Sex-Associated Indels and Candidate Gene Identification in Fujian Oyster (Magallana angulata)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Sex Identification
2.2. DNA Extraction and Whole-Genome Resequencing
2.3. Detection and Quality Filtering of Indels
2.4. Identification of Sex-Association Variants
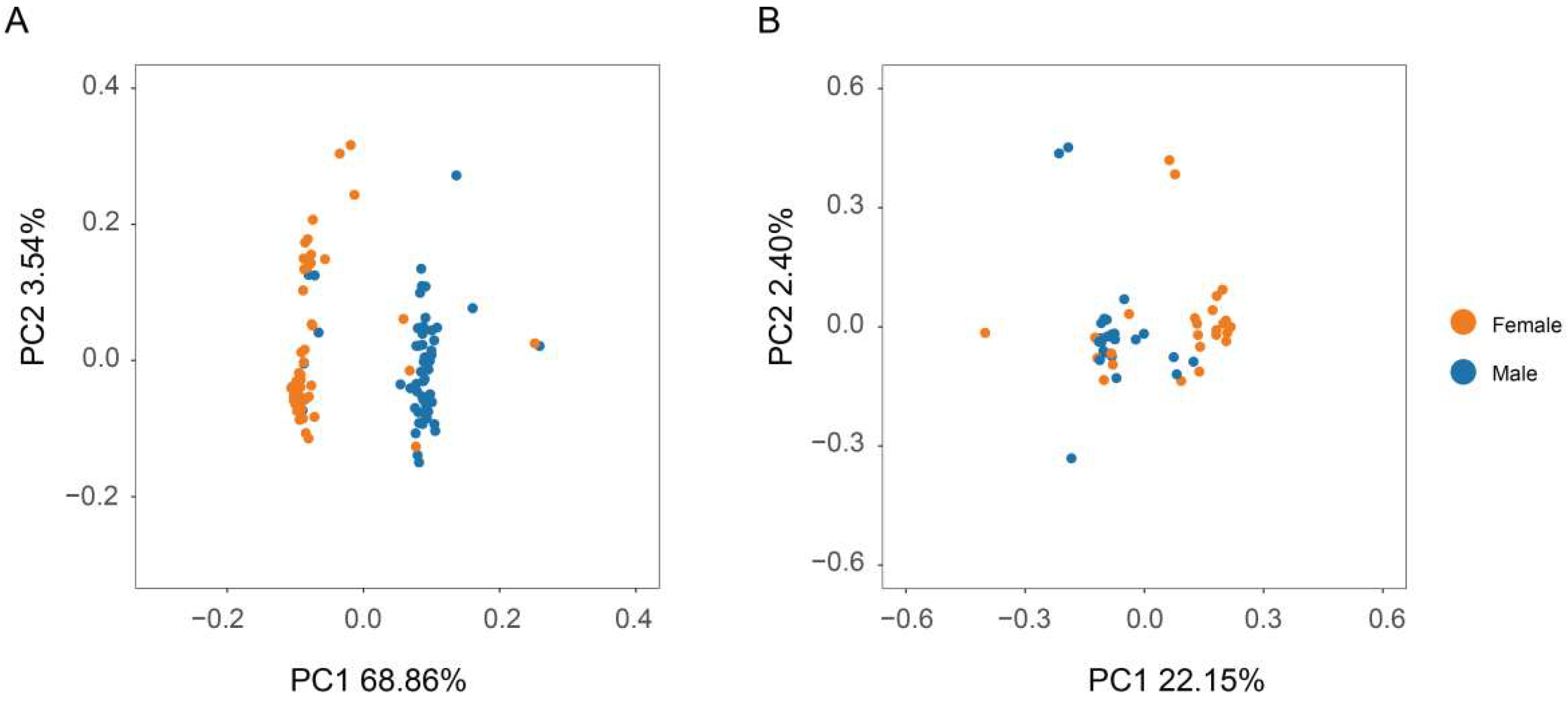
2.5. Principal Component Analysis (PCA)
2.6. Marker Development and Validation
2.7. Functional Annotation and Candidate Gene Identification
2.8. Comparative Analysis of Key Candidate Genes
3. Results
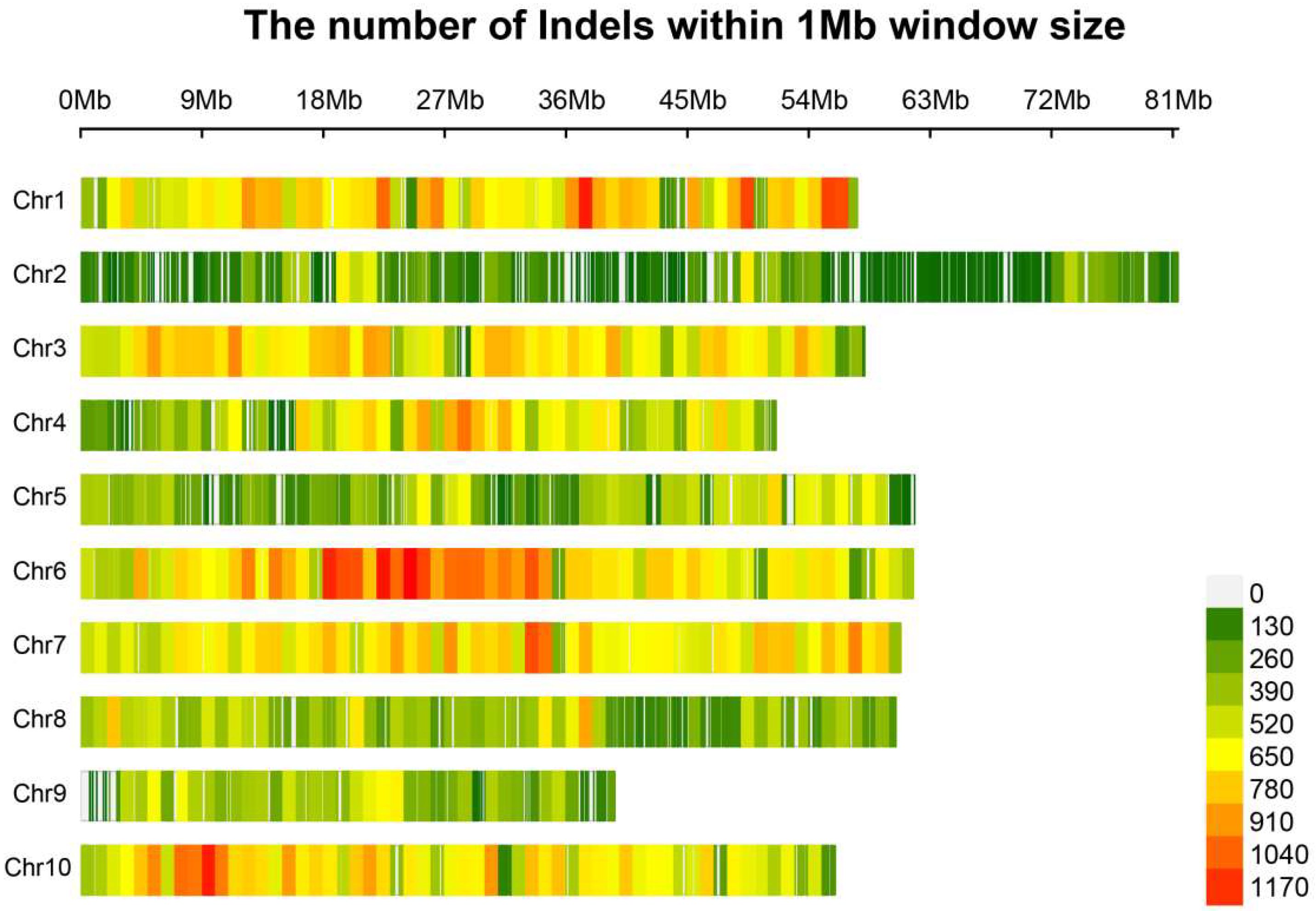
3.1. Resequencing and Variant Detection
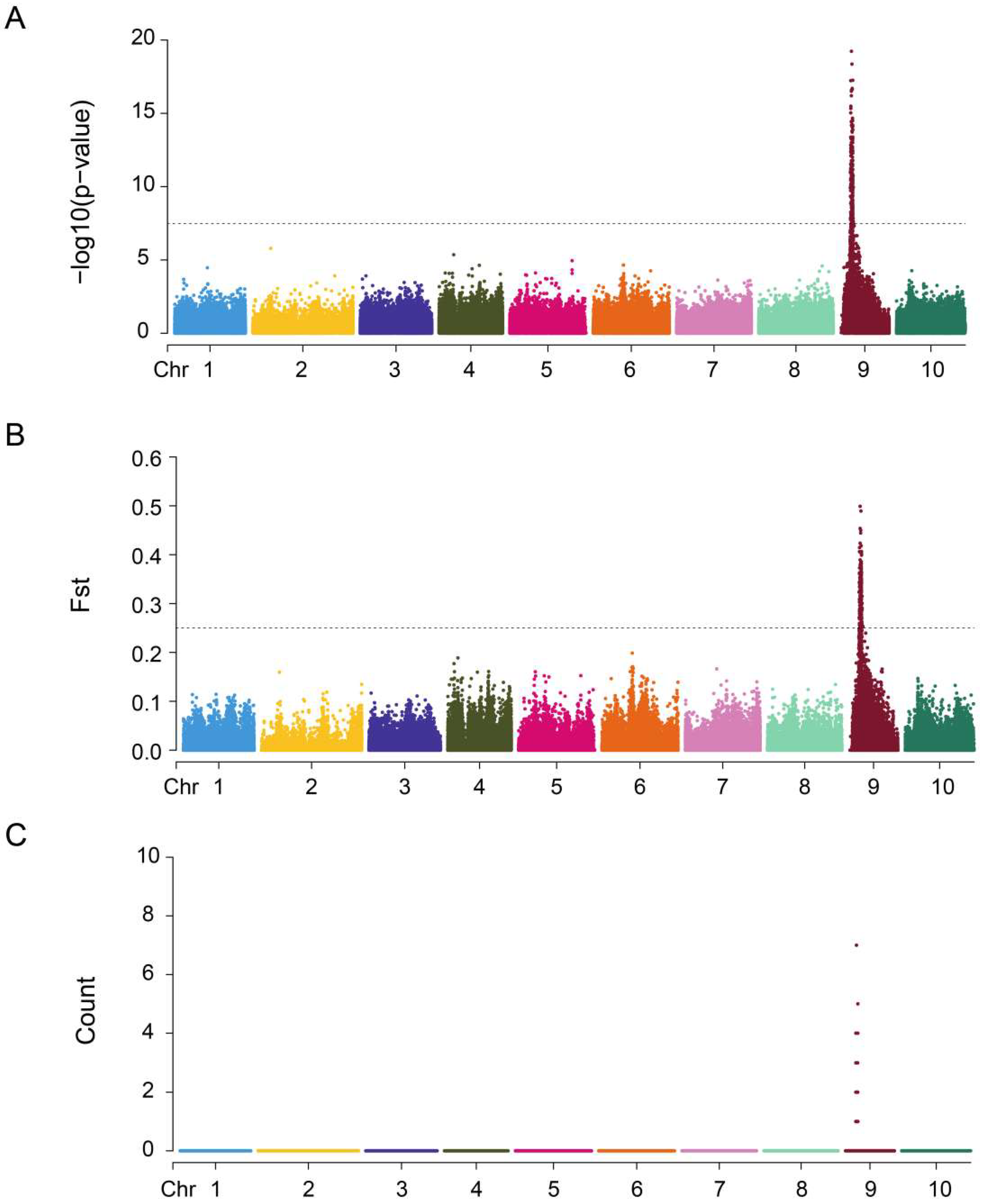
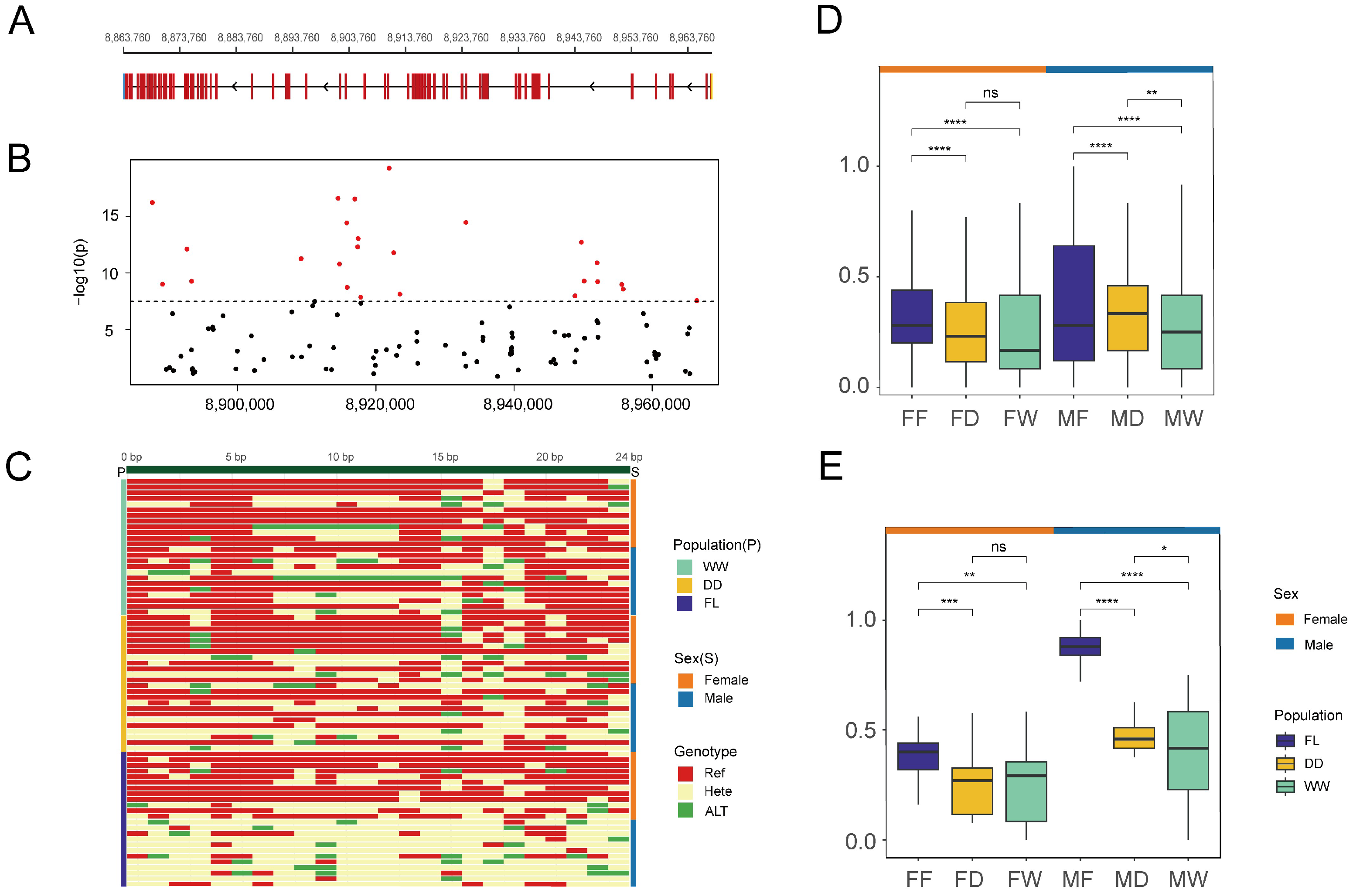
3.2. Identification and Localization of Sex-Associated Indels
3.3. Development and Validation of Sex-Linked Indel Markers
3.4. Functional Annotation of Candidate Sex-Associated Indels
3.5. Variation in Sex-Associated Indels in the PKD1L1 Gene Across Populations
