Abstract
The turnip and the Chinese cabbage belong to the Brassica rapa subspecies, yet they have evolved marked differences in morphology. The turnip has a distinct swelled taproot, while the Chinese cabbage has a big leafy head. The turnip’s taproot is developed mainly by the hypocotyl. To explore the taproot formation, we firstly compared the vascular structure of the hypocotyl during the early developmental stages of the turnip and the Chinese cabbage, finding that there were observable differences in the number of xylem cells and the cell-wall lignification in the hypocotyl vascular tissues after the transition from primary to secondary growth. Laccases (LAC) play an important role in lignification by polymerizing monolignols in the cell wall, however, it is not clear whether differences in the lignification levels in the hypocotyl xylem cell walls are related to the genetic variations of the LAC gene family, between the turnip and the Chinese cabbage. Therefore, we systematically characterized the LAC genes from the turnip and the Chinese cabbage, and 27 LAC genes were identified in each. These LAC genes can be divided into six groups, and each LAC in the turnip is closely adjacent to that in the Chinese cabbage. Gene structure, conserved motif, and chromosomal localization were highly conserved between the turnip and the Chinese cabbage. We also compared the expression pattern of the laccases in the different tissues and hypocotyl’s early development stage, and the results clearly showed the different profiles between the turnip and the Chinese cabbage. Following a comprehensive analysis of these results, we predicted that LAC17.1 and LAC17.3 are two candidate genes that participate in the regulation of lignin synthesis during taproot formation. Our results provide a valuable clue for uncovering the regulation mechanism of the lower lignification level in the turnip’s hypocotyl and fundamental information for further studies of the LAC gene family in Brassica rapa.
1. Introduction
Brassica plants belong to the Brassicaceae family, including the six most cultivated species. According to the triangle of U theory, these six plants can be divided into two groups, the ancestral diploid species, B. rapa (AA, n = 10) B. nigra (BB, n = 8), and B. oleracea (CC, n = 9) as well as three derived ones, B. juncea (AABB, n = 18), B. napus (AACC, n = 19), and B. carinata (BBCC, n = 17) [1]. These plants are valuable resources of vegetables and oil, collectively contributing 12% of the world’s edible vegetable-oil production [2]. The Chinese cabbage (Brassica rapa var. pekinensis) and the turnip (Brassica rapa var. rapa) are two important vegetables in China. The main edible part of the Chinese cabbage is its leafy head, while that of the turnip is its taproot. The taproot is developed mainly from the hypocotyl and, also, partially from the root [3]. The taproot of the turnip is the main organ for edible and medicinal purposes. Thus, the development of hypocotyls directly affects the quality and application value of plants. Previous studies show lignification could affect the tuber and tuberous root formation [3], and our results also demonstrated that gibberellic acid (GA) could induce lignin deposition in the hypocotyl xylem and resulted in the inhibition of taproot formation [4]. Therefore, it is necessary to figure out the lignification process of xylem tissue.
Laccases (LACs) are widely found from bacteria to high plants [5,6,7,8,9]. Recently, researchers found that LACs are expressed in lignified cells of plants [10] and catalyze lignin formation by polymerizing monolignols [11]. Many studies have shown the relationship between LACs and cell lignification [12,13]. AtLAC4, AtLAC8, AtLAC15, AtLAC11, and AtLAC17 of the LAC family are expressed in highly lignified tissues, such as the seed coat, stem, root, pollen grain, and cell wall. Genetic experiments proved that LACs played a critical and nonredundant role in lignin polymerization during vascular development [14,15]. In cotton, LACs affected cell elongation, lignification, and pigmentation, and are closely related to the economic value of cotton fiber [16]. LACs were also found involved in other aspects of plants life, such as biotic or abiotic stress [17,18,19,20,21].
In this study, we compared the hypocotyl vascular structure at the early development stages between the turnip and the Chinese cabbage through free-hand section. We found that the xylem cells in the turnip’s hypocotyl expanded rapidly once the vascular development was transformed from primary growth to secondary growth, and the lignification of the xylem in the turnip significantly decreased, which was opposite to the Chinese cabbage, implying a correlation between lignification and taproot formation. In order to explore the lignification of the xylem during taproot formation, we researched and identified 27 LAC genes in both the turnip and the Chinese cabbage. We analyzed the phylogeny, gene structure, conserved motif, chromosomal localization, and the expression pattern in different tissues and the early hypocotyl development stages. Understanding the LAC gene family genome-wide and clarifying the expression profiles during the lignification of xylem cells will be beneficial to optimize and improve the quality and economic value of the turnip through bioengineering technology.
2. Materials and Methods
2.1. Plant Growth, Sample Collection, and Preparation
Chinese cabbage seeds (Brassica rapa var. pekinensis) were obtained from Liangpin Agricultural Science and Technology Development Co., Ltd. (Harbin, Heilongjiang Province, China), and turnip seeds (Brassica rapa var. rapa) were obtained from Lhasa, Tibet, China (KTRG-B17). The seeds were planted in plastic pots containing humus soil and were grown in a greenhouse at 22 °C with a 16 h light/8 h dark cycle at the Kunming Institute of Botany, Chinese Academy of Sciences. From the fourth day after sowing, the hypocotyls were collected every other day and were cut freehand into slices. Hypocotyls from plants growing for 8 days, 15 days, 22 days, 29 days, and 36 days were collected, respectively, and were frozen rapidly in liquid nitrogen for RNA extraction.
