Abstract
This study developed a dual-antibody sandwich ELISA detection method for Photobacterium damselae, an important pathogen in aquaculture, based on two outer membrane proteins of outer membrane protein C (OmpC) and β-barrel assembly machinery A (BamA) from the strain of P. damselae XP11. By optimizing the reaction concentrations of the capture antibody of rabbit anti-OmpC or anti-BamA and the HRP-labeled detection antibody of rabbit anti-BamA, it was found that using 1.0 μg/mL of rabbit anti-OmpC or 0.9 μg/mL of rabbit anti-BamA as capture antibodies, and 0.90 μg/mL of HRP-labeled rabbit anti-BamA as the detection antibody, could specifically detect different isolates of P. damselae. The detection limit of this method for the supernatant protein of P. damselae disrupted by ultrasound was 0.2 μg/mL. Repeatability tests showed that the coefficient of variation for detecting 25 strains of bacteria was below 9.1%. Compared with the OmpC-BamA sandwich ELISA detection method, the BamA–BamA combination exhibited better specificity. The results of this study provide an important reference for the rapid detection of P. damselae and other bacterial pathogens in aquaculture.
Key Contribution:
For the first time, a sandwich ELISA for P. damselae detection using two anti-Omps was developed. Both anti-OmpC and anti-BamA antibodies functioned effectively as capture antibodies, but anti-BamA provided superior detection specificity.
1. Introduction
Photobacterium damselae is an important bacterial pathogen in the aquaculture industry [], capable of infecting various economically important fish and crustaceans, causing highly lethal diseases and resulting in significant economic losses to aquaculture []. Originally classified within the genera Vibrio or Listonella, P. damselae, a Gram-negative bacterium, was later reclassified into the genus Photobacterium [,,]. The bacterium, which appears rod-shaped or coccobacillary, lacks distinct host specificity [], and its virulence is closely related to its strong hemolytic activity [] and potent protease activity []. P. damselae can cause severe infections, including soft tissue infections [] (such as cellulitis and necrotizing fasciitis) and bacteremia [], with a high mortality rate post-infection, making it one of the most dangerous pathogens in marine aquaculture [,]. In recent years, its zoonotic potential has been increasingly revealed, posing a potential threat to human health and further highlighting the urgent need for rapid and accurate detection methods []. Traditional detection of P. damselae involves bacterial culture and molecular techniques [,,] and a recently published colloidal gold detection method for P. damselae showing promising application prospects []. However, there have been no reports on the development of a sandwich detection method for P. damselae using two antibodies. This study innovatively targets two outer membrane proteins (Omps) of OmpC and BamA from a strain of P. damselae XP11 and, based on the expression of these 2 Omps and the preparation of rabbit antisera, aims to establish a highly sensitive and specific sandwich ELISA detection method for P. damselae.
2. Materials and Methods
2.1. Bacterial Strains
A total of 25 bacterial strains were employed in this study. Specifically, 7 strains of P. damselae were utilized, including strain XP11, which was used for the expression of outer membrane proteins in this study. Additionally, there were 4 commercially sourced strains (XP12, XP13, XP14, and XP15) and 2 strains isolated in this laboratory (XP16 and XP17). Two strains of P. phosphoreum, one strain of P. leiognathi, and one strain of genus Photobacterium (P1, P2, P3, and P4) were purchased from the Shanghai Microbial Culture Collection Center and the Henan Provincial Engineering Research Center for Industrial Microbial Strains. The remaining 12 strains of aquatic pathogens were isolated and preserved in our laboratory (Table 1).

Table 1.
The 25 strains of bacteria used in this study.
2.2. Construction of Two E. coli Expression Vectors
The complete genome sequence of P. damselae XP11 (GenBank accession number CP099544) was downloaded from GenBank. The full-length gene sequences of BamA and OmpC were obtained from this genome sequence and subjected to immunogenicity analysis using Bcepred and IEDB software version 3.0 (https://webs.iiitd.edu.in/raghava/bcepred/bcepred_submission.html (accessed on 7 January 2024); http://www.iedb.org). Immunogenic fragments of BamA and OmpC were selected, and a 6× His-Tag sequence was linked to their 3′ ends. Subsequently, NdeI and EcoRI restriction sites were added to the 5′ and 3′ ends of the fused sequences, respectively. The lengths of the 2 gene fragments were 648 bp and 567 bp, and the sequences were synthesized by Wuhan Pujian Company. The pET28b vector and the BamA-His and OmpC-His sequences were double-digested with NdeI and EcoRI, followed by ligation to construct the recombinant expression plasmids pET28b-BamA and pET28b-OmpC (Figure 1). The plasmid DNA (1–2 μL) was separately transformed into pre-prepared E. coli competent cells. Positive clones were identified through colony PCR amplification, double digestion, and DNA sequencing.
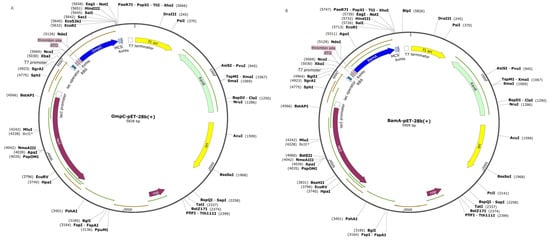
Figure 1.