4. Discussion
4.1. Mapping the Sex-Determining Region in Fujian Oyster
4.2. Development and Validation of Sex-Linked Molecular Markers
4.3. Identification and Functional Implications of Candidate Genes
4.4. Domestication-Driven Enrichment of Male-Linked Variants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martínez, P.; Viñas, A.M.; Sánchez, L.; Díaz, N.; Ribas, L.; Piferrer, F. Genetic architecture of sex determination in fish: Applications to sex ratio control in aquaculture. Front. Genet. 2014, 5, 340. [Google Scholar] [CrossRef]
- Li, X.-Y.; Mei, J.; Ge, C.-T.; Liu, X.-L.; Gui, J.-F. Sex determination mechanisms and sex control approaches in aquaculture animals. Sci. China Life Sci. 2022, 65, 1091–1122. [Google Scholar] [CrossRef]
- Xie, J.; Ning, Y.; Han, Y.; Su, C.; Zhou, X.; Wu, Q.; Guo, X.; Qi, J.; Ge, H.; Ke, Y. Identification of SNPs and Candidate Genes Associated with Growth Using GWAS and Transcriptome Analysis in Portuguese Oyster (Magallana angulata). Fishes 2024, 9, 471. [Google Scholar] [CrossRef]
- Qin, Y.; Li, R.; Liao, Q.; Shi, G.; Zhou, Y.; Wan, W.; Li, J.; Ma, H.; Zhang, Y.; Yu, Z. Comparison of biochemical composition, nutritional quality, and metals concentrations between males and females of three different Crassostrea sp. Food Chem. 2023, 398, 133868. [Google Scholar] [CrossRef] [PubMed]
- Nicolini, F.; Ghiselli, F.; Luchetti, A.; Milani, L. Bivalves as emerging model systems to study the mechanisms and evolution of sex determination: A genomic point of view. Genome Biol. Evol. 2023, 15, evad181. [Google Scholar] [CrossRef] [PubMed]
- Collin, R. Phylogenetic patterns and phenotypic plasticity of molluscan sexual systems. Integr. Comp. Biol. 2013, 53, 723–735. [Google Scholar] [CrossRef]