2.2. Free-Hand Section
The hypocotyls of the turnip and the Chinese cabbage were sampled every other day from the fourth day after planting and were washed with distilled water and cut freehand into sections. The thin sections were stained with Safranin O-Fast Green. After staining, the slides were checked under a Leica DM1000 microscope.
2.3. Identification of the LAC Proteins in Turnip and Chinese Cabbage
To identify the LAC proteins in the turnip and the Chinese cabbage, we first downloaded the Arabidopsis thaliana LAC proteins from TAIR (https://www.arabidopsis.org, accessed on 21 November 2020), which have been reported previously [6]. These Arabidopsis proteins were then queried, and Protein BLAST was used search for LAC proteins of Chinese cabbage and turnip against the Brassica rapa var. pekinensis protein database at NCBI (https://www.ncbi.nlm.nih.gov/, GenBank accession: GCA_008629595.1, accessed on 22 November 2020) or the turnip protein database from the genome assembly data, which is deposited in the Genome Sequence Archive in the BIG Data Center under accession numbers CRA005412 and GWHBFXQ00000000. In total, 27 Chinese cabbage LAC proteins and 27 turnip LAC proteins were identified and named BraLAC1–BraLAC22 and BrrLAC1–BrrLAC22, respectively, according to their Arabidopsis thaliana orthologs. These proteins were reconfirmed as belonging to the LACCASE superfamily, by analyzing them online with Interpro (https://www.ebi.ac.uk/interpro/, accessed on 22 November 2020) and CD-search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 22 November 2020). The relative molecular weight, isoelectric point, hydrophobicity, and other physical and chemical properties of the predicted amino acid were analyzed by the ProtParam online tool (http://expasy.org/tools, accessed on 22 November 2020).
2.4. Phylogenetic, Motif Analysis, and Gene Exon/Intron Structure Determination
The amino-acid-sequence alignments of the BraLACs and BrrLACs were performed by ClustalW, for the MEGA 7.0 software (available online https://megasoftware.net/, accessed on 25 November 2020), with default settings. Consensus trees were generated with 1000 bootstrap replicates. Conserved motifs were analyzed online on the MEME Suite (http://meme-suite.org/, accessed on 25 November 2020) with the maximum 15 motifs. The Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn/, accessed on 25 November 2020) online tool was used to analyze gene structure analysis, mainly the distribution of introns and exons.
2.5. Phylogenetic-Tree Construction of LACs among Different Species
The LAC amino acid sequences of 17 Arabidopsis thaliana, 27 Oryza sativa, and 52 Populus trichocarpa were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/, accessed on 28 November 2020). An un-rooted neighbor joining (NJ) was constructed using the distance-based adjacency method, in MEGA 7.0 software with 1000 bootstrap replicates. The classification of BrrLACs and BraLACs into a subfamily was based on that of LACs in Arabidopsis.
2.6. Chromosome Location of LAC Genes in Turnip and Chinese Cabbage
The chromosome location data of the Chinese cabbage was download from NCBI (https://www.ncbi.nlm.nih.gov/, accessed on 28 November 2020), and the chromosome location data of the turnip was from our database. The chromosome location maps of the BrrLACs and BraLACs were generated with TBtools [22].
2.7. Total RNA Isolation and Gene Expression of BrrLACs and BraLACs
The expression levels of BrrLACs and BraLACs were evaluated by the quantitative real-time PCR (qPCR) method. The hypocotyl materials of the turnip and the Chinese cabbage were sampled in five growth periods (8, 15, 22, 29, 36 days) after sowing. Total RNA was extracted using TRIzolTM (Invitrogen, Carlsbad, CA, USA) and treated with DNase I to erase genome DNA. After checking the quality and quantity of total RNA by 1% agrose gel electrophoresis and a NanoDrop™ 1000 Spectrophotometer (Thermo Fisher, Waltham, MA, USA), 1 µg of RNA was used to synthesize cDNA with M-MLV reverse transcriptase (Promega, Madison, WI, USA). A total of 20 μL reaction mixture was used, containing 10 μL SYBR GreenER qPCR SuperMix (Thermo Fisher, USA), 0.4 μL of each primer (10 μM), 1 μL of 10-fold dilution of cDNA, and 0.4 μL ROX Reference Dye. RT-qPCR was performed on the StepOnePlus™ Real-Time PCR System (Applied Biosystems, Waltham, MA, USA). Tubulin-α gene was used as the internal reference gene. The relative expression levels of BrrLACs and BraLACs were analyzed with the 2−ΔΔCT method [23]. The primers used for qPCR are shown in Supplementary Table S1.
2.8. Statistical Analysis
All the experimental data were calculated and analyzed using ANOVA and SPSS 17.0 software (SPSS Inc., Chicago, DE, USA, available online https://www.ibm.com/analytics/spss-statistics-software accessed on 22 November 2020). Graphs of the standard curves were plotted using GraphPad Prism (GraphPad Prism Software, Inc., San Diego, CA, USA), and the heat map was constructed in R Studio 1.4 (RStudio, Inc., Boston, MA, USA, available online https://www.rstudio.com accessed on 22 November 2020).
