Abstract
Fruit traits are important in pepper (Capsicum annuum L.) and affect its quality and yield. These traits are controlled by quantitative trait loci (QTLs). In this study, we identified many major QTLs that control fruit length (Ftl), fruit diameter (Ftd), fruit shape (Fts), fruit weight (Ftw) and locule number (Lcn) in the F2 and F2:3 populations developed from the QTL mapping of GS6 (P1) and Qiemen (P2). A total of 111 simple sequence repeats and insertion/deletion markers were utilized to construct a linkage map with 12 linkage groups over a length of 1320.72 cM. An inclusive composite interval mapping analysis indicated that many QTLs were detected and included ftl2.1, ftd2.1, fts1.1, ftw2.1 and lcn1.1. As a novel QTL, lcn1.1 was located between HM1112 and EPMS709, and the genetic distance was 3.18 cM covering 60 predicted genes. Within the region, we identified Capana01g004285 as a candidate gene by functional annotation and expression analysis and found that it encodes the BREVIS RADIX (BRX) protein. Knockdown of CaBRX through the virus-induced gene silencing approach in GS6 reduced the number of locules and influenced the expressions of genes related to flower and locule development, suggesting that CaBRX plays an important function in the development of locules.
1. Introduction
Pepper (Capsicum spp.) is a member of the Solanaceae family and is a vital vegetable globally [1]. Approximately 31 species of Capsicum are recognized in the genus, and five have been domesticated. The cultivated species include Capsicum annuum L., Capsicum chinense Jacq., Capsicum frutescens L., Capsicum baccatum L., and Capsicum pubescens Ruiz. & Pavon [2]. C. annuum is the oldest domesticated crop in the Americas and also the most widely grown crop in the world [3]. Pepper fruits are widely used in the food and chemical industries owing to their unique pungency and rich nutrients [3]. A total of 38.03 million tons were produced worldwide in 2019 [4]. The yield is the typical quantitative trait and is influenced by many factors, including fruit dimensions, the number of fruits, and fruit weight [5]. In spite of the effects of the environment, the traits of fruit are largely determined by their heredity. Recent advances in quantitative trait loci (QTLs) mapping have identified many genes that contribute to the control of traits in fruit, such as plant height, fruit weight, fruit shape and locule number in pepper [6,7].
As the major QTL of plant height in peppers, ph3.1 explained more than 29% of the total phenotypic variation in two experiments. In the QTL analysis of fruit weight (Ftw), fw2.1 was found to be the main QTL that affects fruit weight and is located on chromosome 2 [6]. Since pepper and tomato (Solanum lycopersicum L.) belong to the Solanaceae family, many researchers compared the QTLs of pepper with those of tomato to promote research on pepper. Zygier et al. showed that fw2.1 explained 62% of the trait variation and is tightly linked to Ovate, which is related to fruit shape (Fts) in tomato [8]. fw4.1 and fw4.2 were identified as major fruit weight QTLs on chromosome 4 [8]. Tomato fw2.2, the first vegetable fruit weight QTL that was cloned, had a significant effect in tomato fruit, although it is not effective in pepper [8,9]. fs3.1 is the major QTL related to fruit shape, which explained 60% of the phenotypic variation on chromosome 3 in pepper [6]. fs2.1, similar to fw2.1, also localized to Ovate. CaOvate, the Ovate-like gene, is likely to be involved in the development of fruit shape [10]. Chunthawodtiporn et al. indicated three candidate genes (Big Brother, Ovate, and KLUH/CYP78A5) on chromosomes 1, 2, and 3 that may have major effects on fruit size in pepper [11]. In addition, pepper fs3.1 and fs10.1 were not consistent with those in tomato, implying that divergent selection occurred during their domestication [8]. Two fruit length (Ftl) QTLs (fl2.1 and fl3.1) and four fruit diameter (Ftd) QTLs (fd1.1, fd2.1, fd4.1 and fd11.2) in pepper may correspond to tomato QTLs [5]. Moreover, Arjun et al. identified two fruit length QTLs designated paufl2.1 and paufl2.2, which both explained 21.78% of the phenotypic variation [12].
The locule number (Lcn) is one of the important agronomic traits that contributes to fruit size [13]. In pepper, there are few studies related to locules, which grow from flower carpels [14]. Nine Nlo (number of fruit locules) QTLs were identified between ‘Yolo Wonder’ (YW) and ‘Criollo de Morelos 334’ (CM334), including Nlo2.1, Nlo8.1, Nlo12.1 and Nlo12.2 [7]. Nlo2.1 might correspond to lcn2.1, which is the major locule number QTL on chromosome 2 and explained 30% of the phenotypic variation in tomato fruit [7]. The meristem plays essential roles in growth and developmental processes, particularly those that affect the number of carpels [15]. WUSCHEL (WUS) and CLAVATA3 (CLV3) proteins negatively regulate the function of meristems in Arabidopsis thaliana [16]. Additionally, AGAMOUS (AG) activates CRABS CLAW (CRC) in the nectaries and carpels, while it represses the expression of WUS in floral stem cells [15,17]. In tomato, the locule number (lc) locus is found on chromosome 2, while the fasciated (fas) locus is on chromosome 11 [18]. Subsequent studies have shown that lc is a gain-of-function mutation of SlWUS, while fas is a partial loss-of-function mutation of SlCLV3 in tomato [19]. Additionally, exogenous GA signals influence SlYABBY2b to regulate the development of locules [14].
Polymorphic molecular markers and high-density genetic maps play vital roles in QTL mapping [20]. Simple sequence repeats (SSRs) and insertion/deletion (InDel) markers are extensively used in mapping programs owing to their high throughput, good genome coverage and cost effectiveness [21,22]. In previous studies, multiple genetic maps were constructed for important agricultural traits, including yield and traits related to resistance [5,7,23]. Despite advances in technology, the research on locule number related to QTL mapping in pepper is still limited compared with that in other Solanaceous crops.
In this study, 111 markers were used to construct a linkage map in F2 populations derived from a cross between the small-fruited pepper cultivar GS6 (C. annuum) and the large-fruited pepper cultivar Qiemen (C. annuum). Several fruit-related QTLs were detected in the two populations (F2 and F2:3), including ftl2.1, ftd2.1, fts1.1, ftw2.1, and lcn1.1. As a novel QTL, lcn1.1 was identified on chromosome 1 based on two years of trials and analysis. In the major locule number QTL, the candidate gene Capana01g004285, which encodes the BREVIS RADIX (BRX) protein, displayed variable expression in the tissues of two parents. In addition, a virus-induced gene silencing (VIGS) approach was utilized to further explore its function in the regulation of locule number. Knockdown of CaBRX reduced the locule number and affected the expression levels of related genes in GS6. Together, our results suggest that CaBRX contributes to the development of locules in C. annuum.
