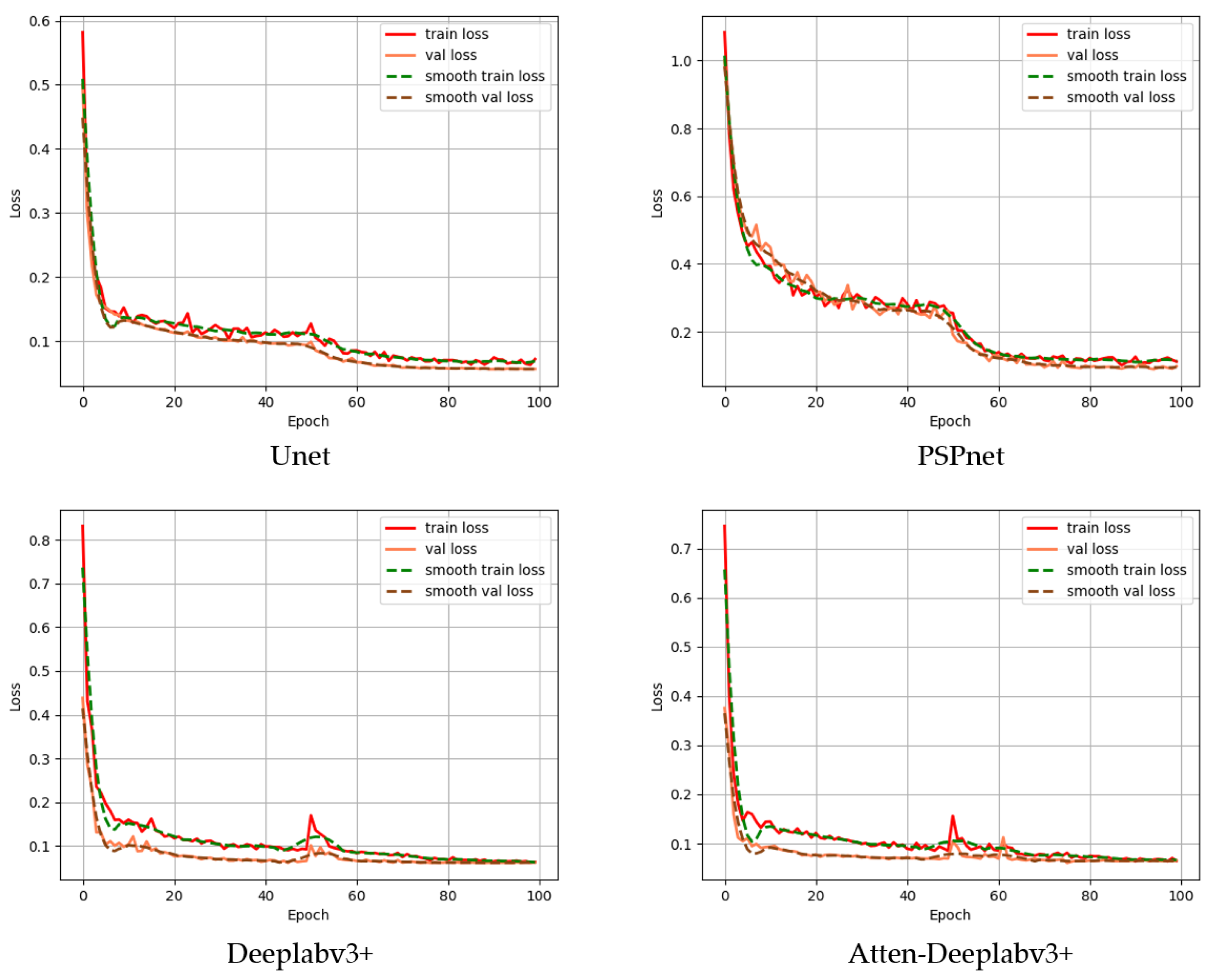
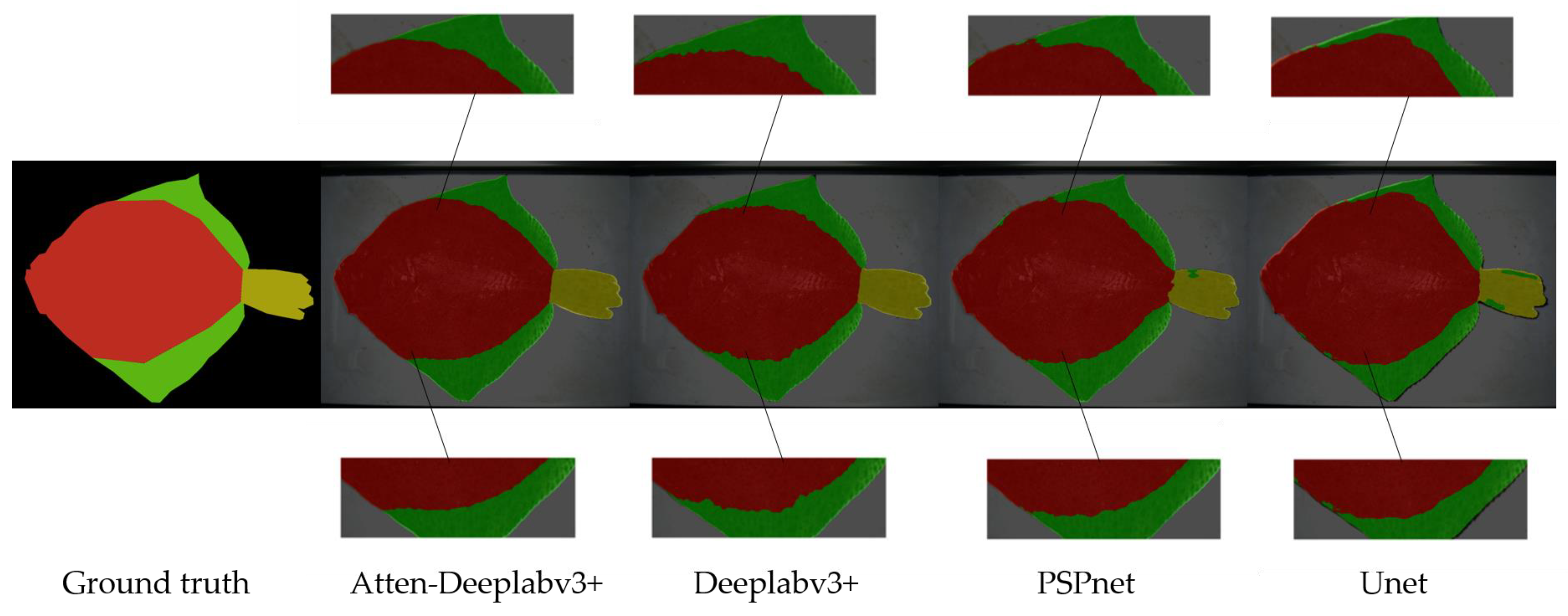
3.1. Semantic Segmentation
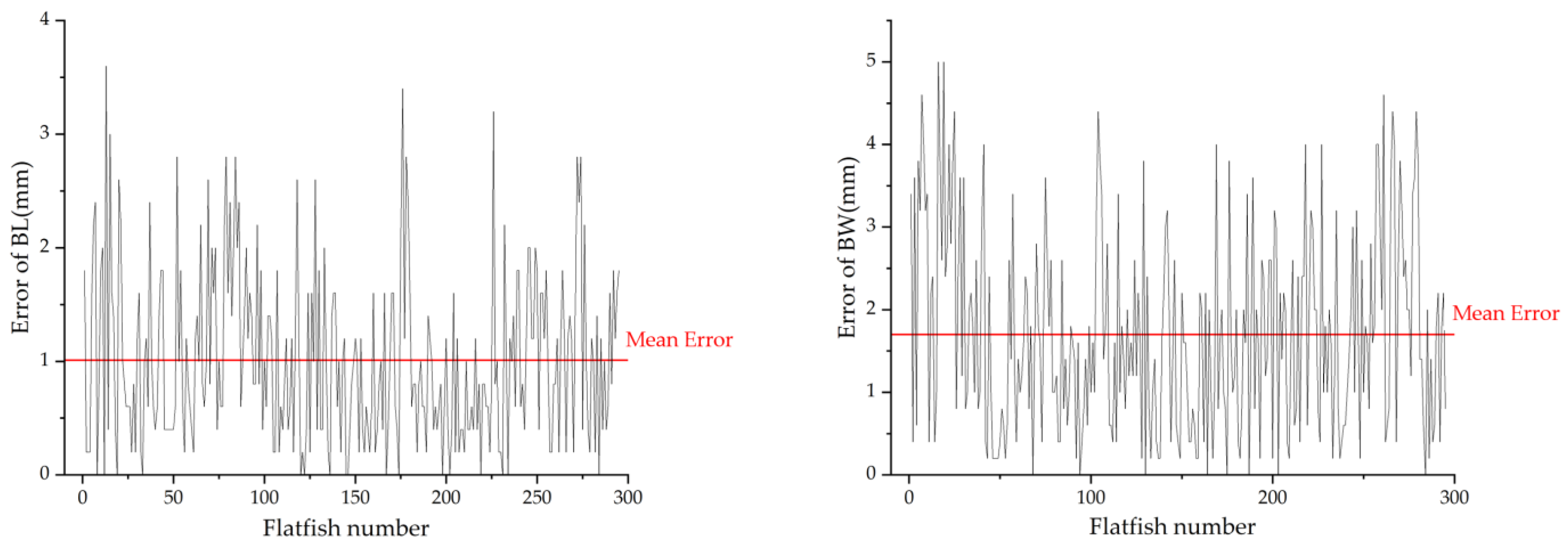
3.2. The Performance of BL and BW Estimation Algorithm
3.3. The Performance of the Injection Position Estimation Model
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sarvenaz, K.T.; Sabine, S. Nutritional Value of Fish: Lipids, Proteins, Vitamins, and Minerals. Rev. Fish. Sci. Aquac. 2018, 26, 243–253. [Google Scholar] [CrossRef]
- Ninawe, A.S.; Dhanze, J.R.; Dhanze, R.; Indulkar, S.T. Fish Nutrition and Its Relevance ro Human Health; CRC Press: London, UK, 2020; pp. 10–26. [Google Scholar]
- FAO. Fishery and Aquaculture Statistics; Global Aquaculture Production 1950–2020 (FishstatJ); FAO Fisheries and Aquaculture Department: Rome, Italy, 2022; Available online: http://www.fao.org/fishery/statistics/software/fish-stati/enStatistics (accessed on 23 October 2022).
- Statistics Bureau of the People’s Republic of China. China Fishery Statistical Yearbook; China Statistics Press: Beijing, China, 2021.
