Abstract
Mango (Mangifera indica L.) belongs to the genus Mangifera and family Anacardiaceae, and is an important tropical fruits. Artificial cross breeding (hand pollination) is an important method for breeding new mango cultivars. It is easy to produce false hybrids in the process of artificial pollination breeding. Therefore, it is necessary to establish rapid and accurate molecular detection methods to identify the authenticity of hybrids. Mango ‘Jinhuang’ and ‘Renong No.1′ and 65 individual plants of their F1 hybrids were used as experimental materials, eight SSRs (simple sequence repeats) primer pairs with polymorphism in parents were used to identify the F1 hybrids by capillary electrophoresis. The results showed that PCR product size (bp) for eight primers ranged from 108 bp (ES55) to 176 bp (ES63) in 65 samples. A total of 62 true hybrids were identified from 65 hybrid progenies, and the true hybrid rate was 95.38%. A total of 18 alleles were amplified by eight SSRs, seven SSR loci showed binary segregations, whereas only one SSR locus ES83 showing ab:ac:bb:bc segregation fitted to the expected segregation ratio of 1:1:1:1. The value of expected heterozygosity (He), ranged from 0.34 to 0.62, whereas the value of observed heterozygosity ranged from 0.44 to 0.81. Chi-square test showed that eight SSR loci were in accordance with Mendel’s segregation law. The results of cluster analysis showed that the parents and 62 true hybrids could be classified into two categories at 0.58: the first category contained 27 offspring, clustered with ‘Jinhuang’ and showed a maternal genetic tendency. The second category contained 35 offspring, clustered with ‘Renong No.1′ and showed a partial paternal genetic tendency. DNA fingerprint of hybrids from ‘Jinhuang’ × ‘Renong No.1′ cross were constructed using eight SSR primers for variety protection.
1. Introduction
Mango (Mangifera indica L.) is a large evergreen tree belonging to the genus Mangifera and family Anacardiaceae. It is the fifth most economically important fruit crop, following banana, grape, apple, and orange [1]. Mango is originated from India, Myanmar, Thailand, Indonesia, the Philippines, and other south Asia countries, and is now widely distributed throughout the tropical and subtropical countries and regions at latitudes between 28° and 30° north and south [2]. China is one of the major mango producing countries in the world, with a total area of 343,333 hectares and an output of 3.31 million metric tons in 2020 (statistical data from Ministry of Agriculture and Rural Affairs). Mango is an important horticultural crop in Hainan, Guangxi, Guangdong, Yunnan, Fujian, and Sichuan provinces [3].
Mango industry in China is mainly based on some commercial cultivars, which are introduced from Taiwan province of China, and other countries including the USA, and Australia [4]. The main cultivars such as ‘Tainong No.1′, ‘Guifei’, and ‘Jinhuang’ are easily affected by low temperature and rainy weather during the flowering period, and consequently bear seedless fruits resulting in low yield [5]. ‘Keitt’ accounts for more than 80% of production in the area of Jinsha river dry hot valley, and is susceptible to bacterial black spot and physiological disorders [6,7]. In addition, new cultivars with independent intellectual property rights are still lacking. All these problems block the sustainable and healthy development of mango industry in China.
Many major mango cultivars were selected from the seedling selections populations of desired maternal parents, such as ‘Yuexi No. 1′, ‘Guire No. 82′, and ‘Panxi Hong’, which were bred in the 20th century [3]. ‘Eldon’, ‘Glenn’, ‘Lippens’, ‘Smith’, ‘Tommy Atkins’, and ‘Zill’ are considered to be the progeny of ‘Haden’ [8]. Artificial cross breeding (hand pollination) is an important method for breeding new mango cultivars [9]. The advantages of cross breeding include low cost, wide separation of offspring, and the combination of excellent traits from parents. Mango is a perennial plant with highly heterozygous genotypes, due to the cross-flowered fruiting. The cycle of mango artificial hybrid breeding is long, and open pollination and multi-varieties control pollination processes easily produce false hybrids, and are difficult to identify hybrid paternity.