2. Materials and Methods
2.1. Plant Materials, Mapping Population
Two pepper parental lines C. annuum cv. GS6 (P1, female) and C. annuum cv. Qiemen (P2, male) were used to develop segregating populations. The GS6 line is fasciculate and small with a smaller locule number, while Qiemen is a bell pepper with a larger locule number. Thirty F1 and 200 F2 individuals were cultivated in the greenhouse with normal water and fertilizer management at Northwest A&F University, Yangling, China (34.28′ N, 108.07′ E), in the spring of 2018. The 186 F2:3 populations were generated by selfing pollination from F2 and were cultivated in the greenhouse, under the same management as above described, in the spring of 2019.
2.2. Statistical Analysis of the Phenotyping Data
The phenotypic analysis of fruit-related traits was scaled as described by IPGRI [24]. The fruit weight was weighed in a balance that was accurate to 0.01 g. Fruit length and fruit diameter were carefully measured using a digital caliper. The fruit shape index was calculated by the ratio of fruit length to fruit diameter. The locule number was counted after the fruit had been cut across. More than 20 fruits were measured in the parental line and F1, respectively. We randomly selected three to five mature fruits from each individual plant to calculate the average value in F2. Fruits from the ten plants of each plant progeny was measured in the F2:3 population. The correlation analysis of these traits was performed based on multiple statistical properties containing the mean, standard deviation, variance, skewness, and kurtosis using SPSS 22.0 (IBM, Inc., Armonk, NY, USA).
2.3. Linkage Map Construction
The polymorphic markers were screened from 687 SSR markers and 642 InDel markers in parental lines, which covered the whole Capsicum genome based on the NCBI database (https://www.ncbi.nlm.nih.gov/, accessed on 1 August 2018) and published data [22,25,26,27]. Fifty SSR and sixty-one InDel markers were used to analyze the genotype of F2 populations, excluding poorly amplified and hardly separated markers. Young leaves from all the plants were collected and rapidly frozen at −80 °C for subsequent genomic DNA extraction using a modified CTAB method, in which 20% of 5 M NaCl in alcohol (v/v) was used instead of isoamyl alcohol [28]. The linkage map was constructed by QTL IciMapping 4.0 software (CAAS, Beijing, China) using the Kosambi function [29]. The markers are listed in Table S1.
2.4. QTLs Mapping
QTL analyses associated with fruit traits were identified by QTL IciMapping 4.0 software (CAAS, Beijing, China) using the ICIM-ADD method. A minimum logarithm of odds (LOD) threshold was set to 3 to scan the QTLs using a 1 cM scanning step. The p value for entering variables (PIN) was set to 0.001 [30]. The iQTLs that were identified were named with abbreviations for the traits and chromosome position.
2.5. Candidate Genes Selection and Analysis
Based on Zunla-1 reference genomes of pepper, the predicted genes in the lcn1.1 QTL region were analyzed using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 10 December 2019) to predict the annotation. The annotation details of the genes are shown in Table S2. The expression patterns of the predicted genes were obtained from the public RNA-Seq data at different fruit developmental stages (PepperHub, http://pepperhub.hzau.edu.cn/, accessed on 12 December 2019) [31]. The neighbor-joining (NJ) phylogenetic tree was constructed by MEGA X as previously described [32]. The coding sequences of CaBRX and CaGRF were respectively amplified from pepper cv. GS6 and Qiemen. Multiple sequence alignments were blasted against the Zunla-1 genome using DNAMAN (Lynnon Biosoft, San Ramon, CA, USA).
2.6. RNA Extraction and qRT-PCR Analysis of the Candidate Gene
The flowers of cv. GS6 and Qiemen were collected at the flower bud formation stage (Flower-1), slightly opened stage (Flower-2) and fully opened stage (Flower-3). The fruits from parental lines were collected at the immature stage (Fruit-1), color-changed stage (Fruit-2) and the red-ripened stage (Fruit-3). All the samples were frozen and extracted for RNA using an RNAprep pure extraction kit (Tiangen Biotech, Xi’an, China). The reverse transcription for qRT-PCR was performed using a cDNA Synthesis Kit (Vazyme, Nanjing, China). qRT-PCR was analyzed on QuantStudio®5 (Life Technologies, Paisley, UK) as previously described [32]. CaUBI3 was used for normalization, and the data were calculated using the 2−ΔΔCT method. The measurements were performed in triplicate. The specific primers are listed in Table S3.
2.7. VIGS of Candidate Gene in Pepper
The VIGS primers were obtained from the Sol Genomics Network (http://vigs.solgenomics.net/, accessed on 1 August 2020). A specific 342 bp fragmentof CaBRX was cloned from GS6 fruit and inserted into the pTRV2 vector, as well as the specific 374 bp of CaGRF. The vector pTRV1, pTRV2:00 (empty vector, as a negative control), pTRV2:CaPDS (phytoene desaturase gene, as a positive control with a bleaching phenotype), pTRV2:CaBRX, and pTRV2:CaGRF were integrated into Agrobacterium strain GV3101 and injected into full extended cotyledons leaves of GS6 as previously described [32]. The plants were placed in a temperature-controlled growth chamber under 22 °C/18 °C in 16 h day and 8 h night until the fruit matured. The locule number was measured when the fruits were mature.
2.8. Statistical Analysis
The data were measured by one-way analysis of variance (ANOVA) with Tukey’s test (p < 0.05) using SPSS 22.0 software (IBM, Inc., Armonk, NY, USA) to determine significant differences. All values were measured as means ± SE (standard error) with three biological replicates.