3. Results
3.1. Hypocotyl Anatomical Characteristics during Early Developmental Stage in Turnip and Chinese Cabbage
The turnip and the Chinese cabbage belong to the Brassica rapa subspecies, but they display distinct phenotypes. The Chinese cabbage has a big leafy head, while the turnip develops a swelling taproot, mainly from the hypocotyl. In order to explore whether there are differences in the hypocotyl vascular structure, we dissected and compared the hypocotyls of the turnip and the Chinese cabbage, collected during the early developmental stages, by freehand section. According to the characteristics of the vascular structure from the slice sections, we divided the early development period into three stages: Stage I represents primary growth, Stage II represents initial secondary growth, and Stage III represents secondary growth (Figure 1). In Stage I, no obvious differences were observed in the morphology and vascular structure of the hypocotyls between the turnip and the Chinese cabbage. In Stage II, the degree of xylem lignification in the turnip and the Chinese cabbage both gradually increased, and the lignified cells mainly distributed the center of the xylem. The edge of the xylem cell group in the Chinese cabbage was clear, while that in the turnip appeared as a zigzag pattern. With the development of hypocotyl in Stage III, the hypocotyl diameter of the turnip significantly increased compared with the Chinese cabbage, while the hypocotyl vascular structures were distinctly different between the turnip and the Chinese cabbage. Moreover, the lignification in the xylem of the Chinese cabbage was further enhanced, while the lignification of the xylem in the turnip hypocotyl sharply decreased. As a result, parenchyma cells occupied most portions of the taproot xylem, and only a few lignified cells were distributed radially in the xylem tissue and the middle of the vascular tissue. These results implied that lignification of the xylem cells may correlate with taproot formation in the turnip.
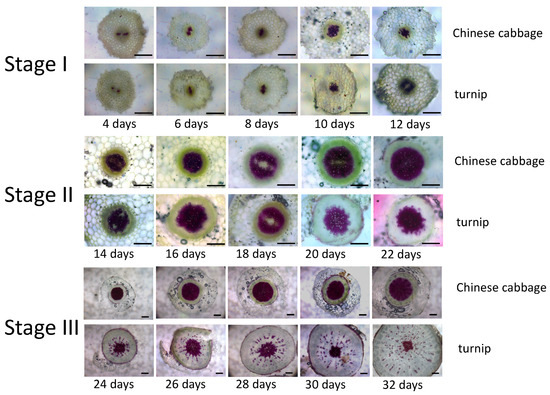
Figure 1.
Changes of hypocotyl vascular structure during early development stage in turnip and Chinese cabbage. The free-hand sections of hypocotyl were made from 4–32-day-old seedlings and stained with Safranin O-Fast Green. Bar = 15 µm.
3.2. Identification of LACCASE (LAC) in Turnip and Chinese Cabbage
We have shown the significant differences in the lignification patterns of the xylem, between the turnip and the Chinese cabbage. Previous studies have shown that plant LACs play important roles in lignin biosynthesis. LACs catalyze the last step of monolignols oxidation and polymerization in lignin synthesis, by directly using O2 to oxidize all types of monolignols. To explore the possible different roles of LACs in lignin biosynthesis between the turnip and the Chinese cabbage, we firstly searched out 27 LAC proteins from the turnip and the Chinese cabbage, named BrrLACs and BraLACs, respectively, by blasting the Arabidopsis LAC proteins against the genomes of the turnip and the Chinese cabbage. As shown in Table 1, the CDS length of the LAC ranged from 1623 bp to 1776 bp in the turnip and the Chinese cabbage. Most homologous LAC genes in the turnip and the Chinese cabbage have the same CDS length, except for LAC1, LAC4.1, and LAC8.1. The computed molecular masses of LAC proteins ranged from 60.2 kDa to 65.9 kDa in the turnip and 60.1 kDa to 66.0 kDa in the Chinese cabbage, and the theoretical isoelectric points (pI) ranged from 5.99 to 9.71 in the turnip and 5.96 to 9.73 in the Chinese cabbage. The grand average of hydropathicity (GRAVY) was negative for most of the BraLAC and BrrLAC proteins ranging from −0.301 to −0.034, indicating that they were hydrophilic. Only BraLAC7 and BrrLAC7 proteins were hydrophobic, possessing a positive GRAVY value. The predicted-protein instability index indicated that most LAC proteins were stable (instability index < 40), while BrrLAC3.1, BrrLAC3.2, BrrLAC9, and BrrLAC13 in the turnip, and BraLAC3.1, BraLAC3.2, BraLAC9, and BraLAC13 in the Chinese cabbage showed low protein stability (instability index > 40) (Table 1).

Table 1.
Physical and chemical properties of the predicted proteins of BrrLAC genes.
3.3. Phylogenetic Analysis
To assess the evolutionary relationship among the 27 BrrLACs in the turnip and the 27 BraLACs in the Chinese cabbage, we further searched the LAC proteins of three other species (Arabidopsis thaliana, Oryza sativa, and Populus trichocarpa) from the NCBI database. A phylogenetic tree of 150 LAC proteins from these species was constructed by the neighbor-joining method, with 1000 bootstrap replicates, using MEGA 7.0 software. These LAC proteins were clustered into seven groups: Group I to Group VII (Figure 2). BrrLACs and BraLACs were clustered together in all groups, and they were most closely clustered with LACs in Arabidopsis, except for BrrLAC22 and BraLAC22. Interestingly, LAC22 in the turnip and the Chinese cabbage was clustered with PtrLAC4.1 and PtrLAC4.2 in Poplar, suggesting they may have the similar functions. Group III and Group IV contained LACs from the turnip, Chinese cabbage, Arabidopsis, and Poplar but not from rice, while Group VII contained LACs only from rice, suggesting these LACs might have a distinct function in monocotyledons and dicotyledons.