Construction of expression plasmids for pET-28a-OmpC (A) and pET-28a-BamA (B).
2.3. Prokaryotic Expression Purification and Identification of Two Omps
Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added into the cultured E. coli suspension (OD600nm = 0.4) at a final concentration of 1 mM. The bacterial culture was divided into two aliquots: one was induced at 37 °C with shaking at 200 rpm for 4 h, while the other was induced at 16 °C with shaking at 200 rpm overnight. After centrifugation at 12,000× g for 10 min, the supernatant was discarded, and the pellet was resuspended in PBS buffer. The cells were lysed by sonication for 20 min (90 W, 3 s on/3 s off), followed by centrifugation at 12,000× g for 10 min. The supernatant was collected and labeled as “NPE” (Non-denatured Protein Extract). The pellet was resuspended in 100 μL of PBS (pH 7.5, containing 8 M urea) and labeled as “DPE” (Denatured Protein Extract). Protein expressions of NPE and DPE were analyzed by SDS-PAGE.
The supernatant containing OmpC after sonication and centrifugation was mixed with Ni-NTA resin and incubated at 4 °C with shaking for 30 min. The resin was washed sequentially with PBS buffer 1 (pH 7.5; W1), PBS buffer 2 (pH 7.5, 10 mM imidazole; W2), and PBS buffer 3 (pH 7.5, 30 mM imidazole; W3). Finally, the protein was eluted with PBS elution buffer (pH 7.5, 300 mM imidazole). The eluted OmpC was dialyzed and analyzed by SDS-PAGE. For BamA purification, the inclusion body pellet was mixed with Ni-NTA resin and incubated with shaking for 30 min. The mixture was loaded onto a nickel column, and the bottom cap was removed before centrifugation at 700× g for 2 min at 4 °C to collect the flow-through. The column was washed with 6 mL of W1 under the same conditions, and the wash was discarded. This washing step was repeated 3 times. Finally, 3 mL of elution buffer was added to the column, and the eluate was collected after centrifugation. The eluted fractions were pooled, dialyzed, and analyzed by SDS-PAGE.
2.4. Preparation and Titer Determination of Rabbit Polyclonal Antibodies
Polyclonal antisera against OmpC and BamA were prepared in rabbits as described in reference []. The ELISA titers of the antisera were determined following reference []. Briefly, ELISA plates were coated with 0.1 μg OmpC or BamA overnight at 4 °C. After washing 3 times with PBST, the plates were blocked with 5% skim milk (200 μL/well) at 37 °C for 2 h. Serial dilutions of the antisera (1:2000–1:128,000) were added (100 μL/well), followed by incubation at 37 °C for 1 h. After washing, HRP-conjugated goat anti-rabbit IgG (Thermo fisher, Shanghai, China; 1:4000) was added (100 μL/well) and incubated at 37 °C for 1 h. After washing, 100 μL of OPD substrate (o-Phenylenediamine) was added for color development at 37 °C for 15 min in the dark. The reaction was stopped, and the OD450nm was measured.
2.5. Preparation of Whole-Cell and Sonicated Supernatant Protein Antigens of P. damselae
P. damselae XP11 was cultured overnight in LB medium at 30 °C. The cells were washed 3 times with PBS (4 °C, 5000 rpm, 5 min) and resuspended in coating buffer to a concentration of 5 × 105 CFU/mL. Additionally, 24 strains listed in Table 1 were cultured, and their concentrations were adjusted similarly. Half of each bacterial suspension was sonicated for 20 min (90 W, 3 s on/3 s off), and the supernatant was collected after centrifugation (4 °C, 10,000 rpm, 10 min). The protein concentration was determined using the Bradford assay and adjusted to 1 μg/mL.
2.6. Horseradish Peroxidase (HRP) Labeling of Rabbit Anti-BamA IgG
Purified rabbit anti-OmpC and anti-BamA IgG were respectively used as capture antibodies, while only rabbit anti-BamA was HRP-labeled as the detection antibody. The labeling was performed using the sodium periodate method as described in reference []. Briefly, 5 mg of HRP was dissolved in 1 mL of distilled water, and the antibody solution was dialyzed to the optimal pH. Freshly prepared 0.1 M sodium periodate (0.2 mL) was added, and the mixture was stirred at room temperature in the dark for 20 min. The oxidized HRP was dialyzed overnight at 4 °C against 1 mM sodium acetate buffer (pH 4.4). The HRP solution was then mixed with the IgG, purified by G200 chromatography, and treated with 5 mg/mL sodium borohydride for 30 min at room temperature. After overnight incubation at 4 °C, the conjugate was further purified using Sephadex G200. The concentrations of enzyme (OD403nm × 0.4) and IgG [(OD280nm − OD403nm × 0.3)] × 0.62] were calculated, and the enzyme-to-protein ratio (E/P) was determined as 1–2 (effective labeling).