3. Results
3.1. Phenotypic Variations of Fruit Traits
As shown in Figure S1 and Table 1, two parents have significant differences in phenotype, including Ftl, Ftd, Fts, Ftw and Lcn. GS6 contains small fruit (2.72 g) with a shape index of 2.80 and has 3.00 Lcn. The fruits of Qiemen are large (114.79 g) and have 3.81 Lcn, with a shape index of 0.92. In the F2 and F2:3 populations, the Ftl is skewed toward Qiemen, while the Ftw, Ftd and Lcn are skewed toward GS6. The Fts of F2:3 (1.89) is in the middle of parental lines. The frequency distributions of these traits in the F2 and F2:3 populations conformed to an approximately continuous normal distribution, which indicates that these traits are quantitative (Figure 1). Correlation analyses were used to reveal the link between each other and showed that Lcn significantly positively correlated with Ftw (0.277 and 0.146) and Ftd (0.312 and 0.250) in the F2 and F2:3 populations, respectively. Lcn negatively correlated with Ftl (−0.171 and −0.210) and Fts (−0.435) in both experiments (Table 2). The correlations were significant (p < 0.05) or very significant (p < 0.01), indicating that Lcn could impact other traits.

Table 1.
Statistical analysis of the phenotypes of fruit-related traits in pepper.
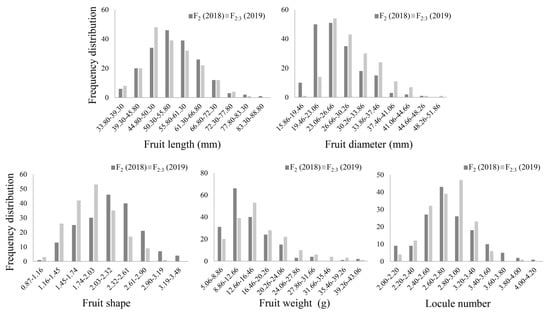
Figure 1.
Frequency distributions of fruit length (Ftl), fruit diameter (Ftd), fruit shape (Fts), fruit weight (Ftw) and locule number (Lcn) in the F2 and F2:3 populations.

Table 2.
Correlations among fruit-related traits in the F2 and F2:3 populations.
3.2. Genetic Linkage Map
A total of 687 SSR markers and 642 InDel markers were used to screen the polymorphism in F2 populations. A total of 58 SSR markers (9.48%) and 75 InDel markers (11.68%) with clear bands and polymorphism were selected to construct a linkage map using the QTL IciMapping 4.0 software. A total of 111 markers were successfully assigned to 12 linkage groups, which spanned a length of 1320.72 cM with an average distance of 11.90 cM (Figure 2, Table 3). In particular, the longest linkage group was linked with chromosome 1 and covered 17 markers over a distance of 196.50 cM. The linkage groups in chromosomes 2 and 5 had a lower marker density without gaps.

Figure 2.
Distribution of QTLs on the linkage map of pepper (Capsicum annuum L.) based on 111 SSR and InDel markers. The lengths of the map in centimorgans (cM) are on the left side of the chromosome, and the names of markers and QTLs are on the right side.

Table 3.
Statistics of the linkage map in pepper.
3.3. QTL Analysis for Fruit Traits
Combined with the phenotypic data and genetic linkage map, we performed a whole-genome scan to detect the QTLs with inclusive composite interval mapping (ICIM) methods using QTL IciMapping 4.0 software. In total, 17 fruit-related QTLs were identified on six chromosomes, including 3 Ftl QTLs, 5 Ftd QTLs, 3 Fts QTL, 4 Ftw QTLs, and 2 Lcn QTLs (Table 4, Figure 2). Five QTLs were detected in both the F2 and F2:3 populations, including ftl2.1, ftd2.1, fts1.1, ftw2.1 and lcn1.1, proving the reliability of these QTLs (Figure 3). ftl3.1, ftd4.1, ftd10.1, fts2.1, fts10.1 and ftw4.1 were detected in the F2 population, while ftl6.1, ftd1.1, ftd10.2, ftw3.1, ftw10.1 and lcn3.1 were identified in the F2:3 population. The results could have been affected by environmental differences.

Table 4.
QTLs identified in the F2 and F2:3 families (2018 and 2019).
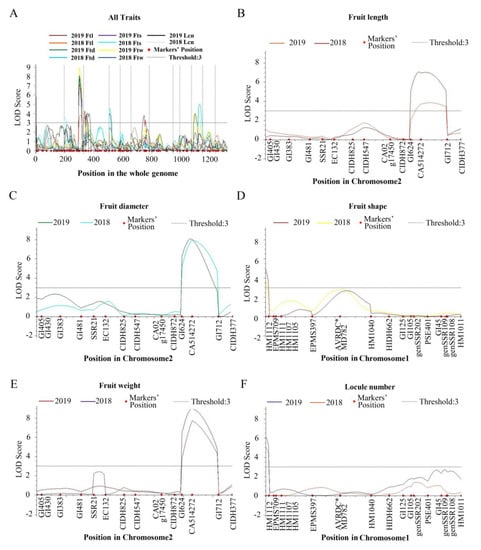
Figure 3.
The distribution of fruit-related QTLs detected in the F2 and F2:3 populations. (A) All traits on the whole genome. (B–F) Fruit length (Ftl), fruit diameter (Ftd), fruit shape (Fts), fruit weight (Ftw) and locule number (Lcn) on different chromosomes. These QTLs are indicated by differently colored lines. The LOD score threshold was set to 3.
Three Ftl QTLs were distributed on chromosomes 2, 3 and 6. ftl2.1 was mapped to the region of 105.98-123.72 cM of chromosome 2 with a logarithm of odds (LOD) score of 3.84 in the F2 population. In the F2:3 population, ftl2.1 was located between 99.78 cM and 105.98 cM and explained 16.26% of the phenotypic variation with an LOD value of 3.78. In the F2 population, ftl3.1 was located between PSE048 (0 cM) and SSR30 (13.63 cM) on chromosome 3, which explained 9.82% of the phenotypic variance with a 3.60 LOD value. ftl6.1 was located between genSSR3420 (60.45 cM) and GI1469 (90.57 cM) on chromosome 6 with a contribution rate of 7.54% in the F2:3 population.
Five Ftd QTLs were identified on chromosomes 1, 2, 4 and 10. ftd1.1 was detected between HM1112 and EPMS709 on chromosome 1 in the F2:3 population, which explained 4.46% of the phenotypic variation. In the F2 and F2:3 populations, ftd2.1 explained 13.28% and 14.26% of the phenotypic variation with a higher LOD score (7.96 and 8.07, respectively), and the genetic distance ranged from 99.78 cM to 123.72 cM. ftd4.1 was detected in the F2 population and explained 8.26% of the phenotypic variation. The QTL was located between PSE087 and CIDH671 covering 19.88 cM on chromosome 4 with an LOD value of 4.5. The QTL ftd10.1 explained 11.16% of the phenotypic variation in the F2 population and was located between GI2361 (48.61 cM) and GI2434 (77.63 cM) on chromosome 10. ftd10.2 was located between GI2241 (14.48 cM) and CAMS179 (38.50 cM) in the F2:3 population with 9.98% of the phenotypic variation.