Molecular marker technology is widely used to identify the authenticity of hybrids. Simple sequence repeat (SSR), also known as microsatellite, is a DNA sequence with a few (1~6) nucleotides as tandem repeats, which is widely distributed in eukaryotic genomes. Compared with restriction fragment length polymorphism (RFLP) and random amplified polymorphic DNA (RAPD), SSR molecular markers have the advantages of good repeatability, high polymorphism, high resolution, and easy operation [10,11]. Therefore, SSR molecular markers are widely used for variety identification in diverse fruit species including grape [12], pummelo [13], and litchi [14]. In mango, SSR and expressed sequence tag-derived SSR (EST-SSR) markers have been undertaken for mapping, parentage assignment and verification, and genetic diversity, as well as inferring phylogenetic relationships between species and varieties [15,16,17,18]. Traditional SSR molecular technology uses polyacrylamide gel electrophoresis and silver staining to observe and count the results of SSR markers. This technology has the disadvantages of large workload, low precision, small amount of analysis, and being time-consuming. In addition, it cannot afford the detection of large quantities of samples. Therefore, SSR fluorescent labeling technology has been developed and successfully applied to the identification of a variety of grapes [19], bayberry [20], watermelon [21], and other crops [22,23] due to its high efficiency, accuracy, sensitivity, and automation. At present, only a few literatures have reported the application of SSR fluorescent labeling technology to identify the authenticity of mango hybrids [24], genetic diversity and genetic relatedness of cultivars [25,26,27], confirmation of maternal or zygotic origin of embryogenic cultures [28].
Artificial cross breeding (hand pollination) is an important method for breeding new mango cultivars. It is easy to produce false hybrids during hybridization, which makes it difficult to distinguish the authenticity of the hybrid offspring. Therefore, it is necessary to establish rapid and accurate molecular detection technique for identifying the authenticity of hybrids. In this study, F1 hybrid population of ‘Jinhuang’ and ‘Renong No.1′ were used as experimental material, the authenticity of 65 F1 progenies was identified by SSR fluorescence marker technology in order to screen out true hybrids with good parental traits. Our results would lay a foundation for the follow-up mango cross breeding and molecular marker-assisted breeding.
2. Materials and Methods
2.1. Plant Materials
In 2010, we have carried out cross breeding in the orchard of Jinhe base in Panzhihua city, Sichuan province. Artificial hybrid pollination was performed by using ’Jinhuang’ as the female parent and ‘Renong No.1′ as the male parent. When the fruits were ripe, the seeds of the hybrid fruits were collected and sowed in Zhanjiang. 65 F1 individuals from the cross of ‘Jinhuang’ × ‘Renong No.1′ were used for evaluating the segregation of SSR genotypes, which were grown in the National Tropical Fruit Tree Germplasm Resource nursery (Mango branch nursery), South Subtropical Crops Research Institute, Chinese Academy of Tropical Agricultural Sciences, Zhanjiang, Guangdong Province. Young leaves were collected, treated with liquid nitrogen, and stored at −80 °C for later use.
2.2. DNA Extraction
Genomic DNA was extracted from young leaves by using Qiagen DNeasy®® Plant extraction Kits (Hilden, Germany) according to the manufacturer’s instructions. DNA quality was checked by electrophoresis on 1% (w/v) agarose gel. The concentration of DNA was determined using a spectrophotometer of Nano Drop1000 v3.3.1 (Nanodrop, USA) by measuring the absorption at 260 and 280 nm. DNA samples were diluted to a final concentration of 20 ng/µL and stored at −20 °C.
2.3. The Choice of SSR
SSR markers were selected from the reference reported by Luo et al. [29]. About 34 SSR primer pairs with different amplification bands between ‘Jinhuang’ and ‘Renong No.1′ were selected for hybrid identification and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). A total of 26 primer pairs were discarded because of their low level of polymorphism between parents and six progenies or other shortcomings identified during the preliminary amplification tests. Finally, eight pairs of SSR marker primers with polymorphism between parents and easy to amplify and distinguish bands were screened. The primer information is shown in Table 1.

Table 1.
Characteristics of eight EST-SSR markers in this study.
2.4. PCR Amplication and SSR Analysis
Fluorescence-based M13-tailed PCR assay was used to amplify microsatellites. PCR amplification was performed in a 20 µL reaction volume containing 2 µL DNA (20 ng/µL), 10 µL 2 × Taq PCR Mix, 0.1 µL forward primer (10 pmol/µL), 0.4 µL tail primer, 0.5 µL reverse primer (10 pmol/µL), and 7 µL nuclease free water. The M13 tail (sequence, TGTAAAACGACGGCCAGT) was added to the 5′ end of each forward primer pair, with an annealing temperature of 53 °C. The tail was alternatively labeled with the following four dyes: PET (red),NED (yellow), HEX (green), and FAM (blue). A total of eight SSRs were employed for 65 hybrid populations.