Figure 2.
Phylogenetic analysis of laccase proteins from turnip and Chinese cabbage. Phylogenetic tree was constructed using laccase proteins from turnip (27), Chinese cabbage (27), Arabidopsis thaliana (17), Oryza sativa (27), and Populus trichocarpa (52). Phylogenetic tree was constructed using MEGA 7.0, by the neighbor-joining method, with 1000 bootstrap replicates.
3.4. Gene Structure and Conserved Motif Analysis of LACs in Turnip and Chinese Cabbage
We performed the conserved motif scanning of LAC proteins in the turnip and the Chinese cabbage, arranging them according to the LAC phylogenetic tree (Figure 3A). The number and arrangement of the conserved motif of LAC proteins are shown in Figure 3B. There were 15 conserved motifs to be found in most LAC proteins, except for BrrLAC8.1/8.2 and BraLAC8.1/8.2, which lacked motif 5, as well as BrrLAC9 and BraLAC9, which lacked motif 1 and motif 5. All these LAC proteins had a relatively conserved motif in their C-terminus. In most cases, plants have three copper-binding domains. To explore whether the lack of motif 5 in BraLAC8.1/8.2 as well as the lack of motif 1 and motif 5 in LAC9 would affect the three copper-binding domains, we performed conserved-domain research through the Pfam data. As the results indicated (Supplementary Data), all LAC proteins in the Chinese cabbage and the turnip had three copper-binding domains.

Figure 3.
Phylogenetic, conserved motif, and gene architecture analysis of turnip and Chinese cabbage. (A) The phylogenetic tree of turnip and Chinese cabbage laccase proteins. (B) Conserved motif in laccase proteins. (C) Exon–intron architecture of turnip and Chinese cabbage laccase genes. Exons are represented by light blue boxes, and introns are represented with black lines.
Further, the exon–intron structures of BrrLACs and BraLACs were analyzed in GSDS. As shown in Figure 3C, the intron number of LACs in the turnip and the Chinese cabbage varied from 3 to 6. Most orthologous genes of LACs from the turnip and the Chinese cabbage had an equal number and approximate length of exons and introns. However, there are some exceptions. For examples, BraLAC4.1 had six introns while BrrLAC4.1 had five introns; BraLAC8.1 had six introns while BrrLAC8.1 had five introns; the first intron length in BraLAC10 was longer than that in BrrLAC10; the fifth intron length in BraLAC16 was longer than that in BrrLAC16; the third intron length in BrrLAC17.3 was longer than that in BraLAC17.3; and the third intron length in BrrLAC8.1 was longer than that in BraLAC8.1. Differences in intron numbers and lengths might affect the genes’ expression in the post-transcriptional level.
3.5. Chromosomal Localization
To determine the genome organization and distribution of BrrLACs and BraLACs on the chromosomes of the turnip and the Chinese cabbage, a gene chromosomal location map was constructed (Figure 4). The results showed that the 27 BrrLACs in the turnip were unevenly distributed in eight chromosomes in the turnip, and chromosome A02/05/10 contained the most (>5). Several BrrLAC genes were clustered in the chromosomes, such as BrrLAC8.2/10/11.1/12.1, BrrLAC3.2/4.2/5.1/6, and BrrLAC11.2/12/2/14. BraLACs in the Chinese cabbage had a similar chromosomal distribution compared with the BrrLACs in the turnip. Strangely, BraLAC13 was not in chromosome A07 as it was for BrrLAC13, isntead located in chromosome A10.

Figure 4.
Chromosomal locations of LAC genes for turnip (A) and Chinese cabbage (B). The chromosome numbers are shown on the left side of each chromosome. Locations of LACs on chromosomes are indicated according to the location of the genes and length of chromosomes. Chromosomal distances are given in bp.