The fruit shape-related QTL fts1.1 was located between HM1112 and EPMS709 on chromosome 1, which explained 7.99% and 12.23% of the phenotypic variation with an LOD value of 3.77 and 4.98, respectively. fts2.1 was detected in the F2 population with an LOD score of 3.54 and explained 8.91% of the phenotypic variation. fts10.1 was located between 48.61 cM and 77.63 cM on chromosome 10 with a contribution rate of 9.58% in the F2 population.
We found four QTLs that were related to fruit weight in the F2 and F2:3 populations, namely ftw2.1, ftw3.1, ftw4.1 and ftw10.1. In the F2 and F2:3 populations, ftw2.1 was detected on chromosome 2 and explained 15.19% and 16.43% of the phenotypic variation, which was located between CA514272 (105.98 cM) and GI712 (123.72 cM) with higher LOD scores (7.71 and 8.93, respectively). In the F2:3 population, ftw3.1 and ftw10.1 explained 5.86% and 11.88% of the phenotypic variation, respectively. ftw3.1 was mapped between GI1059 (17.64 cM) and genSSR1930 (33.50 cM) on chromosome 3. The marker interval of ftw10.1 was the same as that of ftd10.2. In the F2 population, ftw4.1 explained 9.52% of the phenotypic variation covering 19.88 cM on chromosome 4.
In the QTL analysis of locule number, we detected lcn1.1 and lcn3.1 on chromosomes 1 and 3, respectively. lcn3.1 was located between GI1059 (17.64 cM) and genSSR1930 (33.50 cM) with a contribution rate of 8.81% in the F2:3 population. As a novel major-effect QTL, lcn1.1 was located between HM1112 and EPMS709 covering 3.18 cM in F2 and F2:3 populations, which explained 11.98% and 14.31% of the phenotypic variation, respectively. The LOD scores of lcn1.1 ranged from 5.04 to 6.19. The results indicated that lcn1.1 may play a crucial part in the development of locule.
3.4. Prediction and Analysis of Candidate Genes for the Locule Number
The QTLs with a high contribution rate (PVE > 10%) in the experiment were primarily concentrated on chromosomes 1, 2 and 10. Compared with previous studies, lcn1.1 was the first QTL reported that was associated with locule number to our knowledge. The QTL region of lcn1.1 was located over a genetic distance of 3.18 cM, and the physical interval was approximately 2.52 Mb in the Zunla-1 reference genome, which contained 60 putative predicted genes. The expression profiles of the predicted genes were analyzed in different stages of fruit development based on the public RNA-Seq data [31] (Figure S2). We focused on the genes that are specifically expressed at the seed stage, such as Capana01g004285 and Capana01g004288. The expression of Capana01g004285 was specifically up-regulated in the S4 stage and showed obvious differential expression during the whole stages. The expression levels of Capana01g004288 were abundant in the seed stage. In an annotation analysis, we focused on genes that were specifically annotated and related to development (Table S2). Capana01g004285 and Capana01g004288 were selected for additional research based on their expression profiles and the annotation information analysis.
Capana01g004285 encodes the BREVIS RADIX (BRX) protein and is designated CaBRX. A phylogenetic analysis showed that CaBRX was homologous to SlBRX, PjBRX, SaBRX and AtBRX (Figure 4A). The function of AtBRX in cell proliferation and elongation has been reported, indicating that CaBRX may play roles in plant growth and development. The expression patterns of CaBRX were detected in GS6 and Qiemen (Figure 4B). In GS6, the expression of CaBRX was up-regulated during flowering, and it was down-regulated when the fruit matured. In Qiemen, the transcript levels of CaBRX were higher in the young stage (Flower-1 and Fruit-1). The expression patterns of CaBRX differed between GS6 and Qiemen.
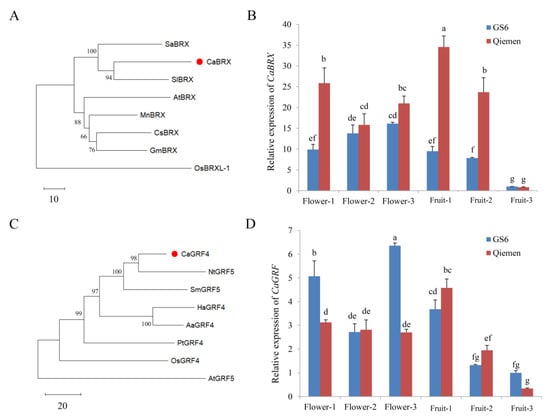
Figure 4.
Analysis of CaBRX and CaGRF. (A) Phylogenetic analysis of BRX from pepper (Capsicum annuum), Arabidopsis thaliana, tomato (Solanum lycopersicum), rice (Oryza sativa), witchweed (Striga asiatica), mulberry tree (Morus notabilis), cucumber (Cucumis sativus), and soybeans (Glycine max). (B) Expression patterns of CaBRX in different stages of GS6 and Qiemen. (C) Phylogenetic analysis of GRF from pepper (C. annuum), A. thaliana, Danshen (Salvia miltiorrhiza), sunflower (Helianthus annuus), tobacco (Nicotiana tabacum), sweet wormwood (Artemisia annua), rice (O. sativa), and poplar (Populus trichocarpa). (D) Expression patterns of CaGRF in different stages of GS6 and Qiemen. Flower-1: flower bud formation stage. Flower-2: slightly opened stage. Flower-3: fully opened stage. Fruit-1: immature stage. Fruit-2: color-changed stage. Fruit-3: red-ripen stage. Data are the mean with SE of three independent replicates. The letters (a–g) show significant differences (Tukey’s test, p < 0.05). SE, standard error.