2.7. Optimization Concentrations of Capture and Detection Antibodies
A checkerboard titration was used to determine the optimal working concentrations of the capture and detection antibodies. The capture antibody (OmpC) was diluted to 2.50, 1.00, and 0.50 μg/mL, while BamA was diluted to 1.80, 0.90, 0.45, and 0.23 μg/mL for ELISA plate coating. The HRP-labeled detection antibody was diluted to 1.80, 0.90, 0.45, and 0.23 μg/mL. The standard sandwich ELISA procedure was followed, and the optimal concentration combination was selected based on the highest signal-to-noise (S/N) ratio.
2.8. Sandwich ELISA for P. damselae Detection
The sandwich ELISA was performed as described in reference []. Briefly, ELISA plates were coated with rabbit anti-OmpC IgG (100 μL/well) overnight at 4 °C. After washing with PBST, the plates were blocked with 200 μL/well of blocking buffer (1% BSA in PBS; pH 7.2) at 37 °C for 2 h. The test antigen (1.0 μg/mL crude outer membrane protein) was added (100 μL/well) and incubated at 37 °C for 1 h. HRP-labeled rabbit anti-BamA IgG (100 μL/well) was then added and incubated at 37 °C for 1 h. After TMB (3,3′,5,5′-Tetramethylbenzidine) substrate incubation for 10 min, the reaction was stopped with 2 M H2SO4, and the OD450nm was measured to calculate the S/N ratio. A similar procedure was used for the sandwich ELISA with rabbit anti-BamA and its HRP-labeled counterpart.
2.9. Sensitivity, Specificity, and Reproducibility of the Sandwich ELISA for P. damselae
2.9.1. Sensitivity Validation
The sonicated bacterial supernatant was serially diluted from 200 μg/mL to 0.02 μg/mL, and the sensitivity was evaluated using the established sandwich ELISA method.
2.9.2. Specificity Validation
To assess specificity, the sandwich ELISA was performed using 7 strains of P. damselae and 18 non-target bacterial supernatants. The method was considered specific if the antibodies only bound to P. damselae antigens without cross-reactivity. Additionally, ELISA plates were coated with 1.0 μg/mL rabbit anti-OmpC or 0.90 μg/mL rabbit anti-BamA, followed by the addition of 1.0 μg/mL XP11 sonicated supernatant. HRP-labeled rabbit anti-BamA was pre-mixed with 25 bacterial strains (105 cfu/mL, v/v = 1) and incubated at 37 °C for 30 min. The mixtures were then centrifugated and the supernatant was added to the coated plates to assess competitive binding. PBS and HRP-labeled rabbit anti-BamA alone served as negative and positive controls, respectively. A strain was deemed cross-reactive if it reduced the S/N ratio to ≤60% of the positive control.
2.9.3. Reproducibility Validation
Seven strains of P. damselae and eighteen non-target bacterial samples were tested in triplicate using the sandwich ELISA. The mean (X) and standard deviation (SD) of the OD450nm values were calculated, and the coefficient of variation (CV) was determined as (SD/X) × 100%. A lower than 10% of CV indicated high reproducibility.
3. Results
3.1. Results of Two Constructed Recombinant Vectors and Its Expression and Purification
The purified target DNA fragments were ligated into the pET-28a plasmid and transformed into E. coli BL21 competent cells. Recombinant plasmids pET-28a-OmpC and pET-28a-BamA were successfully screened. Sequencing confirmed that the inserted fragments were identical to the BamA and OmpC gene sequences (Supplementary Figure S1) of P. damselae XP11 (GenBank accession no. CP099544). SDS-PAGE analysis revealed that BL21-OmpC and BL21-BamA recombinant strains expressed target protein bands at approximately 22 kDa and 24 kDa, respectively, under different induction conditions (No. 1: 16 °C/16 h; No. 2: 37 °C/4 h). No corresponding bands were observed in uninduced controls (N) (Figure 2). Additionally, OmpC was exclusively expressed in the soluble fraction (supernatant), while BamA was detected in both the supernatant and inclusion body pellet, with predominant expression in the inclusion bodies. As shown in Figure 3, the purified OmpC and BamA proteins exhibited clear bands at their expected molecular weights (~22 kDa and ~24 kDa, respectively), confirming high purity.
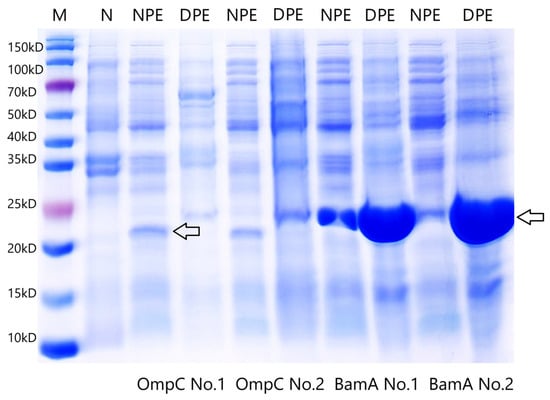
Figure 2.
SDS-PAGE electrophoresis of 2 outer membrane proteins expressed in Escherichia coli. NPE: Non-denatured Protein Extract from the supernatant; DPE: Denatured Protein Extract from the inclusion body; M: marker; N: uninduced strain; No. 1: 16 °C/16 h; No. 2: 37 °C/4 h. Arrows point the expressed proteins of OmpC and BamA.