3.6. Tissue-Specific Expression Patterns of Laccase Genes in Turnip and Chinese Cabbage
It was necessary to determine the expression patterns of LAC genes in the different tissues (leaf, stem, silique, root, and flower) of the turnip and the Chinese cabbage for predicting the possible function of laccases on turnip-taproot formation. As shown in Figure 5A,B most of the LAC genes showed a distinct tissue-specific expression pattern. In the turnip (Figure 5A), 19 out of 27 BrrLACs were highly expressed in stems, such as BrrLAC2, BrrLAC3.1, BrrLAC3.2, BrrLAC4.1, BrrLAC4.2, BrrLAC5.1, BrrLAC5.2, BrrLAC6, BrrLAC7, BrrLAC10, BrrLAC11.1, BrrLAC11.2, BrrLAC12.1, BrrLAC12.2, BrrLAC16, BrrLAC17.1, BrrLAC17.2, BrrLAC17.3, and BrrLAC22; 10 out of 27 BrrLAC genes were highly expressed in siliques, such as BrrLAC1, BrrLAC5.1, BrrLAC5.2, BrrLAC6, BrrLAC13, BrrLAC14, BrrLAC15.1, BrrLAC15.2, BrrLAC17.1, and BrrLAC17.2; and 7 out of 27 BrrLACs were highly expressed in roots (BrrLAC1, BrrLAC3.2, BrrLAC5.1, BrrLAC7, BrrLAC8.1, BrrLAC8.2, and BrrLAC9), while only two BrrLAC genes (BrrLAC6 and BrrLAC17.1) were highly expressed in the leaves, and most of the BrrLAC genes were not detected or expressed in a relatively low level in the flowers. More than half of the LAC genes in the Chinese cabbage had similar tissue-specific expression profiles to those in the turnip (Figure 5A). However, we also observed that the expression patterns of some LAC genes were differentiated between the Chinese cabbage and the turnip. LAC2, LAC4.1, LAC11.2, LAC16, LAC17.1, and LAC17.3 were expressed in the turnip siliques at low level, while they were highly expressed in the Chinese cabbage siliques. LAC8.1 and LAC8.2 were strongly expressed in the turnip root, but they were expressed at a relatively low level in the Chinese cabbage root. The expression of BrrLAC9 in the root and BrrLAC17.2 in the stem of the turnip was significantly higher than those in the Chinese cabbage. In the turnip, LAC11.1 were expressed in the leaf, stem, silique, and root, while LAC11.1 in the Chinese cabbage was only expressed in silique. In addition, we also found that almost all of the LAC genes were not expressed in the flowers in the turnip and the Chinese cabbage (Figure 5A,B).
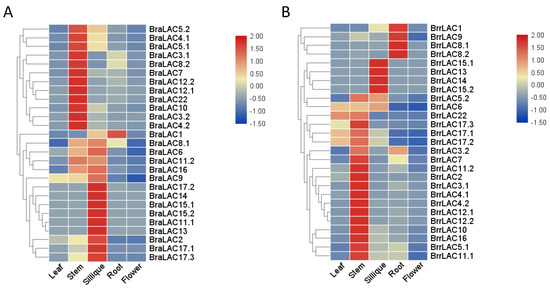
Figure 5.
Transcription pattern of LAC genes across different tissues (leaf, stem, silique, root, and flower) in Chinese cabbage (A) and turnip (B) by RT-qPCR. Leaves, stems, and roots were collected from 30-day-old seedlings, while siliques and flowers were collected from bolting plants. The relative expression level was normalized using TUB gene. Values are the mean and SD of three replicates.
3.7. Expression Analysis of LAC Genes during Early Hypocotyl Development Stage in Turnip and Chinese Cabbage
We have shown that the lignification of hypocotyl during the early development stage was remarkably different between the turnip and the Chinese cabbage. To explore whether LAC genes were responsible for such a difference, we checked the expression trends of the LAC genes in the turnip and the Chinese cabbage. According to the results of freehand slices, five growth points of plants after sowing were selected: 8 days, 15 days, 22 days, 29 days, and 36 days. The qPCR results were presented in Figure 6. In the turnip, the expression level of five LAC genes (BrrLAC 4.1/4.2/10/11.2/12.2) increased continually. Meanwhile, more than half of the LAC genes (BrrLAC2, BrrLAC3.1, BrrLAC3.2, BrrLAC5.1, BrrLAC5.2, BrrLAC6, BrrLAC7, BrrLAC8.1, BrrLAC8.2, BrrLAC11.1, BrrLAC12.1, BrrLAC13, BrrLAC16, BrrLAC17.1, BrrLAC17.3, and BrrLAC17.3) declined their expression when they reached the peak at 22 days, and, further, some of these LAC genes (BrrLAC2, BrrLAC3.1, BrrLAC3.2, BrrLAC6, BrrLAC7, BrrLAC8.1, BrrLAC8.2, BrrLAC11.1, BrrLAC12.1, BrrLAC13, BrrLAC17.1, and BrrLAC17.3) significantly decreased to relatively low levels. In the Chinese cabbage, 13 LAC genes (BraLAC4.1, BraLAC4.2, BraLAC5.1, BraLAC5.2, BraLAC6, BraLAC7, BraLAC11.2, BraLAC12.1, BraLAC12.2, BraLAC15.1, BraLAC16, BraLAC17.1, and BraLAC17.3) showed an increase in the expression level during the early development stages, maintaining a higher level after growing for 22 days. Eight BraLAC genes (BraLAC1, BraLAC2, BraLAC3.2, BraLAC4.1, BraLAC10, BraLAC14, BraLAC15.2, and BraLAC22) showed a rise first and then decreased in expression level. By comparing the expression profiles of LAC genes, three LAC genes (LAC6, LAC17.1, and LAC17.3) have similar trends with the lignin-content variation in the turnip and the Chinese cabbage.

Figure 6.
Transcription patterns of LAC genes in early developmental stages of turnip and Chinese cabbage. The hypocotyl of plants was collected at 8 days, 15 days, 22 days, 29 days, and 36 days after sowing. The relative expression level was normalized using TUB gene. Values are the mean and SE of three replicates.