Capana01g004288 encodes a growth-regulating factor (GRF) protein and is designated CaGRF. Compared with the homologous members, CaGRF was more similar to NtGRF5 (Figure 4C). Overexpression of the rice (Oryza sativa) gene OsGRF4 reduced the formation of tillers and internodes and promoted the development of extensive adventitious roots/shoots on nodes. As shown in Figure 4D, the expression levels of CaGRF significantly increased in the Flower-3 stage of GS6 (6.36-fold). In Qiemen, the expression of CaGRF was up-regulated in the Fruit-1 stage (4.58-fold). As the fruit matured, the transcript levels of CaGRF decreased in GS6 and Qiemen. In addition, the protein sequences of CaBRX did not differ between GS6 and Qiemen compared with Zunla-1 genome, as well as CaGRF (Figure S3).
3.5. VIGS of Candidate Genes in Cv. GS6
To gain insight into the functions of CaBRX and CaGFR, a virus-induced gene silencing (VIGS) assay was performed to reduce the expression levels of these genes in GS6. Quantitative real-time PCR (qRT-PCR) was used to confirm that the genes have been successfully silenced (Figure 5A,B). We detected the locule numbers of fruits, which included TRV2:CaBRX, TRV2:CaGRF and TRV2:00 plants, when the fruit matured. As shown in Figure 5C,D, most of the TRV2:00 and TRV2:CaGRF fruit contained two or three locules, while almost all of TRV2:CaBRX had two locules. The locule number between TRV2:CaGRF and TRV2:00 did not differ significantly. However, the locule number was significantly reduced in TRV2:CaBRX compared with the control plants, which suggested that CaBRX positively regulated the locule number in pepper (Figure 5D). To investigate whether the silencing of candidate genes could affect the genes related to flower and locule development, qRT-PCR was performed to detect the expression levels of CaWUS (homologous gene of SlWUS), CaCLV3 (homologous gene of SlCLV3), CaAG (homologous gene of AtAG) and CaCRC (homologous gene of AtCRC). CaCLV3, CaAG and CaCRC were more highly expressed in the TRV2:CaBRX plants compared with the controls (Figure 5E). The expression of CaWUS was significantly down-regulated in the TRV2:CaBRX plants.
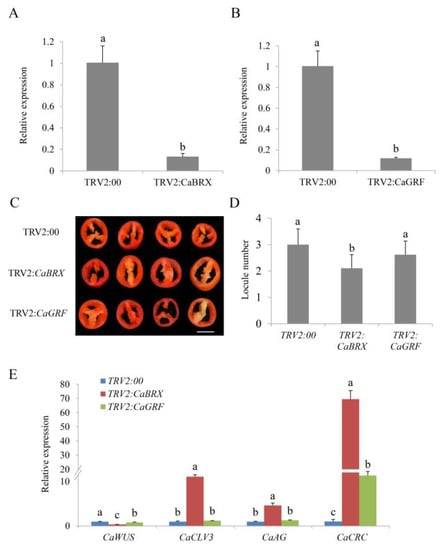
Figure 5.
Silencing of CaBRX reduces the locule number in pepper (Capsicum annuum L.). (A) The expression of CaBRX in CaBRX-silenced and control plants. (B) The expression of CaGRF in CaGRF-silenced and control plants. (C) Appearance of the locule number. (D) The locule number of silenced and control plants. Data are the mean with SE of three independent replicates, and each replicate consists of eight plants. Scale bar: 1 cm. (E) The relative expression of CaWUS, CaCLV3, CaAG and CaCRC in silenced and control plants. The data indicate the mean with SE of three independent replicates. The letters (a–c) indicate significant differences (Tukey’s test, p < 0.05). SE, standard error.
4. Discussion
The agronomic production of pepper is highly important, since the fruits are used in foodstuffs, pigment extraction, food additives, pharmaceuticals, and ornamentals [33,34]. In the past, the traits of fruit, such as fruit weight, fruit length, fruit diameter, fruit shape index, and locule number, have been widely studied using QTL mapping in many species, which are the usual methods for quantitative trait genetic studies [5,7,12]. Many QTLs about fruit and plant developmental traits have been identified and advanced the research of their mechanisms. The QTL analyses of pepper fruit traits include plant height, fruit weight, fruit shape and flowering, among others. The fruit-related QTLs were first studied in 2001 in a cross between two C. annuum genotypes, Maor (bell-type pepper) and Perennial (small-fruited line) [6]. Multiple QTLs were distributed on linkage groups 2, 3, 4, 8 and 10, implying their importance in the control of developmental processes [6]. Subsequently, the fruit-related QTLs were detected in different species using advanced techniques in pepper [8]. Compared with other traits, there are few studies on locule number in pepper. Locules develop from carpels with seeds and cavities [13]. The locule number is an agronomic trait that influences fruit weight and even commercial quality [35].
In this study, 111 markers were utilized to construct the genetic map with a length of 1320.72 cM. Zygier et al. reported that locule number had no effect on the size of pepper fruits based on a correlation analysis [8]. Nevertheless, our data proved that the locule number displayed a significant correlation with other traits. The locule number significantly positively correlated with Ftw and Ftd, whereas it significantly negatively correlated with Ftl and Fts. The results were partially consistent with those of Palloix et al. [7]. Nlo significantly positively correlated with fruit diameter but was significantly negatively correlated with fruit shape.
We identified 17 fruit-related QTLs that contained ftw2.1, ftl2.1, ftd2.1, fts1.1 and lcn1.1 in fruit weight, fruit length, fruit diameter, fruit shape and locule number. Based on the physical positions, ftw2.1 was located between CA514272 and GI712 on 106 cM in the genetic position, which was mapped between 156 Mbp and 165 Mbp. The results consisted of the QTL fw2.1, which was located at approximately 156 Mbp in C. frutescens and C. chinense [8]. fw2.1 co-located with the fruit-shape QTL ovate. Silencing of CaOvate in cv. “Round” led to a more elongated shape, suggesting that CaOvate controls fruit shape in pepper [10]. In our study, the physical position of ftl2.1 was close to that of ftw2.1, as well as ftd2.1, which suggests that this interval played a significant role in fruit traits. In previous studies, the fruit diameter and fruit weight QTLs were correlated at 170 Mbp [11,36]. Additionally, the physical position of ftd2.1 (approximately 153 Mbp) in our study was far apart from FD-2 (1 Mbp to 2 Mbp), which was shown by Han et al. [37]. fts1.1, ftd1.1 and lcn1.1 were located at approximately 294 Mbp. A previous study showed that fs3.1 explained more than 60% of the phenotypic variation at TG130 [5].