Two thermal cycling protocol were used for DNA amplification. The first profile was 94 °C pre-denaturation for 5 min, 32 cycles of 94 °C for 30 s, primer annealing temperature (54–60 °C depend on primers) for 30 s and 72 °C for 30 s, followed by 8 cycles of 94 °C for 30 s, 53 °C for 30 s and 72 °C for 30 s, at last followed by a final extension at 72 °C for 8 min. All the PCR reactions were performed using a Bioer Life Pro Thermal cycler (Bioer Technology, Hangzhou, China). PCR products were detected by electrophoresis on 1% agarose gel. The amplified products labelled with fluorescent dye PET, NED, HEX, or FAM were multiplexed pooled before separation on an automatic 96 capillary automated DNA sequencer (ABI3130 DNA Analyser, Applied Biosystems, Foster City, CA, USA).
The PCR product was denatured and DNA fragment size was assessed on an ABI3130 Capillary-based electrophoresis DNA sequencer. Fragment length was read by Genemapper v4.0 software. The obtained data were further analyzed using the peak scanner software (Applied Biosystems, USA) to determine the exact fragment size for the PCR product.
2.5. Data Analysis
The observed heterozygosity (HO) and expected heterozygosity (He), Chi-square test, and probability values were calculated using PopGen32 software (v1.31) [30]. The dendrogram analysis was constructed using the unweighted pairwise group method with arithmetic means (UPGMA) algorithm. The robustness was assessed by creating 1000 bootstrap replicates of the data and then constructing majority rule consensus trees.
3. Results
3.1. Genetic Identification of Mango Using SSR Markers
A total of 34 pairs of primers with different bands between ‘Jinhuang’ and ‘Renong No.1′ were selected for screening the hybrid identification primers from the mango SSR primers reported by Luo Chun et al. [29]. Among them, eight pairs of SSR marker primers with polymorphism between parents and easy to amplify and distinguish bands were screened. The results of DNA fragment size are shown in Table 2. A total of 18 alleles were amplified by eight primer pairs. PCR product size (bp) for eight primers ranged from 108 bp (ES55) to 176 bp (ES63) in 65 samples, and the allele size ranged from 108 bp to 113 bp in ES55 and from 173 bp to 176 bp in ES63, respectively. The genotypes of ES35, ES39, ES55, ES63, ES78, ES83, ES98, and ES172 in ‘Jinhuang’ and ‘Renong No.1′ were analyzed, and at least one gene locus could be segregated in the offspring. The possible locus combinations of hybrid offspring were analyzed according to the genotypes of parents. Only those with multiple SSR loci can be identified as true hybrid progeny (Table 3).

Table 2.
PCR product size (bp) for eight SSR primers obtained from gene scan analysis for ‘Jinhuang’ (♀), ‘Renong No.1′ (♂), and 65 F1 individuals.

Table 3.
Genotypes of eight SSR molecular makers for two parents (‘Jinhuang’♀ and ‘Renong No.1′♂) and their segregation patterns among 65 offsprings.
A false hybrid does not contain a paternal-specific band in F1 genotype. At ES39 locus, the genotype of ‘Jinhuang’ was 152/162, and genotype of ‘Renong No.1′ was 162/162. Therefore, the specific band 162/162 of hybrid F1 individuals amplified at ES39 site was a true hybrid, while specific band 152/152 of JR-59 was a false hybrid (Figure 1). At ES55 locus, the genotype of ‘Jinhuang’ was 108/113, and that of ‘Renong No.1′ was 113/113. Therefore, the true hybrid with specific band 113/113 was amplified at the ES55 locus, while JR13-3 with the specific band 108/108 was a false hybrid (Figure 2). At ES63 locus, the genotype of ‘Jinhuang’ was 173/173, and that of ‘Renong No.1′ was 173/176, therefore, the genotype of true hybrid at this locus was 173/176, while a new locus 179 appeared on JR-44, which was also a false hybrid (Figure 3).
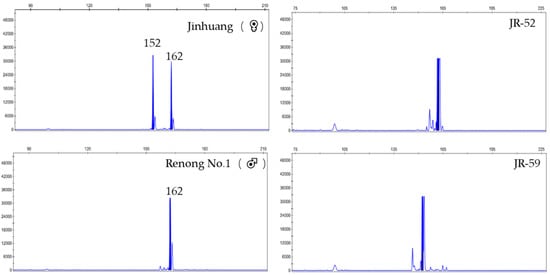
Figure 1.
Capillary electrophoresis results obtained with ES39 for ‘Jinhuang’, ‘Renong No.1′, true hybrid JR-52, and false hybrid JR-59.