4. Discussion
Ther turnip and the Chinese cabbage are both biennial Brassica plants. They are two variants of the same Brassica rapa species, but they have distinct morphological differences. The turnip has a unique swelling and edible taproot, which is mainly developed from its hypocotyl and root [24,25]. By comparing the anatomical structures of the hypocotyl of the turnip and the Chinese cabbage at early stages, we draw two conclusions: (1) the formation of the turnip taproot is the result of the increase in xylem cells; (2) most xylem cells are parenchyma cells with low-level lignification. Several studies have demonstrated that storage root formation is accompanied by the reduction in lignification [26,27,28]. Our previous study also proves that lignification is a restricting factor for taproot formation in the turnip. Therefore, further study on the regulation of lignification in the turnip taproot will be helpful to understand taproot expansion.
Laccases are multi-copper-oxidase enzymes that catalyze the oxidation of different compounds (phenolics and non-phenolics). Laccases participate in multiple physiology, biochemistry, and developmental processes including cell morphology, pigment formation, and second cell-wall biosynthesis. In this study, we comprehensively analyzed the laccase gene family in the turnip and the Chinese cabbage on a genome-wide scale. We comprehensively analyzed the laccase gene family in the turnip and the Chinese cabbage to compare the difference in phylogeny, gene structure, conserved motif, chromosomal localization, tissue-specific expression, and expression profile during the hypocotyl-development stage. Combining with the anatomical results, we predicted that expression regulation of BrrLAC17.1 and BrrLAC17.3 genes plays an important role in controlling the lignification level of the turnip hypocotyl during taproot formation.
Phylogenetic analysis showed that the LAC homologs between the turnip and the Chinese cabbage shared close relationships, and they were classified in the same groups with Arabidopsis. Compared with the homologous gene in Arabidopsis, a lot of LAC genes in the turnip and the Chinese cabbage have two–three copies, which was in line with the theory that Brassica rapa had experienced a whole-genome-triplication (WGT) event, approximately 9–15 million years ago [29,30,31]. Further analysis of the LAC motifs showed that the turnip and the Chinese cabbage shared a very similar protein-conserved sequence, which indicated that there was no obvious difference, at least in the protein function. In gene constructs, most BrrLAC genes and BraLAC genes showed similar alignment and length of exon and intron, but there are some exceptions. For example, there are six introns of LAC4.1 in the Chinese cabbage, while there are five introns in the turnip. The length of the third intron of LAC17.3 in the turnip was longer than that in the Chinese cabbage. In eukaryotes, the pre-mRNA would undergo RNA splicing, meaning removing non-coding introns from RNA transcripts to form mature RNA. As plenty of studies have demonstrated, the number and length of introns could be involved in RNA splicing and affect gene expression [32,33,34]. Therefore, the differences in the exon-intron organization of these LAC genes would be the potential reason responsible for the differential spatiotemporal expression between the turnip and the Chinese cabbage.
Significant differences in lignin content were observed in the hypocotyl vascular structure of the turnip and the Chinese cabbage during the early development stages. The lignification level of the xylem in the vascular tissue in the Chinese cabbage increased gradually, while that in the turnip decreased significantly after the transition from primary to secondary growth. With that in mind, we screened out three LAC genes (LAC6, LAC17.1, and LAC17.3), the expression profiles of which were consistent with the variant trends of the lignin content in the turnip and the Chinese cabbage. In Arabidopsis, peroxidases and laccases fulfil different functions in lignin polymerization. Specifically, AtLAC4, AtLAC11, and AtLAC17 are involved in monolignol polymerization, at least in the vascular tissue in Arabidopsis [14,15]. In Populus, PtrLAC2, PtrLAC3, PtrLAC14, and PtrLAC16 are homologous genes of AtLAC4 in Arabidopsis, having been proven to be active in catalyzing lignin polymerization [35,36,37,38]. Compared with Populus, the turnip and the Chinese cabbage have a closer genetic relationship to Arabidopsis. Therefore, LAC4.1, LAC4.2, LAC11.1, LAC11.2, LAC17.1, LAC17.2, and LAC17.3 are more likely to be involved in lignin synthesis in the turnip and the Chinese cabbage. Furthermore, most of these genes were expressed in the stems of the turnip and the Chinese cabbage, except for BraLAC4.1. Based on these facts, we predicted that LAC17.1 and LAC17.3 were two candidates participating in the regulation of lignin deposition in the hypocotyl during taproot formation in the turnip.
Though the expression pattern of LAC6 in the turnip and the Chinese cabbage had similar variant trends of lignin content, the function of LAC6 and its homolog in other species are not clear. In the phylogenetic tree, LAC6 also has a long distance from other LACs in the turnip, therefore, whether its function is related to turnip formation needs to be further studied.
5. Conclusions
Anatomical results of the turnip and the Chinese cabbage at early stages implied that lignification was involved in turnip-taproot formation. We, thus, analyzed the LAC genes of the turnip and the Chinese cabbage in the conserved motif, chromosome location, and spatiotemporal expression profiles. Our results indicated that LAC17.1 and LAC17.3 are two candidates that participate in the regulation of lignin synthesis during taproot formation. Further studies should be focused on the regulation mechanism of these two genes during the transition from primary to secondary growth in the turnip’s taproot.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae8060522/s1. Supplementary Data: Conserved-domain of LAC proteins in Chinese cabbage and turnip; Table S1: The qPCR primers for LAC genes of turnip and Chinese cabbage.