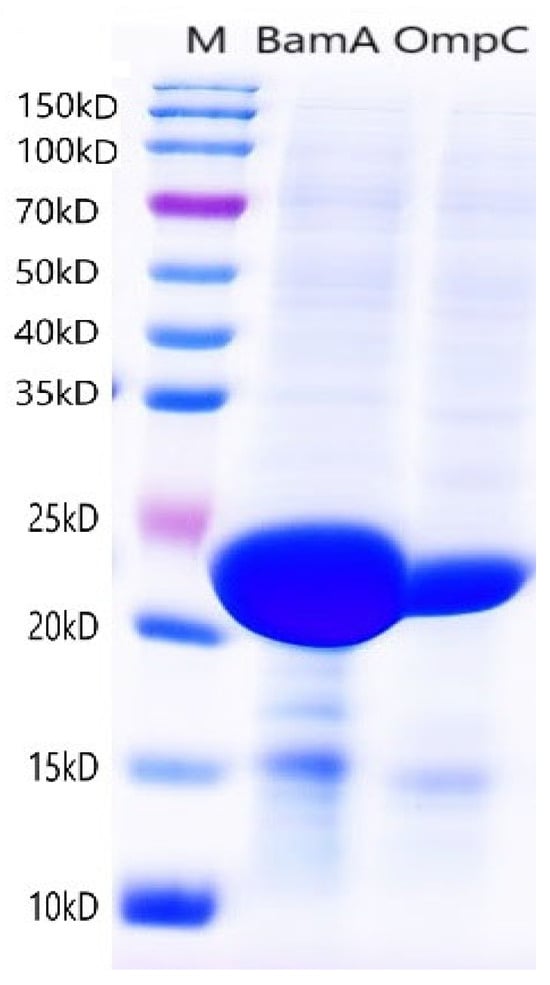
Figure 3.
SDS-PAGE electrophoresis of 2 purified outer membrane proteins; M: marker.
3.2. Titration Results of Rabbit Antisera Against OmpC and BamA
New Zealand white rabbits were immunized with purified OmpC or BamA. After three to four booster immunizations, the titers of the antisera were determined using an indirect ELISA method, with the criterion for a positive titer being an OD450nm value with a signal-to-noise (S/N) ratio of ≥2.0. The results showed that, when coated with 0.1 μg of OmpC and BamA protein per well, the titers of the rabbit antisera against OmpC and BamA were 1:128,000 and 1:64,000, respectively (Table 2).

Table 2.
Indirect ELISA titer of rabbit anti-OmpC/BamA serum.
3.3. Optimal Concentrations of Two Antibodies for Detecting P. damselae via Sandwich ELISA
To determine the optimal detection concentrations for the two antibodies, purified IgG from rabbit anti-OmpC and rabbit anti-BamA were used as capture antibodies at various concentrations. HRP-conjugated IgG against BamA was used as the detection antibody at different concentrations. The sonicated supernatant of P. damselae was used as the sandwich antigen at a concentration of 1 μg/mL. The results indicated that when the capture antibody concentration of anti-OmpC was 2.5 or 1.0 μg/mL, and the detection antibody concentration was 0.9 μg/mL, the S/N values for antigen detection were around 4.9 (Table 3). To conserve the anti-OmpC antibody, this study will use 1.0 μg/mL of anti-OmpC and 0.9 μg/mL of anti-BamA as the capture and detection antibodies, respectively. Similarly, when the capture antibody concentration of anti-BamA was 1.8 or 0.9 μg/mL, and the detection antibody concentration was 1.8 or 0.9 μg/mL, the S/N values exceeded 10 (Table 4). To conserve the anti-BamA antibody, this study will use 0.9 μg/mL of anti-BamA as both the capture and detection antibodies.

Table 3.
Determination of the optimal concentration for capturing and detecting antibodies.

Table 4.
Determination of the optimal concentration for capturing and detecting antibody BamA.
3.4. Sensitivity Results of Sandwich ELISA for Detecting P. damselae
The sensitivity of detecting the sonicated supernatant protein of P. damselae strain XP11 was assessed using two different sandwich ELISA methods. The results indicated that when the concentration of the protein of XP11 was ≥0.2 μg/mL, the S/N ratio was greater than two for both methods, demonstrating that the lowest detection limit for both sandwich ELISA methods is 0.2 μg/mL (Table 5). In terms of numerical values, the method using anti-OmpC as the capture antibody and anti-BamA as the detection antibody exhibited higher sensitivity compared to the method using anti-BamA as both the capture and detection antibodies.

Table 5.
Sensitivity detection results of sandwich ELISA.
3.5. Cross-Reactivity Results of Sandwich ELISA for Detecting P. damselae
When using 1.0 μg/mL of rabbit anti-OmpC as the capture antibody and 0.9 μg/mL of rabbit anti-BamA as the detection antibody, only the strain of B60 showed cross-reaction with an S/N value reaching 60% of the OD450nm of the XP11 strain (left side of Table 6). When 0.9 μg/mL of rabbit anti-BamA as the capture and detection antibody, all other strains were determined as having no cross-reactions with seven strains of P. damselae (right side of Table 6).

Table 6.