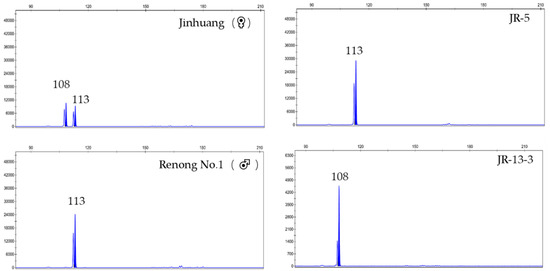
Figure 2.
Capillary electrophoresis results obtained with ES55 in ‘Jinhuang’, ‘Renong No.1′, true hybrid JR-5, and false hybrid JR-13-3.
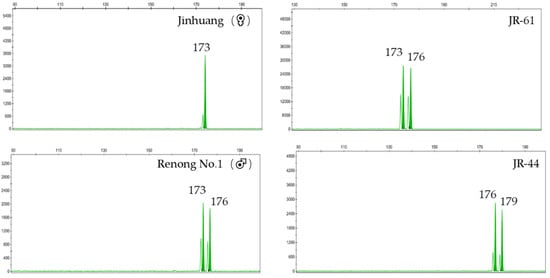
Figure 3.
Capillary electrophoresis results obtained with ES63 in ‘Jinhuang’, ‘Renong No.1′, true hybrid JR-61, and false hybrid JR-44.
Eight SSR primers, ES35, ES39, ES55, ES63, ES78, ES83, ES98, and ES172, were used to mutually verify 65 progenies. The results showed that 62 F1 individuals could amplify parental polymorphic bands at eight loci. It was found that 62 offspring might be true hybrids, one offspring might not be the hybrid offspring of ‘Jinhuang’ and ‘Renong No.1′ or a false hybrid, and two offsprings might be inbred or hybrid, which need further identification. Among them, 59 F1 individuals amplified polymorphic bands of their parents at two to seven loci, which could be identified as true hybrids, and three offspring were identified as true hybrids at only one locus. In total, true hybrids accounted for 95.38% of all F1 hybrids.
3.2. Segregation of SSR Loci
In order to characterize whether SSR alleles were derived from single locus or multiple loci, segregation of SSR genotypes was evaluated by using 65 F1 individuals obtained from the cross of ’ Jinhuang’ × ‘Renong No.1′ (Table 3). Eight SSR loci showed segregation of SSR genotypes in the F1 individuals of ‘Jinhuang’ × ‘Renong No.1′. Among them, seven SSR loci showed binary segregation (a/a:a/b, a/b:b/b, a/a:b/a, a/b:a/a), whereas only one SSR locus ES83 showing ab:ac:bb:bc segregation fitted to the expected segregation ratio of 1:1:1:1. These results indicated that eight SSR loci used in this study were derived from single locus, which can be used to evaluate considerably exact genetic diversity and relatedness of mango cultivars. The expected heterozygosity (He) level, which is the best estimator of genetic diversity in the population, ranged from 0.34 at ES35 and ES78 loci to 0.62 at ES83 loci with an average of 0.40, whereas observed heterozygosity (HO) values ranged from 0.44 to 0.81 with an average of 0.52.
According to the segregation type of co-dominant SSR markers in F1 generation, the segregation bands of different alleles at the same locus of eight SSR markers were sorted out for x2 test. The results showed that there was no partial segregation at eight loci (p < 0.05), which was in accordance with Mendel’s law of segregation.
3.3. Dendrogram Analysis of Parents and Progenies
Eight pairs of EST-SSRs primers were selected for diversity analysis of parents and hybridized progenies. According to SSR amplification results, UPGMA cluster analysis was conducted based on the hybrid parents and 62 hybrid progeny populations. As shown in Figure 4, 62 hybrid progenies were classified into two categories when similarity coefficient was 0.58. Category 1 was clustered together with female parent ‘Jinhuang’, which contained 27 individuals with the proportion of 44%, showing partial maternal inheritance. The Category 2 was clustered together with male parent ‘Renong No.1′, which contained 35 plants with the proportion of 56%, showing partial paternal inheritance. The clustering results showed that most individual plants of this combination had closer genetic relationship with male parent ‘Renong No.1′ and had a tendency of paternal inheritance.