- Park, J.J.; Kim, H.; Kang, S.W.; An, C.M.; Lee, S.-H.; Gye, M.C.; Lee, J.S. Sex ratio and sex reversal in two-year-old class of oyster, Crassostrea gigas (Bivalvia: Ostreidae). Dev. Reprod. 2012, 16, 385. [Google Scholar] [CrossRef]
- Guo, X.; Li, Q.; Wang, Q.Z.; Kong, L.F. Genetic Mapping and QTL Analysis of Growth-Related Traits in the Pacific Oyster. Mar. Biotechnol. 2012, 14, 218–226. [Google Scholar] [CrossRef]
- Han, Z.; Li, Q.; Xu, C.; Liu, S.; Yu, H.; Kong, L. QTL mapping for orange shell color and sex in the Pacific oyster (Crassostrea gigas). Aquaculture 2021, 530, 735781. [Google Scholar] [CrossRef]
- Zhou, X.; Ning, Y.; Xie, J.; Han, Y.; Tang, C.; Su, C.; Wan, Q.; Wu, Q.; Guo, X.; Qi, J.; et al. Correction: Identification of sex-linked markers and genes in Portuguese oyster (Magallana angulata). Front. Mar. Sci. 2025, 12, 1643904. [Google Scholar] [CrossRef]
- Hayashi, K.; Yoshida, H.; Ashikawa, I. Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes. Theor. Appl. Genet. 2006, 113, 251–260. [Google Scholar] [CrossRef]
- Bhangale, T.R.; Rieder, M.J.; Livingston, R.J.; Nickerson, D.A. Comprehensive identification and characterization of diallelic insertion–deletion polymorphisms in 330 human candidate genes. Hum. Mol. Genet. 2005, 14, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Browning, B.L.; Browning, S.R. Genotype Imputation with Millions of Reference Samples. Am. J. Hum. Genet. 2016, 98, 116–126. [Google Scholar] [CrossRef]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.-y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2012, 14, 178–192. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Ginestet, C. ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. Ser. A Stat. Soc. 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Haley, L. Genetics of sex determination in the American oyster. Proc. Natl. Shellfish. Assoc. 1979, 69, 54–57. Available online: https://cir.nii.ac.jp/crid/1570291224717821568 (accessed on 29 July 2025).