- Kalantzi, I.; Rico, A.; Mylona, K.; Pergantis, S.A.; Tsapakis, M. Fish farming, metals and antibiotics in the eastern Mediterranean Sea: Is there a threat to sediment wildlife? Sci. Total Environ. 2021, 764, 142843. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Bruce, T.J.; Jones, E.M.; Cain, K.D. A Review of Fish Vaccine Development Strategies: Conventional Methods and Modern Biotechnological Approaches. Microorganisms 2019, 7, 569. [Google Scholar] [CrossRef] [PubMed]
- Plant, K.P.; LaPatra, S.E. Advances in fish vaccine delivery. Dev. Comp. Immunol. 2011, 35, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Santos, Y.; Garcia-Marquez, S.; Pereira, P.G.; Pazos, F.; Riaza, A.; Silva, R.; El Morabit, A.; Ubeira, F. Efficacy of furunculosis vaccines in turbot, Scophthalmus maximus (L.): Evaluation of immersion, oral and injection delivery. J. Fish Dis. 2005, 28, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Ellis, A. Health and disease in Atlantic salmon farming. In Handbook of Salmon Farming; Stead, S.M., Laird, L.M., Eds.; Springer Praxis: London, UK, 2002; Volume 10, pp. 373–401. [Google Scholar]
- Zhu, Y.; Xu, H.; Jiang, T.; Hong, Y.; Zhang, X.; Xu, H.; Xing, J. Research on the automatic injection technology of Ctenopharynuodon idellus vaccine. Fish. Mod. 2020, 47, 12–19. [Google Scholar] [CrossRef]
- Brudeseth, B.E.; Wiulsrød, R.; Fredriksen, B.N.; Lindmo, K.; Løkling, K.-E.; Bordevik, M.; Steine, N.; Klevan, A.; Gravningen, K. Status and future perspectives of vaccines for industrialised fin-fish farming. Fish Shellfish Immunol. 2013, 35, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Leira, H.L.; Baalsrud, K.J. Operator safety during injection vaccination of fish. Dev. Biol. Stand. 1997, 90, 383–387. [Google Scholar] [PubMed]
- Understanding-Fish-Vaccination. Available online: https://thefishsite.com/articles/understanding-fish-vaccination (accessed on 23 October 2022).
- Vaccination-Aquaculture-Nettsteder-Skala Maskon. Available online: https://en.skalamaskon.no/aquaculture2/vaccination (accessed on 23 October 2022).
- Products-Lumic AS & Strømmeservice AS. Available online: https://lumic.no/en/products/ (accessed on 23 October 2022).
- The Fish Site. Evaluation of Fish Vaccination Machines in Norway. Available online: https://thefishsite.com/articles/evaluation-of-fish-vaccination-machines-in-norway (accessed on 23 October 2022).