Author Contributions
J.W. performed the major part of this study; Y.L. and S.Y. helped to finish the experiments; J.W and C.W. wrote this manuscript; C.W. and Y.Y. designed the experiments. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by a major program of the National Natural Science Foundation of China (No. 31590823), the 13th Five-Year Informatization Plan of the Chinese Academy of Sciences (No. XXH13506), the Yunnan Provincial Science and Technology Department–Kunming Medical University Joint Project (No. 202001AY070001-018), and the Key Program for Basic Research of Yunnan Province (2018FA052).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nagaharu, U.; Nagaharu, N. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Labana, K.; Gupta, M. Importance and origin. In Breeding Oilseed Brassicas; Springer: Berlin/Heidelberg, Germany, 1993; pp. 1–7. [Google Scholar]
- Zierer, W.; Ruscher, D.; Sonnewald, U.; Sonnewald, S. Tuber and Tuberous Root Development. In Annual Review of Plant Biology; Merchant, S.S., Ed.; Springer: Berlin/Heidelberg, Germany, 2021; Volume 72, pp. 551–580. [Google Scholar]
- Liu, Y.Y.; Wen, J.; Ke, X.C.; Zhang, J.; Sun, X.D.; Wang, C.T.; Yang, Y.P. Gibberellin inhibition of taproot formation by modulation of DELLA-NAC complex activity in turnip (Brassica rapa var. rapa). Protoplasma 2021, 258, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Diamantidis, G.; Effosse, A.; Potier, P.; Bally, R. Purification and characterization of the first bacterial laccase in the rhizospheric bacterium Azospirillum lipoferum. Soil Biol. Biochem. 2000, 32, 919–927. [Google Scholar] [CrossRef]
- Polashock, J.J.; Anagnostakis, S.L.; Milgroom, M.G.; Hillman, B.I. Isolation and Characterization of a Virus-Resistant Mutant of Cryphonectria-Parasitica. Curr. Genet. 1994, 26, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Fernandez Larrea, J.; Stahl, U. Isolation and characterization of a laccase gene from Podospora anserina. Mol. Gen. Genet. 1996, 252, 539–551. [Google Scholar]
- Wahleithner, J.A.; Xu, F.; Brown, K.M.; Brown, S.H.; Golightly, E.J.; Halkier, T.; Kauppinen, S.; Pederson, A.; Schneider, P. The identification and characterization of four laccases from the plant pathogenic fungus Rhizoctonia solani. Curr. Genet. 1996, 29, 395–403. [Google Scholar] [CrossRef]
- Savinova, O.S.; Moiseenko, K.V.; Vavilova, E.A.; Chulkin, A.M.; Fedorova, T.V.; Tyazhelova, T.V.; Vasina, D.V. Evolutionary Relationships Between the Laccase Genes of Polyporales: Orthology-Based Classification of Laccase Isozymes and Functional Insight from Trametes hirsute. Front. Microbiol. 2019, 10, 152. [Google Scholar] [CrossRef] [Green Version]
- Bao, W.; O’Malley, D.M.; Whetten, R.; Sederoff, R.R. A laccase associated with lignification in loblolly pine xylem. Science 1993, 260, 672–674. [Google Scholar] [CrossRef]
- Liang, M.; Haroldsen, V.; Cai, X.; Wu, Y. Expression of a putative laccase gene, ZmLAC1, in maize primary roots under stress. Plant Cell Environ. 2006, 29, 746–753. [Google Scholar] [CrossRef]
- O’Malley, D.M.; Whetten, R.; Bao, W.; Chen, C.L.; Sederoff, R.R. The role of of laccase in lignification. Plant J. 1993, 4, 751–757. [Google Scholar] [CrossRef]
- Mayer, A.M.; Staples, R.C. Laccase: New functions for an old enzyme. Phytochemistry 2002, 60, 551–565. [Google Scholar] [CrossRef]
- Berthet, S.; Demont-Caulet, N.; Pollet, B.; Bidzinski, P.; Cezard, L.; Le Bris, P.; Borrega, N.; Herve, J.; Blondet, E.; Balzergue, S.; et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 2011, 23, 1124–1137. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Nakashima, J.; Chen, F.; Yin, Y.B.; Fu, C.X.; Yun, J.F.; Shao, H.; Wang, X.Q.; Wang, Z.Y.; Dixon, R.A. LACCASE Is Necessary and Nonredundant with PEROXIDASE for Lignin Polymerization during Vascular Development in Arabidopsis. Plant Cell 2013, 25, 3976–3987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balasubramanian, V.K.; Rai, K.M.; Thu, S.W.; Hii, M.M.; Mendu, V. Genome-wide identification of multifunctional laccase gene family in cotton (Gossypium spp.); expression and biochemical analysis during fiber development. Sci. Rep. 2016, 6, 34309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Q.; Min, L.; Yang, X.; Jin, S.; Zhang, L.; Li, Y.; Ma, Y.; Qi, X.; Li, D.; Liu, H. Laccase GhLac1 modulates broad-spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and jasmonic acid synthesis. Plant Physiol. 2018, 176, 1808–1823. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.Y.; Lee, C.; Hwang, S.-G.; Park, Y.C.; Lim, H.L.; Jang, C.S. Overexpression of the OsChI1 gene, encoding a putative laccase precursor, increases tolerance to drought and salinity stress in transgenic Arabidopsis. Gene 2014, 552, 98–105. [Google Scholar] [CrossRef]