- Guo, X.; Hedgecock, D.; Hershberger, W.K.; Cooper, K.; Allen, S.K., Jr. Genetic determinants of protandric sex in the Pacific oyster, Crassostrea gigas Thunberg. Evolution 1998, 52, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, P.W.; Hedgecock, D. Sex Determination: Genetic Models for Oysters. J. Hered. 2010, 101, 602–611. [Google Scholar] [CrossRef]
- Santerre, C.; Sourdaine, P.; Marc, N.; Mingant, C.; Robert, R.; Martinez, A.-S. Oyster sex determination is influenced by temperature—First clues in spat during first gonadic differentiation and gametogenesis. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2013, 165, 61–69. [Google Scholar] [CrossRef]
- Sun, D.; Yu, H.; Li, Q. Starvation-induced changes in sex ratio involve alterations in sex-related gene expression and methylation in Pacific oyster Crassostrea gigas. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2023, 267, 110863. [Google Scholar] [CrossRef]
- Sun, D.; Yu, H.; Kong, L.; Liu, S.; Xu, C.; Li, Q. The role of DNA methylation reprogramming during sex determination and sex reversal in the Pacific oyster Crassostrea gigas. Int. J. Biol. Macromol. 2024, 259, 128964. [Google Scholar] [CrossRef]
- Wang, Z.; Ng, C.; Liu, X.; Wang, Y.; Li, B.; Kashyap, P.; Chaudhry, H.A.; Castro, A.; Kalontar, E.M.; Ilyayev, L. The ion channel function of polycystin-1 in the polycystin-1/polycystin-2 complex. EMBO Rep. 2019, 20, e48336. [Google Scholar] [CrossRef]
- Lemos, F.O.; Ehrlich, B.E. Polycystin and calcium signaling in cell death and survival. Cell Calcium 2018, 69, 37–45. [Google Scholar] [CrossRef]
- Field, S.; Riley, K.-L.; Grimes, D.T.; Hilton, H.; Simon, M.; Powles-Glover, N.; Siggers, P.; Bogani, D.; Greenfield, A.; Norris, D.P. Pkd1l1 establishes left-right asymmetry and physically interacts with Pkd2. Development 2011, 138, 1131–1142. [Google Scholar] [CrossRef]
- Deguchi, R.; Takeda, N.; Stricker, S.A. Calcium signals and oocyte maturation in marine invertebrates. Int. J. Dev. Biol. 2015, 59, 271–280. [Google Scholar] [CrossRef]
- Castelli, M.A.; Whiteley, S.L.; Georges, A.; Holleley, C.E. Cellular calcium and redox regulation: The mediator of vertebrate environmental sex determination? Biol. Rev. 2020, 95, 680–695. [Google Scholar] [CrossRef]
- Wilson, C.A.; High, S.K.; McCluskey, B.M.; Amores, A.; Yan, Y.-l.; Titus, T.A.; Anderson, J.L.; Batzel, P.; Carvan III, M.J.; Schartl, M. Wild sex in zebrafish: Loss of the natural sex determinant in domesticated strains. Genetics 2014, 198, 1291–1308. [Google Scholar] [CrossRef]
- Triay, C.; Conte, M.A.; Baroiller, J.-F.; Bezault, E.; Clark, F.E.; Penman, D.J.; Kocher, T.D.; D’Cotta, H. Structure and Sequence of the Sex Determining Locus in Two Wild Populations of Nile Tilapia. Genes 2020, 11, 1017. [Google Scholar] [CrossRef] [PubMed]