- Lee, D.-G.; Yang, Y.-S.; Kang, J.-G. Conveyor belt automatic vaccine injection system (AVIS) for flatfish based on computer vision analysis of fish shape. Aquac. Eng. 2013, 57, 54–62. [Google Scholar] [CrossRef]
- Khankeshizadeh, E.; Mohammadzadeh, A.; Moghimi, A.; Mohsenifar, A. FCD-R2U-net: Forest change detection in bi-temporal satellite images using the recurrent residual-based U-net. Earth Sci. Informatics 2022, 15, 2335–2347. [Google Scholar] [CrossRef]
- Fernandes, A.F.; Turra, E.; de Alvarenga, É.R.; Passafaro, T.L.; Lopes, F.B.; Alves, G.F.; Singh, V.; Rosa, G.J. Deep Learning image segmentation for extraction of fish body measurements. Comput. Electron. Agric. 2020, 170, 105274. [Google Scholar] [CrossRef]
- Liu, B.; Wang, K.; Li, X.; Hu, C. Motion posture parsing of Chiloscyllium plagiosum fish body based on semantic part segmentation. Trans. CSAE 2021, 37, 179–187. [Google Scholar] [CrossRef]
- Li, Y.; Huang, K.; Xiang, J. Measurement of dynamic fish dimension based on stereoscopic vision. Trans. CSAE 2020, 36, 220–226. [Google Scholar] [CrossRef]
- Russell, B.C.; Torralba, A.; Murphy, K.P.; Freeman, W.T. LabelMe: A Database and Web-Based Tool for Image Annotation. Int. J. Comput. Vis. 2008, 77, 157–173. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Chen, L.C.; Papandreou, G.; Schroff, S.; Adam, H. Rethinking atrous convolution for semantic image segmentation. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018. [Google Scholar]
- Ba, J.; Mnih, V.; Kavukcuoglu, K. Multiple object recognition with visual attention. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.L.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted residuals and linear bottlenecks. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Welcome to Python.org. Available online: https://www.python.org/ (accessed on 23 October 2022).
- Home—OpenCV. Available online: http://www.opencv.org/ (accessed on 23 October 2022).
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- The Pascal Visual Object Classes Challenge 2012 (VOC2012) Results. Available online: http://www.pascal-network.org/challenges/VOC/voc2012/workshop/index.html (accessed on 23 October 2022).