P. damselae detection via cross reaction of sandwich ELISA using 24 strains of bacteria.
3.6. Blocking Results of Sandwich ELISA for Detecting P. damselae
First, the ELISA plate was coated with rabbit anti-OmpC or anti-BamA, followed by the addition of 1 μg/mL sonicated supernatant (XP11) for incubation. The results showed that in the rabbit anti-OmpC detection system, six strains of P. damselae and three non-target strains (P2, P3, and B1005) blocked the binding of HRP-conjugated rabbit anti-BamA to XP11 (3.44 × 0.6 = 2.06, left side of Table 7). In the rabbit anti-BamA detection system, three non-target strains (P1, B60, B67) also blocked the binding of HRP-conjugated rabbit anti-BamA to XP11 (5.13 × 0.6 = 3.08, right side of Table 7).

Table 7.
P. damselae detection via blocking sandwich ELISA using 25 strains of bacteria.
3.7. Repeatability Test Results of Sandwich ELISA for Detecting P. damselae
The sonicated supernatant antigens of 25 bacterial strains were detected using sandwich ELISA in three independent replicate experiments. The results showed that when using rabbit anti-OmpC and anti-BamA as the detection and capture antibodies, respectively, only six strains of P. damselae and B60 had average S/N values exceeding 60% of XP11, while the remaining strains were below this threshold. The coefficient of variations (CVs) for all strains in the three repeated tests was less than 9.1% (left side of Table 8). Similarly, when using rabbit anti-BamA as both detection and capture antibodies, only six strains of P. damselae had average S/N values exceeding 60% of XP11, while the CVs of remaining strains below this threshold were within 9.22% (right side of Table 8).

Table 8.
Repeatability test results of P. damselae detection using rabbit anti-OmpC or rabbit anti-BamA as detection antibodies, and HRP-labeled rabbit anti-BamA as capture antibody.
4. Discussion
In recent years, with the continuous expansion of aquaculture scale, the economic losses caused by pathogenic bacteria have become increasingly severe. As a crucial aquaculture pathogen, P. damselae has become a focus for the development of rapid detection methods to control and prevent diseases. This study addresses the issue of severe cross-reactivity in traditional indirect ELISA methods due to the use of whole-cell antigens [,]. Focusing on the sea cucumber-derived P. damselae strain XP11, we selected two characteristic outer membrane proteins, OmpC and BamA, through gene sequence comparison. After successfully expressing and purifying these two proteins, we innovatively established a sandwich ELISA detection system based on two Omps, aiming to develop a new immunological detection method for P. damselae.
The detection method for P. damselae was comprehensively evaluated through cross-reaction, blocking experiments, and repeatability testing, thereby providing a reference for the immunological detection techniques of P. damselae and other aquaculture pathogens. Prior to constructing the recombinant expression plasmid, we selected OmpC and BamA genes from over a dozen Omp genes. Through NCBI comparison analysis, we found that the gene sequences of OmpC and BamA have a similarity of over 97% among different isolates of P. damselae, while the sequence similarity between different species within the same genus is below 78%, and the similarity with closely related Vibrio species is below 67%. This comparison result provides a theoretical basis for the specific detection of P. damselae using rabbit anti-Omp antibodies.
Our study found that OmpC expressed in E. coli is present in the supernatant after sonication and centrifugation, indicating that the protein is expressed in a soluble form. In contrast, the BamA protein primarily exists in the pellet after sonication and centrifugation in the form of inclusion bodies. This difference may be related to the hydrophilicity and spatial structure differences of the two Omps. OmpC has a β-barrel structure and relies on host chaperone proteins such as SurA to assist in folding []. Moreover, the synergistic action of SurA and Skp chaperone proteins can promote the transmembrane transport of β-barrel proteins like OmpC [], which explains why OmpC is mainly expressed in a soluble form in prokaryotic systems. BamA contains transmembrane domains, and the lack of post-translational modification enzymes in prokaryotic expression systems, including protein disulfide isomerase (PDI) and endoplasmic reticulum chaperone proteins (such as BiP), may lead to the exposure of hydrophobic domains in the expressed proteins [,]. Our study finding that purified BamA has very low water solubility confirms this inference.
The sandwich ELISA cross-reactivity experiment results showed that the minimum detection limit for XP11 sonicated crude protein supernatant by both antibody combinations was 0.2 μg/mL, demonstrating high sensitivity (Table 5). In this study, the detection limit was calculated using the formula LOD = Sb + 3σ, where Sb represents the mean optical density (OD) of the negative controls and σ denotes the standard deviation (SD). For the five trials on the left side of Table 5, the mean OD (Sb) of the negative controls was 0.064, with an SD (σ) of 0.0102, resulting in an LOD of 0.095. Similarly, for the trials on the right side, the mean OD was 0.174, with an SD of 0.0162, yielding an LOD of 0.223. Based on these calculations: When using 1.0 μg/mL anti-OmpC and 0.9 μg/mL anti-BamA, the sonicated crude protein supernatant of XP11 exhibited an OD of 0.09, suggesting a protein concentration close to 0.02 μg/mL. When using 0.9 μg/mL anti-BamA, the OD values of 0.36 and 0.11 corresponded to estimated protein concentrations between 0.2 μg/mL and 0.02 μg/mL, with 0.223 serving as an intermediate reference point.