Palloix et al. identified nine locule number QTLs that were distributed on three chromosomes (chromosomes 2, 8 and 12) and five small unassigned linkage groups [7]. These QTLs contained Nlo2.1, Nlo12.1, NloLG22.1 and NloLG25.1; NloLG25.1 explained a relatively higher amount of variation (13%). In tomato, the locule number exerted significant effects on fruit size, which contributed to large fruit [38]. Three tomato QTLs responsible for locule number were identified on chromosome 2 (lcn2.1 and lcn2.2) and chromosome 11 (lcn11.1). lcn2.1 and lcn11.1 corresponded to the locule number (lc) and fasciated (fas) loci, respectively [9,18]. lc and WUSCHEL (WUS) co-regulated the development of shoot apical meristems in tomato [13]. fas encodes a YABBY-like transcription factor that controls the carpel number [39]. The pepper QTL Nlo2.1 might be orthologous to lcn2.1 in tomato [7]. In this study, the Lcn QTLs lcn1.1 and lcn3.1 were identified on chromosomes 1 and 3. Considering the differences in markers and cultivars, as well as the environmental impact, we focused more closely on lcn1.1, the first mapping of Lcn on chromosome 1, which was repeated in both experiments. lcn1.1 explained 11.98% and 14.31% of the phenotypic variation in F2 and F2:3 populations, respectively.
To confirm the candidate gene in lcn1.1, we screened the results with related protein annotation and differential expression patterns. Finally, CaBRX (BREVIS RADIX) and CaGRF (growth-regulating factor) were found to be candidate genes that may have major effects on the development of locules. BRX family proteins are nuclear-localized transcription factors [40]. CaBRX was homologous with AtBRX, which regulates cell proliferation and elongation [40,41]. BRX family proteins contain conserved domains with 55 amino acids, which can mediate protein–protein interactions [42]. Compared with monocotyledons, the evolutionary rates of BRX family in dicotyledons are accelerated and could lead to higher functional diversification [41]. Additionally, several domains in BRX could be encoded by the related BRX-like (BRXL) genes, implying that function overlapped in the BRX family [41]. CaGRF is a member of the growth-regulating factors (GRFs) family. GRFs contain highly conserved QLQ and WRC domains [43]. The QLQ domain mediates the interaction with GRF-interacting factors (GIFs) [44]. The rice protein OsGRF4, is homologous to CaGRF and positively regulates grain shape and panicle length while it negatively regulates seed shattering [45]. OsGRF4 is encoded by the major QTL of grain size, grain length and width 2 (GLW). OsGIF1 interacts with OsGRF4 to improve grain size. Furthermore, microRNA396 (an miRNA) targets GRF to participate in floral organogenesis, suggesting that the miR396-OsGRF4-OsGIF1 pathway may determine the grain size in rice [46]. In summary, we hypothesized that CaBRX or CaGRF affected cell proliferation and fruit shape during fruit development to regulate the number of locules. In this study, the expression patterns of CaBRX and CaGRF differed between Qiemen and GS6, indicating that the genes may play important roles in the development of flowers and fruit. VIGS assays were performed to explore the function of candidate genes in both Qiemen and GS6, however we failed to get result in Qiemen. The differences of locule number were observed in GS6. The locule number was significantly reduced in CaBRX-silenced fruits, while there was no difference between TRV2:CaGRF and TRV2:00. Moreover, the expression levels of CaAG and CaCRC were significantly up-regulated in TRV2:CaBRX plants. As a MADS domain transcription factor, AtAG activates AtCRC in nectaries and carpels [17]. In addition, AtAG represses the expression of AtWUS, which was consistent with our results [15]. In A. thaliana, AtWUS and AtCLV3 form a negative feedback in the regulation of meristems [16]. In tomato, silencing of SlWUS decreases the locule number, while SlCLV3 RNAi lines contain more locule numbers [19,47]. In TRV2:CaBRX plants, the expression of CaWUS was significantly down-regulated compared with that of the controls. Conversely, silencing of CaBRX increased the expression level of CaCLV3. These results indicate that the locule number decreased by silencing of CaBRX.
5. Conclusions
In this study, we constructed a genetic map that was 1,320.72 cM long in pepper. In total, 17 fruit-related QTLs were identified, including ftl2.1, ftd2.1, fts1.1, ftw2.1 and lcn1.1. In the locule number major region lcn1.1, CaBRX was the candidate gene that could possibly be involved in the development of locules. Silencing of CaBRX reduced the locule number and altered the expression of genes related to the development of flowers and locules, including CaWUS, CaCLV3, CaAG and CaCRC, indicating that CaBRX plays an important role in pepper fruit.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae8020146/s1: Figure S1. Fruit morphology and locule phenotype of GS6 (A) and Qiemen (B). Scale bar: 1 cm; Figure S2. Heatmap of 60 predicted genes at different fruit development stages. FST0 and FST1: the fruit were collected on 3 and 7 days after flowering. G: pericarp. ST: seed and placenta. S: seed. T: placenta. 1-11: the fruit were collected on 10, 15, 20, 25, 30, 35, 40, 45, 50, 55 and 60 days after flowering. The candidate genes are indicated by red arrows; Figure S3. Multiple sequence alignment of CaBRX (A) and CaGRF (B) in Qiemen and GS6; Table S1. Primer sequence of markers; Table S2. Annotation information of candidate genes in lcn1.1; Table S3. Primers used in this study.
Author Contributions
Data curation, writing—original draft preparation, writing—review and editing, X.M.; resources, data curation, Y.-M.Q.; formal analysis, Y.L.; validation, Y.-N.Y. Writing—review and editing, resources, supervision, project administration, funding acquisition, Z.-H.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key R&D Program of China (No. 2016YFD0101900) and the National Natural Science Foundation of China (No. 31772309, No. 31860556).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data that support the results are included in this paper and its Supplementary Materials Files.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira-Dias, L.; Vilanova, S.; Fita, A.; Prohens, J.; Rodríguez-Burruezo, A. Genetic diversity, population structure, and rela-tionships in a collection of pepper (Capsicum spp.) landraces from the Spanish centre of diversity revealed by genotyping-by-sequencing (GBS). Hortic. Res. 2019, 6, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Park, M.; Yeom, S.-I.; Kim, Y.-M.; Lee, J.M.; Lee, H.-A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.-T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef] [PubMed]
- FAO. FAOSTAT Statistics Database. 2019. Available online: http://www.fao.org/faostat/ (accessed on 1 August 2020).