- Huang, T.-W.; Hwang, J.-N.; Rose, C.S. Chute based automated fish length measurement and water drop detection. In Proceedings of the International Conference on Acoustics, Speech and Signal Processing (ICASSP), Shanghai, China, 20–25 March 2016. [Google Scholar]
- Lee, C.; Li, J.; Zhu, S. Automated size measurement and weight estimation of body-curved grass carp based on computer vision. In Proceedings of the American Society of Agricultural and Biological Engineers Annual International Meeting, Virtual, 12–16 July 2021. [Google Scholar]
- Lee, D.-G.; Cha, B.-J.; Park, S.-W.; Kwon, M.-G.; Xu, G.-C.; Kim, H.-J. Development of a vision-based automatic vaccine injection system for flatfish. Aquac. Eng. 2013, 54, 78–84. [Google Scholar] [CrossRef]


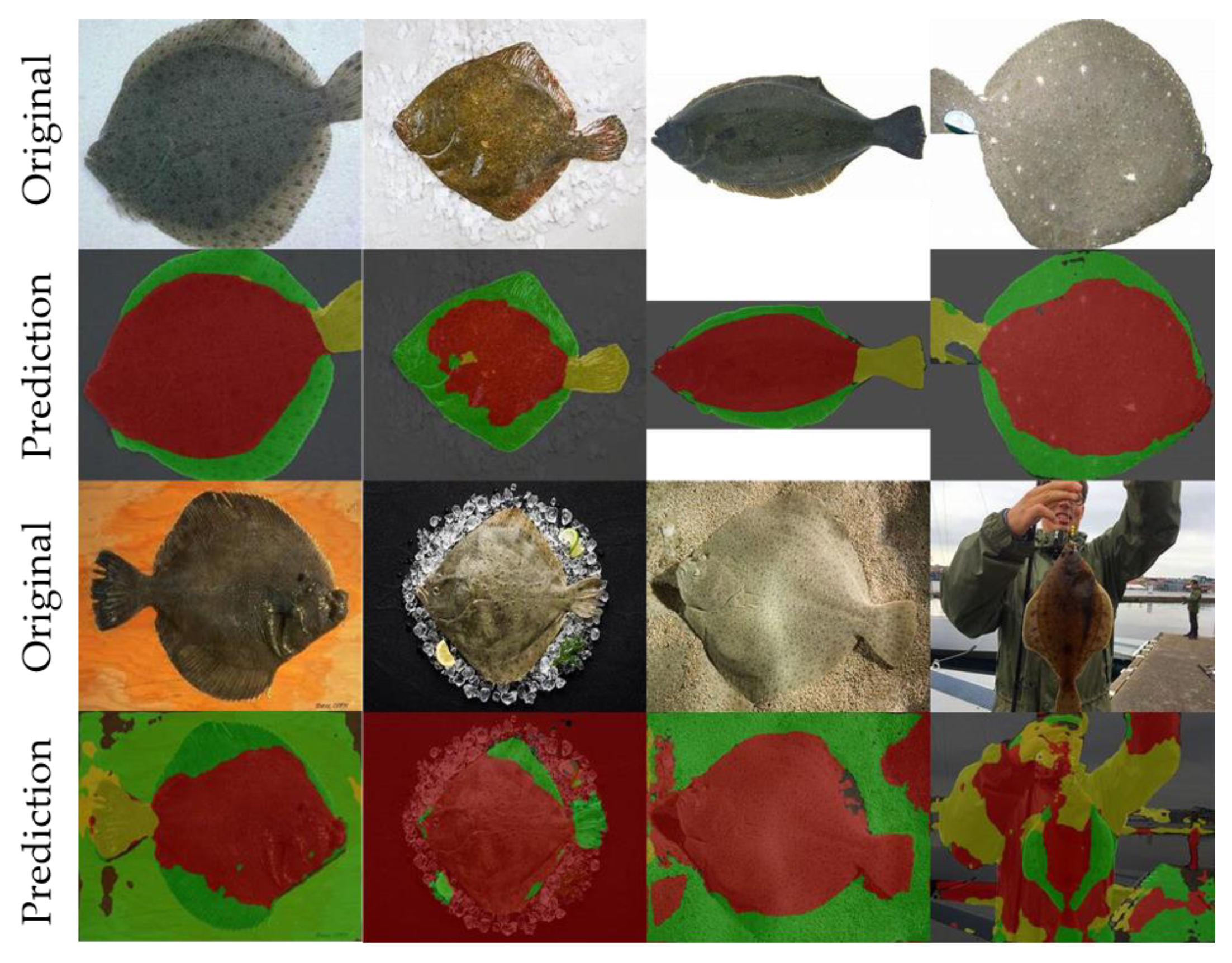

| Networks | IoU | |||
|---|---|---|---|---|
| Background | Fish Body | Pectoral Fin | Caudal Fin | |
| Unet | 99.2% | 94.4% | 81.1% | 90.3% |
| PSPnet | 99.2% | 94.2% | 82.0% | 89.9% |
| Deeplabv3+ | 99.3% | 95.5% | 82.9% | 90.5% |
| Atten-Deeplabv3+ | 99.3% | 96.5% | 85.8% | 91.7% |
| Networks | PA | Training Time | |||
|---|---|---|---|---|---|
| Background | Fish Body | Pectoral Fin | Caudal Fin | ||
| Unet | 99.6% | 97.3% | 89.7% | 94.4% | 1 h 50 min |
| PSPnet | 99.6% | 97.1% | 90.1% | 94.3% | 2 h |
| Deeplabv3+ | 99.6% | 97.7% | 90.7% | 95.3% | 2 h 16 min |
| Atten-Deeplabv3+ | 99.7% | 98.2% | 92.8% | 96.2% | 2 h 27 min |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, W.; Li, C.; Wu, K.; Zhu, S.; Ye, Z.; Li, J. A Method for Estimating the Injection Position of Turbot (Scophthalmus maximus) Using Semantic Segmentation. Fishes 2022, 7, 385. https://doi.org/10.3390/fishes7060385
Luo W, Li C, Wu K, Zhu S, Ye Z, Li J. A Method for Estimating the Injection Position of Turbot (Scophthalmus maximus) Using Semantic Segmentation. Fishes. 2022; 7(6):385. https://doi.org/10.3390/fishes7060385
Chicago/Turabian StyleLuo, Wei, Chen Li, Kang Wu, Songming Zhu, Zhangying Ye, and Jianping Li. 2022. "A Method for Estimating the Injection Position of Turbot (Scophthalmus maximus) Using Semantic Segmentation" Fishes 7, no. 6: 385. https://doi.org/10.3390/fishes7060385
APA StyleLuo, W., Li, C., Wu, K., Zhu, S., Ye, Z., & Li, J. (2022). A Method for Estimating the Injection Position of Turbot (Scophthalmus maximus) Using Semantic Segmentation. Fishes, 7(6), 385. https://doi.org/10.3390/fishes7060385