The homologous dual-antibody system based on anti-BamA antibodies could specifically identify different isolates of P. damselae, with no cross-reactions with other bacteria, including four species from the same genus of Photobacterium (Table 6, right). The heterologous antibody combination of OmpC–BamA also showed good specificity, with only the strain of B60 showing a weak cross-reaction (Table 6 left). However, the blocking experiment indicated that both the OmpC–BamA and the BamA–BamA sandwich ELISA methods had weak cross-reactions with three strains (Table 7). It is worth mentioning that the high consistency between the cross-reactivity and repeatability test results (Table 6 and Table 8) further demonstrated the potential application value of the sandwich ELISA methods established in this study.
In ELISA pathogen detection, the threshold for determination is typically set at 50% to 70% of the OD value of the positive control or the signal-to-noise ratio between positive and negative controls (P/N value) []. In this study, we used a threshold between different samples and negative controls of S/N ≥ 60% of the positive control for sandwich ELISA detection of cross-reacting strains and an S/N value ≤ 60% of the positive control for blocking experiments. The results showed that these thresholds could effectively distinguish between positive and negative samples. Additionally, some studies have used statistical methods to set the threshold at 2 to 3 times the standard deviation (SD) above the average OD value of the negative control to increase reliability []. However, this method was not adopted in our study as it was found to significantly increase the risk of cross-reactions and reduce the specificity of ELISA detection. Moreover, the applicability of the threshold needs to be adjusted in combination with factors such as the detection method, sample type, and kit design [].
Furthermore, the sensitivity and specificity of the kit also affect the threshold setting—high-sensitivity kits may require higher thresholds to reduce false positives, while high-specificity kits can set lower thresholds to minimize false negatives. For example, when the S/N threshold was adjusted to 2.85, the dual-antibody combinations in Table 8 could specifically detect P. damselae, thereby reducing false-positive results. The complexity of the antigen samples, such as whole-cell antigens being more likely to contain interfering substances than sonicated supernatants, may also lead to false-positive or false-negative results []. Finally, the repeatability and precision of the experiment also affect the threshold setting. If the coefficient of variation (CV) is large, the threshold needs to be adjusted more flexibly [].
Setting a fixed value (such as 60%) as the determination standard in this study has certain limitations, as it does not consider the continuity and variability of the data. For example, one sample has an S/N value of 59% of the positive control, while another sample has 61%, and a 2% difference leads to a positive or negative determination. To avoid misjudgments near the threshold, many laboratories have attempted to set a “gray zone” (for example, when the threshold is 60%, the range of 55% to 65% is considered a gray zone), and samples in the gray zone are retested or validated. In a future study, it is possible to combine the continuity of data, variability, and gray zone setting, and increase the number of repeated experiments to improve the accuracy and reliability of detection results.
Although this study investigated the specificity of the detection method using 24 pathogen strains, it did not include many reported aquatic pathogens, which may have somewhat limited the comprehensive evaluation of the kit’s detection specificity. To improve the practical applicability of our findings for diagnostic kit development, we plan to significantly expand the spectrum of tested aquatic pathogens in future research.
5. Conclusions
This study has developed a sandwich ELISA detection method based on two expressed Omps. The results showed that the sandwich ELISA method based on anti-BamA had stronger specificity for detecting P. damselae compared to the sandwich ELISA method using anti-OmpC and anti-BamA. Moreover, this study has broken through the limitation of non-specific cross-reactions that often occur in traditional ELISA detection of whole cells, providing a reliable solution for the rapid detection of aquaculture pathogens. Therefore, the sandwich ELISA method based on anti-BamA in this study is likely to lead to the development of a standardized P. damselae detection kit and offers a valuable solution for the rapid detection technology research of bacterial pathogens.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes10090439/s1, Supplementary Figure S1: Comparison of BamA (A) and OmpC (B) sequences between two expressed plasmids and two gene fragments.
Author Contributions
Z.C.: sampling, data curation, writing—original draft. W.H.: funding acquisition; Q.Y.: methodology, resources; S.G.: funding acquisition, methodology, project administration, resources, software, validation, writing—original draft and editing. All authors have read and agreed to the published version of the manuscript.
Funding
The publication of this article was funded by the Fujian Aquatic Seed Industry Innovation and Industrialization Project (No. 2021FJSCZY03), Bureau of Ocean and Fisheries of Fujian province.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Acknowledgments
Yang Chen and Zhen Yang (JiMei University) are greatly acknowledged for their technical assistance.
Conflicts of Interest
The authors declare no conflicts of interest.
Correction Statement
This article has been republished with a minor correction to the Institutional Review Board Statement/Data Availability Statement. This change does not affect the scientific content of the article.