- Rao, G.U.; Ben Chaim, A.; Borovsky, Y.; Paran, I. Mapping of yield-related QTLs in pepper in an interspecific cross of Capsicum annuum and C. frutescens. Theor. Appl. Genet. 2003, 106, 1457–1466. [Google Scholar] [CrossRef]
- Ben Chaim, A.; Paran, I.; Grube, R.C.; Jahn, M.; Van Wijk, R.; Peleman, J. QTL mapping of fruit-related traits in pepper (Capsicum annuum). Theor. Appl. Genet. 2001, 102, 1016–1028. [Google Scholar] [CrossRef]
- Palloix, A.; Barchi, L.; Lefebvre, V.; Sage-Palloix, A.-M.; Lanteri, S. QTL analysis of plant development and fruit traits in pepper and performance of selective phenotyping. Theor. Appl. Genet. 2009, 118, 1157–1171. [Google Scholar]
- Zygier, S.; Chaim, A.B.; Efrati, A.; Kaluzky, G.; Borovsky, Y.; Paran, I. QTLs mapping for fruit size and shape in chromosomes 2 and 4 in pepper and a comparison of the pepper QTL map with that of tomato. Theor. Appl. Genet. 2005, 111, 437–445. [Google Scholar] [CrossRef]
- Van Der Knaap, E.; Tanksley, S.D. The making of a bell pepper-shaped tomato fruit: Identification of loci controlling fruit morphology in Yellow Stuffer tomato. Theor. Appl. Genet. 2003, 107, 139–147. [Google Scholar] [CrossRef]
- Tsaballa, A.; Pasentsis, K.; Darzentas, N.; Tsaftaris, A.S. Multiple evidence for the role of an Ovate-like gene in determining fruit shape in pepper. BMC Plant Biol. 2011, 11, 46. [Google Scholar] [CrossRef] [Green Version]
- Chunthawodtiporn, J.; Hill, T.; Stoffel, K.; Van Deynze, A. Quantitative Trait Loci Controlling Fruit Size and Other Horticultural Traits in Bell Pepper (Capsicum annuum). Plant Genome 2018, 11, 160125. [Google Scholar] [CrossRef] [Green Version]
- Arjun, K.; Dhaliwal, M.S.; Jindal, S.K.; Fakrudin, B. Mapping of fruit length related QTLs in interspecific cross (Capsicum annuum L. × Capsicum galapagoense Hunz.) of chilli. Breed. Sci. 2018, 68, 219–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muños, S.; Ranc, N.; Botton, E.; Bérard, A.; Rolland, S.; Duffé, P.; Carretero, Y.; Le Paslier, M.-C.; Delalande, C.; Bouzayen, M.; et al. Increase in Tomato Locule Number Is Controlled by Two Single-Nucleotide Polymorphisms Located Near WUSCHEL. Plant Physiol. 2011, 156, 2244–2254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Sun, M.-H.; Qi, M.-F.; Xing, J.; Xu, T.; Liu, H.-T.; Li, T.-L. Alteration of SlYABBY2b gene expression impairs tomato ovary locule number and endogenous gibberellin content. J. Zhejiang Univ. Sci. B 2018, 19, 445–457. [Google Scholar] [CrossRef]
- Liu, X.; Kim, Y.J.; Müller, R.; Yumul, R.E.; Liu, C.; Pan, Y.; Cao, X.; Goodrich, J.; Chen, X. AGAMOUS Terminates Floral Stem Cell Maintenance in Arabidopsis by Directly Repressing WUSCHEL through Recruitment of Polycomb Group Proteins. Plant Cell 2011, 23, 3654–3670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.; Jürgens, G.; Laux, T. The Stem Cell Population of Arabidopsis Shoot Meristems Is Maintained by a Regulatory Loop between the CLAVATA and WUSCHEL Genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-Y.; Baum, S.F.; Alvarez, J.; Patel, A.; Chitwood, D.H.; Bowman, J.L. Activation of CRABS CLAW in the Nectaries and Carpels of Arabidopsis. Plant Cell 2005, 17, 25–36. [Google Scholar] [CrossRef] [Green Version]
- Barrero, L.S.; Cong, B.; Wu, F.; Tanksley, S.D. Developmental characterization of the fasciated locus and mapping of Arabidopsis candidate genes involved in the control of floral meristem size and carpel number in tomato. Genome 2006, 49, 991–1006. [Google Scholar] [CrossRef]
- Chu, Y.; Jang, J.; Huang, Z.; van der Knaap, E. Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct. 2019, 3, e00142. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.-F.; Wang, G.-Y.; Dong, T.-T.; Chen, B.; Du, H.-S.; Li, C.-B.; Zhang, F.-L.; Zhang, H.-Y.; Xu, Y.; Wang, Q.; et al. High-density genetic map construction and QTL mapping of first flower node in pepper (Capsicum annuum L.). BMC Plant Biol. 2019, 19, 167. [Google Scholar] [CrossRef]
- Sugita, T.; Semi, Y.; Sawada, H.; Utoyama, Y.; Hosomi, Y.; Yoshimoto, E.; Maehata, Y.; Fukuoka, H.; Nagata, R.; Ohyama, A. Development of simple sequence repeat markers and construction of a high-density linkage map of Capsicum annuum. Mol. Breed. 2013, 31, 909–920. [Google Scholar] [CrossRef]
- Li, W.; Cheng, J.; Wu, Z.; Qin, C.; Tan, S.; Tang, X.; Cui, J.; Zhang, L.; Hu, K. An InDel-based linkage map of hot pepper (Capsicum annuum). Mol. Breed. 2015, 35, 1–10. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, Y.; Zhang, Z.; Cao, Y.; Yu, H.; Ma, W.; Zhang, B.; Wang, R.; Gao, J.; Wang, L. Fine mapping of the major anthracnose resistance QTL AnRGO5 in Capsicum chinense ‘PBC932’. BMC Plant Biol. 2020, 20, 189. [Google Scholar] [CrossRef] [PubMed]
- IPGRI; AVRDC; CATIE. Descriptors for Capsicum (Capsicum spp.); International Plant Genetic Resources Institute: Rome, Italy; The Asian Vegetable Research and Development Center: Taipei, Taiwan; Centro Agronómico Tropical de Investigación y Enseñanza: Turrialba, Costa Rica, 1995. [Google Scholar]