- Ping, X.K.; Wang, T.Y.; Lin, N.; Di, F.F.; Li, Y.Y.; Jian, H.J.; Wang, H.; Lu, K.; Li, J.N.; Xu, X.F.; et al. Genome-Wide Identification of the LAC Gene Family and Its Expression Analysis Under Stress in Brassica napus. Molecules 2019, 24, 1985. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Li, G.; Zheng, K.J.; Zhu, X.B.; Ma, J.J.; Wang, D.M.; Tang, K.Q.; Feng, X.X.; Leng, J.T.; Yu, H.; et al. The Soybean Laccase Gene Family: Evolution and Possible Roles in Plant Defense and Stem Strength Selection. Genes 2019, 10, 701. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.Q.; Luo, L.; Wang, X.X.; Shen, Z.G.; Zheng, L.Q. Comprehensive Analysis of Rice Laccase Gene (OsLAC) Family and Ectopic Expression of OsLAC10 Enhances Tolerance to Copper Stress in Arabidopsis. Int. J. Mol. Sci. 2017, 18, 209. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Y.; Basset, N.; Petrasch, S.; Zhang, N.W.; Bucher, J.; Shen, S.X.; Zhao, J.J.; Bonnema, G. What makes turnips: Anatomy, physiology and transcriptome during early stages of its hypocotyl-tuber development. Hortic. Res.-Engl. 2019, 6, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.W.; Zhao, J.J.; Lens, F.; de Visser, J.; Menamo, T.; Fang, W.; Xiao, D.; Bucher, J.; Basnet, R.K.; Lin, K.; et al. Morphology, Carbohydrate Composition and Vernalization Response in a Genetically Diverse Collection of Asian and European Turnips (Brassica rapa subsp rapa). PLoS ONE 2014, 9, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, V.; Sergeeva, L.; Ligterink, W.; Aloni, R.; Zemach, H.; Doron-Faigenboim, A.; Yang, J.; Zhang, P.; Shabtai, S.; Firon, N. Gibberellin Promotes Sweetpotato Root Vascular Lignification and Reduces Storage-Root Formation. Front. Plant Sci. 2019, 10, 1320. [Google Scholar] [CrossRef]
- Wang, G.L.; Que, F.; Xu, Z.S.; Wang, F.; Xiong, A.S. Exogenous gibberellin enhances secondary xylem development and lignification in carrot taproot. Protoplasma 2017, 254, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Que, F.; Wang, G.L.; Feng, K.; Xu, Z.S.; Wang, F.; Xiong, A.S. Hypoxia enhances lignification and affects the anatomical structure in hydroponic cultivation of carrot taproot. Plant Cell Rep. 2018, 37, 1021–1032. [Google Scholar] [CrossRef]
- Wang, X.W.; Wang, H.Z.; Wang, J.; Sun, R.F.; Wu, J.; Liu, S.Y.; Bai, Y.Q.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, U1035–U1157. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.; Wu, J.; Wang, X.W. Genome triplication drove the diversification of Brassica plants. Hortic. Res.-Engl. 2014, 1, 14024. [Google Scholar] [CrossRef] [Green Version]
- Beilstein, M.A.; Nagalingum, N.S.; Clements, M.D.; Manchester, S.R.; Mathews, S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 18724–18728. [Google Scholar] [CrossRef] [Green Version]
- Sena, J.S.; Giguere, I.; Boyle, B.; Rigault, P.; Birol, I.; Zuccolo, A.; Ritland, K.; Ritland, C.; Bohlmann, J.; Jones, S.; et al. Evolution of gene structure in the conifer Picea glauca: A comparative analysis of the impact of intron size. BMC Plant Biol. 2014, 14, 95. [Google Scholar]
- Heyn, P.; Kalinka, A.T.; Tomancak, P.; Neugebauer, K.M. Introns and gene expression: Cellular constraints, transcriptional regulation, and evolutionary consequences. Bioessays 2015, 37, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Swinburne, I.A.; Miguez, D.G.; Landgraf, D.; Silver, P.A. Intron length increases oscillatory periods of gene expression in animal cells. Gene Dev. 2008, 22, 2342–2346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranocha, P.; Chabannes, M.; Chamayou, S.; Danoun, S.; Jauneau, A.; Boudet, A.M.; Goffner, D. Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol. 2002, 129, 145–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, S.F.; Fan, C.F.; Li, X.H.; Li, Y.; Hu, J.; Li, C.F.; Luo, K.M. LACCASE14 is required for the deposition of guaiacyl lignin and affects cell wall digestibility in poplar. Biotechnol. Biofuels 2020, 13, 197. [Google Scholar] [CrossRef]
- Bryan, A.C.; Jawdy, S.; Gunter, L.; Gjersing, E.; Sykes, R.; Hinchee, M.A.; Winkeler, K.A.; Collins, C.M.; Engle, N.; Tschaplinski, T.J.; et al. Knockdown of a laccase in Populus deltoides confers altered cell wall chemistry and increased sugar release. Plant Biotechnol. J. 2016, 14, 2010–2020. [Google Scholar] [CrossRef]
- Liu, Y.D.; Cao, S.; Liu, X.T.; Li, Y.; Wang, B.; Sun, Y.; Zhang, C.; Guo, X.R.; Li, H.; Lu, H. PtrLAC16 plays a key role in catalyzing lignin polymerization in the xylem cell wall of Populus. Int. J. Biol. Macromol. 2021, 188, 983–992. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).