References
- Rivas, A.J.; Lemos, M.L.; Osorio, C.R. Photobacterium damselae subsp. damselae, a bacterium pathogenic for marine animals and humans. Front. Microbiol. 2013, 4, 283. [Google Scholar]
- Morick, D.; Blum, S.E.; Davidovich, N.; Zemah-Shamir, Z.; Bigal, E.; Itay, P.; Rokney, A.; Nasie, I.; Feldman, N.; Flecker, M.; et al. Photobacterium damselae subspecies damselae Pneumonia in Dead, Stranded Bottlenose Dolphin, Eastern Mediterranean Sea. Emerg. Infect. Dis. 2023, 29, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Love, M.; Teebken-Fisher, D.; Hose, J.E.; Farmer, J.J., 3rd; Hickman, F.W.; Fanning, G.R. Vibrio damsela, a Marine Bacterium, Causes Skin Ulcers on the Damselfish Chromis punctipinnis. Science 1981, 214, 1139–1140. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.K.; Sutton, D.C.; Fuerst, J.A.; Reichelt, J.L. Evaluation of the genus Listonella and reassignment of Listonella damsela (Love et al.) MacDonell and Colwell to the genus Photobacterium as Photobacterium damsela comb. nov. with an emended description. Int. J. Syst. Bacteriol. 1991, 41, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, G.; Lafay, B.; Ruimy, R.; Breittmayer, V.; Nicolas, J.L.; Gauthier, M.; Christen, R. Small-subunit rRNA sequences and whole DNA relatedness concur for the reassignment of Pasteurella piscicida (Snieszko et al.) Janssen and Surgalla to the genus Photobacterium as Photobacterium damsela subsp. piscicida comb. nov. Int. J. Syst. Bacteriol. 1995, 45, 139–144. [Google Scholar] [CrossRef]
- Essam, H.M.; Abdellrazeq, G.S.; Tayel, S.I.; Torky, H.A.; Fadel, A.H. Pathogenesis of Photobacterium damselae subspecies infections in sea bass and sea bream. Microb. Pathog. 2016, 99, 41–50. [Google Scholar] [CrossRef]
- White, D.M.; Valsamidis, M.A.; Kokkoris, G.D.; Bakopoulos, V. The effect of temperature and challenge route on in vitro hemocyte phagocytosis activation after experimental challenge of common octopus, Octopus vulgaris (Cuvier, 1797) with either Photobacterium damselae subsp. damselae or Vibrio anguillarum O1. Microb. Pathog. 2023, 174, 105955. [Google Scholar] [CrossRef]
- Barca, A.V.; Vences, A.; Terceti, M.S.; do Vale, A.; Osorio, C.R. Low salinity activates a virulence program in the generalist marine pathogen Photobacterium damselae subsp. damselae. mSystems 2023, 8, e0125322. [Google Scholar] [CrossRef]
- Pedersen, K.; Skall, H.F.; Lassen-Nielsen, A.M.; Bjerrum, L.; Olesen, N.J. Photobacterium damselae subsp. damselae, an emerging pathogen in Danish rainbow trout, Oncorhynchus mykiss (Walbaum), mariculture. J. Fish Dis. 2009, 32, 465–472. [Google Scholar]
- Osorio, C.R.; Vences, A.; Matanza, X.M.; Terceti, M.S. Photobacterium damselae subsp. damselae, a generalist pathogen with unique virulence factors and high genetic diversity. J. Bacteriol. 2018, 200, e00002-18. [Google Scholar]
- Weawsawang, W.; Homsombat, T.; Nuanmanee, S.; Saleetid, N.; Thawonsuwan, J.; Pumchan, A.; Hirono, I.; Kondo, H.; Unajak, S. Characterization of Photobacterium damselae subsp. damselae isolated from diseased Asian seabass (Lates calcarifer) and the preliminary development of a formalin-killed cell vaccine. J. Fish Dis. 2024, 47, e13987. [Google Scholar] [PubMed]
- Su, F.J.; Chen, M.M. Protective Efficacy of Novel Oral Biofilm Vaccines against Photobacterium damselae subsp. damselae Infection in Giant Grouper, Epinephelus lanceolatus. Vaccines 2022, 10, 207. [Google Scholar]
- Hayano, S.; Masaki, T.; Tadakuma, R.; Kashima, M. Photobacterium damselae subsp. damselae bacteraemia in a patient with liver cirrhosis. BMJ Case Rep. 2021, 14, e242580. [Google Scholar]
- Eissa, I.A.M.; Derwa, H.I.; Ismail, M.; El-Lamie, M.; Dessouki, A.A.; Elsheshtawy, H.; Bayoumy, E.M. Molecular and phenotypic characterization of Photobacterium damselae among some marine fishes in Lake Temsah. Microb. Pathog. 2018, 114, 315–322. [Google Scholar] [CrossRef]
- Labella, A.; Manchado, M.; Alonso, M.C.; Castro, D.; Romalde, J.L.; Borrego, J.J. Molecular intraspecific characterization of Photobacterium damselae ssp. damselae strains affecting cultured marine fish. J. Appl. Microbiol. 2010, 108, 2122–2132. [Google Scholar]