- Taslima, K.; Khan, M.G.; McAndrew, B.J.; Penman, D.J. Evidence of two XX/XY sex-determining loci in the Stirling stock of Nile tilapia (Oreochromis niloticus). Aquaculture 2021, 532, 735995. [Google Scholar] [CrossRef]
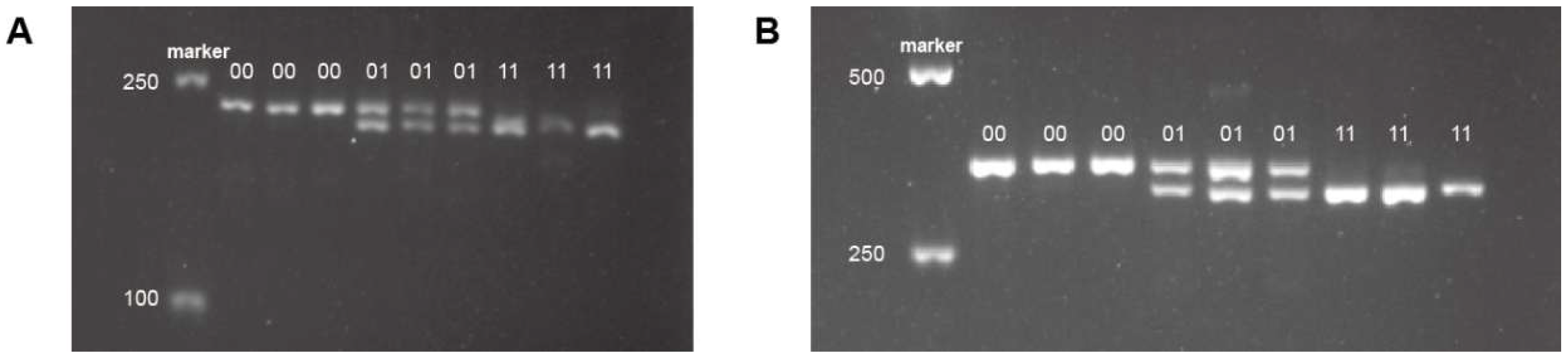
| Genotype | FO-3 | FO-4 | ||
|---|---|---|---|---|
| Male | Female | Male | Female | |
| 0/0 | 4 | 25 | 8 | 27 |
| 0/1 | 22 | 5 | 22 | 3 |
| 1/1 | 4 | 0 | 0 | 0 |
| Rank | Chromosome | Position | GWAS | FST | Male Frequency | Female Frequency | Genomic Region | Gene Name |
|---|---|---|---|---|---|---|---|---|
| 1 | 9 | 8,921,925 | 19.23 | 0.36 | 85.71% | 10.34% | intron | PKD1L1 |
| 2 | 9 | 9,358,715 | 18.36 | 0.44 | 85.71% | 8.62% | intron | LOC128164001 |
| 3 | 9 | 10,038,488 | 17.26 | 0.34 | 87.50% | 15.52% | - | - |
| 4 | 9 | 8,410,372 | 17.23 | 0.41 | 87.50% | 8.62% | upstream_gene | 5-HTRL |
| 5 | 9 | 9,574,321 | 16.70 | 0.34 | 85.71% | 12.07% | intron | SCP |
| 6 | 9 | 9,595,103 | 16.63 | 0.49 | 85.71% | 6.90% | upstream_gene | SCP |
| 7 | 9 | 8,914,529 | 16.57 | 0.45 | 83.93% | 8.62% | intron | PKD1L1 |
| 8 | 9 | 8,916,956 | 16.51 | 0.50 | 85.71% | 6.90% | intron | PKD1L1 |
| 9 | 9 | 8,887,643 | 16.20 | 0.33 | 87.50% | 10.34% | intron | PKD1L1 |
| 10 | 9 | 8,469,050 | 15.49 | 0.33 | 83.93% | 12.07% | intron | TWIST2 |
| 11 | 9 | 8,454,181 | 15.34 | 0.41 | 80.36% | 8.62% | - | - |
| 12 | 9 | 8,412,554 | 15.03 | 0.36 | 85.71% | 10.34% | upstream_gene | 5-HTRL |
| 13 | 9 | 8,412,558 | 15.03 | 0.36 | 85.71% | 10.34% | upstream_gene | 5-HTRL |
| 14 | 9 | 9,800,638 | 14.67 | 0.38 | 80.36% | 8.62% | upstream_gene | CCKRa |
| 15 | 9 | 9,800,641 | 14.52 | 0.39 | 80.36% | 8.62% | upstream_gene | CCKRa |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.; Ning, Y.; Li, L.; Wan, Q.; Li, S.; Yao, Y.; Tang, C.; Wu, Q.; Guo, X.; Qi, J.; et al. Sex-Associated Indels and Candidate Gene Identification in Fujian Oyster (Magallana angulata). Fishes 2025, 10, 438. https://doi.org/10.3390/fishes10090438
Han Y, Ning Y, Li L, Wan Q, Li S, Yao Y, Tang C, Wu Q, Guo X, Qi J, et al. Sex-Associated Indels and Candidate Gene Identification in Fujian Oyster (Magallana angulata). Fishes. 2025; 10(9):438. https://doi.org/10.3390/fishes10090438
Chicago/Turabian StyleHan, Yi, Yue Ning, Ling Li, Qijuan Wan, Shuqiong Li, Ying Yao, Chaonan Tang, Qisheng Wu, Xiang Guo, Jianfei Qi, and et al. 2025. "Sex-Associated Indels and Candidate Gene Identification in Fujian Oyster (Magallana angulata)" Fishes 10, no. 9: 438. https://doi.org/10.3390/fishes10090438
APA StyleHan, Y., Ning, Y., Li, L., Wan, Q., Li, S., Yao, Y., Tang, C., Wu, Q., Guo, X., Qi, J., Ke, Y., Ge, H., & Cai, M. (2025). Sex-Associated Indels and Candidate Gene Identification in Fujian Oyster (Magallana angulata). Fishes, 10(9), 438. https://doi.org/10.3390/fishes10090438