- Tan, S.; Cheng, J.; Zhang, L.; Qin, C.; Nong, D.-G.; Li, W.-P.; Tang, X.; Wu, Z.-M.; Hu, K.-L. Construction of an Interspecific Genetic Map Based on InDel and SSR for Mapping the QTLs Affecting the Initiation of Flower Primordia in Pepper (Capsicum spp.). PLoS ONE 2015, 10, e0119389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-F.; Sun, H.-H.; Xu, Y.; Chen, B.; Yu, S.-C.; Geng, S.-S.; Wang, Q. Development of a large number of SSR and InDel markers and construction of a high-density genetic map based on a RIL population of pepper (Capsicum annuum L.). Mol. Breed. 2016, 36, 1–10. [Google Scholar] [CrossRef]
- Guo, G.; Zhang, G.; Pan, B.; Diao, W.; Liu, J.; Ge, W.; Gao, C.; Zhang, Y.; Jiang, C.; Wang, S. Development and Application of InDel Markers for Capsicum spp. Based on Whole-Genome Re-Sequencing. Sci. Rep. 2019, 9, 3691. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.C.; Flores-Vergara, M.A.; Krasynanski, S.; Kumar, S.; Thompson, W. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef]
- Kosambi, D.D. The Estimation of Map Distances from Recombination Values. Ann. Eugen. 1943, 12, 172–175. [Google Scholar] [CrossRef]
- Li, H.; Ribaut, J.-M.; Li, Z.; Wang, J. Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor. Appl. Genet. 2008, 116, 243–260. [Google Scholar] [CrossRef]
- Liu, F.; Yu, H.; Deng, Y.; Zheng, J.; Liu, M.; Ou, L.; Yang, B.; Dai, X.; Ma, Y.; Feng, S.; et al. PepperHub, an Informatics Hub for the Chili Pepper Research Community. Mol. Plant 2017, 10, 1129–1132. [Google Scholar] [CrossRef]
- Ma, X.; Gai, W.; Qiao, Y.; Ali, M.; Wei, A.; Luo, D.; Li, Q. Identification of CBL and CIPK gene families and functional char-acterization of CaCIPK1 under Phytophthora capsici in pepper (Capsicum annuum L.). BMC Genom. 2019, 20, 775. [Google Scholar] [CrossRef]
- Park, M.; Lee, J.-H.; Han, K.; Jang, S.; Han, J.; Lim, J.-H.; Jung, J.-W.; Kang, B.-C. A major QTL and candidate genes for capsaicinoid biosynthesis in the pericarp of Capsicum chinense revealed using QTL-seq and RNA-seq. Theor. Appl. Genet. 2019, 132, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Lee, H.-Y.; Ro, N.-Y.; Hur, O.-S.; Lee, J.-H.; Kwon, J.-K.; Kang, B.-C. QTL mapping and GWAS reveal candidate genes controlling capsaicinoid content in Capsicum. Plant Biotechnol. J. 2018, 16, 1546–1558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Illa-Berenguer, E.; Van Houten, J.; Huang, Z.; Van Der Knaap, E. Rapid and reliable identification of tomato fruit weight and locule number loci by QTL-seq. Theor. Appl. Genet. 2015, 128, 1329–1342. [Google Scholar] [CrossRef]
- Chunthawodtiporn, J. Breeding for Phytophthora Capsici Resistance: Study in QTL Analysis for Disease Resistance and Horti-Cultural Traits in Bell Pepper (Capsicum annuum). Ph.D. Thesis, University of California, Davis, CA, USA, 2016. [Google Scholar]
- Han, K.; Jeong, H.-J.; Yang, H.-B.; Kang, S.-M.; Kwon, J.-K.; Kim, S.; Choi, D.; Kang, B.-C. An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum). DNA Res. 2016, 23, 81–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lippman, Z.; Tanksley, S.D. Dissecting the Genetic Pathway to Extreme Fruit Size in Tomato Using a Cross Between the Small-Fruited Wild Species Lycopersicon pimpinellifolium and L. esculentum var. Giant. Heirloom. Genetics 2001, 158, 413–422. [Google Scholar] [CrossRef]
- Cong, B.; Barrero, L.S.; Tanksley, S.D. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 2008, 40, 800–804. [Google Scholar] [CrossRef]
- Mouchel, C.F.; Briggs, G.C.; Hardtke, C.S. Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev. 2004, 18, 700–714. [Google Scholar] [CrossRef] [Green Version]
- Briggs, G.C.; Mouchel, C.F.; Hardtke, C.S. Characterization of the Plant-Specific BREVIS RADIX Gene Family Reveals Limited Genetic Redundancy Despite High Sequence Conservation. Plant Physiol. 2006, 140, 1306–1316. [Google Scholar] [CrossRef] [Green Version]
- Beuchat, J.; Li, S.; Ragni, L.; Shindo, C.; Kohn, M.H.; Hardtke, C.S. A hyperactive quantitative trait locus allele of Arabidopsis BRX contributes to natural variation in root growth vigor. Proc. Natl. Acad. Sci. USA 2010, 107, 8475–8480. [Google Scholar] [CrossRef] [Green Version]
- Choi, D.; Kim, J.H.; Kende, H. Whole Genome Analysis of the OsGRF Gene Family Encoding Plant-Specific Putative Transcription Activators in Rice (Oryza sativa L.). Plant Cell Physiol. 2004, 45, 897–904. [Google Scholar] [CrossRef] [Green Version]
- Piya, S.; Liu, J.; Burch-Smith, T.; Baum, T.J.; Hewezi, T. A role for Arabidopsis growth-regulating factors 1 and 3 in growth–stress antagonism. J. Exp. Bot. 2020, 71, 1402–1417. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Zhang, W.; Wang, Y.; He, Q.; Shu, F.; Liu, H.; Wang, J.; Wang, J.; Yuan, L.; Deng, H. OsGRF4 controls grain shape, panicle length and seed shattering in rice. J. Integr. Plant Biol. 2016, 58, 836–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Gao, F.; Xie, K.; Zeng, X.; Cao, Y.; Zeng, J.; He, Z.; Ren, Y.; Li, W.; Deng, Q.; et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 2016, 14, 2134–2146. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Qi, M.; Sun, M.; Liu, Y.; Liu, Y.; Xu, T.; Li, Y.; Li, T. Tomato Transcription Factor SlWUS Plays an Important Role in Tomato Flower and Locule Development. Front. Plant Sci. 2017, 8, 457. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).