- Carraro, R.; Dalla Rovere, G.; Ferraresso, S.; Carraro, L.; Franch, R.; Toffan, A.; Pascoli, F.; Patarnello, T.; Bargelloni, L. Development of a real-time PCR assay for rapid detection and quantification of Photobacterium damselae subsp. piscicida in fish tissues. J. Fish Dis. 2018, 41, 247–254. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, J.; Gao, Y.; Zhou, D.; Zhang, B.; Li, X.; Zhang, J. Development of a Colloidal Gold Immunochromatographic Strip for Specific Detection of Photobacterium damselae subsp. piscicida. J. Fish Dis. 2025, 48, e14062. [Google Scholar] [CrossRef]
- He, L.; Wu, L.; Tang, Y.; Lin, P.; Zhai, S.; Xiao, Y.; Guo, S. Immunization of a novel outer membrane protein from Aeromonas hydrophila simultaneously resisting A. hydrophila and Edwardsiella anguillarum infection in European eels (Angullia angullia). Fish Shellfish Immunol. 2020, 97, 300–312. [Google Scholar] [CrossRef]
- Guo, S.; He, L.; Wu, L.; Xiao, Y.; Zhai, S.; Yan, Q. Immunization of a novel bivalent outer membrane protein simultaneously resisting Aeromonas hydrophila, Edwardsiella anguillarum and Vibrio vulnificus infection in European eels (Angullia angullia). Fish Shellfish Immunol. 2020, 97, 46–57. [Google Scholar] [CrossRef]
- Regidi, S.; Ravindran, S.; Vijayan, A.L.; Maya, V.; Sreedharan, L.; Varghese, J.; Ramaswami, K.; Gopi, M. Effect of lyophilization on HRP-antibody conjugation: An enhanced antibody labeling technology. BMC Res. Notes 2018, 11, 596. [Google Scholar] [CrossRef]
- Shen, C.; Chang, S.; Luo, Q.; Chan, K.C.; Zhang, Z.; Luo, B.; Xie, T.; Lu, G.; Zhu, X.; Wei, X.; et al. Structural basis of BAM-mediated outer membrane β-barrel protein assembly. Nature 2023, 617, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Golchin, M.; Mollayi, S.; Mohammadi, E.; Eskandarzade, N. Development of a diagnostic indirect ELISA test for detection of Brucella antibody using recombinant outer membrane protein 16 kDa (rOMP16). Vet. Res. Forum. 2022, 13, 387–391. [Google Scholar] [PubMed]
- Dogra, V.; Verma, S.; Singh, G.; Wani, A.H.; Chahota, R.; Dhar, P.; Verma, L.; Sharma, M. Development of OMP based indirect ELISA to gauge the antibody titers in bovines against Pasteurella multocida. Iran. J. Vet. Res. 2015, 16, 350–356. [Google Scholar] [PubMed]
- Li, G.; He, C.; Bu, P.; Bi, H.; Pan, S.; Sun, R.; Zhao, X.S. Single-Molecule Detection Reveals Different Roles of Skp and SurA as Chaperones. ACS Chem. Biol. 2018, 13, 1082–1089. [Google Scholar] [CrossRef]
- Heinz, E.; Lithgow, T. A comprehensive analysis of the Omp85/TpsB protein superfamily structural diversity, taxonomic occurrence and evolution. Fron. Microbiol. 2014, 5, 370. [Google Scholar] [CrossRef]
- Humes, J.R.; Schiffrin, B.; Calabrese, A.N.; Higgins, A.J.; Westhead, D.R.; Brockwell, D.J.; Radford, S.E. The Role of SurA PPIase Domains in Preventing Aggregation of the Outer-Membrane Proteins tOmpA and OmpT. J. Mol. Biol. 2019, 431, 1267–1283. [Google Scholar] [CrossRef]
- Zaydman, M.A.; Brestoff, J.R.; Jackups, R., Jr. Using information theory to optimize a diagnostic threshold to match physician-ordering practice. J. Biomed. Inform. 2021, 117, 103756. [Google Scholar] [CrossRef]
- Oluka, G.K.; Namubiru, P.; Kato, L.; Ankunda, V.; Gombe, B.; Cotten, M.; The COVID-19 Immunoprofiling Team; Musenero, M.; Kaleebu, P.; Fox, J.; et al. Optimisation and Validation of a Conventional ELISA and Cut-offs for Detecting and Quantifying Anti-SARS-CoV-2 Spike, RBD, and Nucleoprotein IgG, IgM, and IgA Antibodies in Uganda. Front. Immunol. 2023, 14, 1113194. [Google Scholar] [CrossRef]
- Ritchie, B.M.; Connors, J.M.; Sylvester, K.W. Comparison of an IgG-Specific Enzyme-Linked Immunosorbent Assay Cutoff of 0.4 Versus 0.8 and 1.0 Optical Density Units for Heparin-Induced Thrombocytopenia. Clin. Appl. Thromb. Hemost. 2017, 23, 282–286. [Google Scholar] [CrossRef]
- Li, X.; Li, G.; Teng, Q.; Yu, L.; Wu, X.; Li, Z. Development of a Blocking ELISA for Detection of Serum Neutralizing Antibodies Against Newly Emerged Duck Tembusu Virus. PLoS ONE 2012, 7, e53026. [Google Scholar] [CrossRef]
- Cheng, X.; Liu, Q.; Shi, J.; Zhou, C.; Su, X.; Dong, Z.; Xing, W.; Lu, H.; Pan, C.; Li, X.; et al. Reliability and Reproducibility of the Diameters Method in Rapid Determination of Acute Infarct Volume in Magnetic Resonance Diffusion-Weighted Imaging. Cerebrovasc. Dis. 2020, 49, 575–582. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).