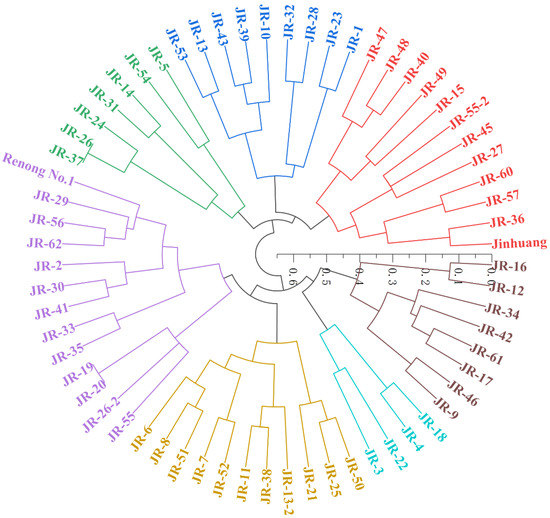
Figure 4.
Dendrogram of parents and progenies of ‘Jinhuang’ × ‘Renong No.1′ derived from UPGMA cluster analysis based on eight SSRs makers. Different colors represent offsprings clustered in different categories.
3.4. Development of Hybrids DNA Fingerprinting
To identify the different hybrid offsprings and provide a molecular basis for variety conservation, fingerprints of 62 true hybrids from ‘Jinhuang’ × ‘Renong No.1′ cross were developed using the PCR amplification loci from eight SSR primers (Table 4). About 18 amplified loci generated by using eight primer pairs were unable to distinguish the offsprings of JR-19 and JR-20, JR-26 and JR-37, indicating that the genetic backgrounds of JR-19 and JR-20, JR-26 and JR-37 were relatively close.

Table 4.
Fingerprints of 62 true hybrids from ‘Jinhuang’ × ‘Renong No.1′ cross using alleles detected with eight SSR primers.
4. Discussion
SSR markers have been widely used to differentiate mango cultivars and identify genetic diversity [17,26], distinguish some synonymous (identical SSR genotypes with different cultivar names) and homonymous (different SSR genotypes with the same cultivar name) accessions [31], genetic relatedness [16], and so on. All these SSR markers will be used in the future studies of genetic diversity, genetic map construction, and marker-assisted selection and utilized for breeding programs in mango. In previous study, we carried out transcriptome analysis of ‘Zill’ mango fruits [32], and 93 EST-SSR markers were developed based on transcriptome sequence data [29]. In this study, 34 pairs of primers with different bands between ‘Jinhuang’ and ‘Renong No.1′ were selected for the screening of hybrid identification primers from 93 SSR markers, of which eight pairs of SSR marker primers with polymorphism between parents and easy to amplify and distinguish bands were screened.
Cross breeding is an important way to breed new mango varieties. However, the difficulties in artificial hybridization, parent compatibility selection, and population construction of target traits have limited the progress of breeding new high quality mango varieties. At present, SSR molecular markers have been widely used in hybrid identification of various crops [14,33,34]. Eight SSR primers showed higher polymorphism between parents in combination with polyacrylamide gel electrophoresis were used to identify authenticity of ‘Jinhuang’ × ‘Irwin’ hybrids, and a total of 184 F1 seedlings were identified as true hybrids [35]. These results showed that it was feasible to identify the authenticity of fruit tree hybrids by SSR molecular markers. However, the traditional SSR molecular marker technology has many disadvantages, such as heavy workload, low precision, time-consuming, and incapability for the detection of a large number of samples. Compared with polyacrylamide gel electrophoresis detection, fluorescent capillary electrophoresis has many advantages, such as higher resolution, as fine as 1 bp. In addition, electrophoresis results are displayed in a peak plot corresponding to the size of the amplified fragments, which can be directly collected to achieve digitization of the results in a form, which is easy to record, analyze, and save. The application of high-throughput, multi-PCR, and ten-fold capillary electrophoresis detection technology has improved the detection efficiency by 33-fold [36]. Briefly, SSR fluorescence labeled capillary electrophoresis detection method has the advantages of high-throughput, sensitiveness, cost-effectiveness, and high accuracy. Wang et al. [20] used eight polymorphic SSR markers to identify hybrids of 117 bayberry hybrid progenies, and successfully identified 83 true hybrids. Cheng et al. [21] screened a pair of SSR primer for seed purity identification of watermelon hybrid ‘Wunong No.8’ by using fluorescent marker SSR technology, and the purity was 99%, which was consistent with the results of morphological identification of ‘Wunong No.8’ in the field. Lei et al. [37] used 17 pairs of SSR primers to identify true and false hybrids of two F1 hybrid populations in tea, and successfully identified 41 and 35 true hybrids, with hybrid rates of 85.42% and 79.55%, respectively.
However, when SSR molecular markers were used to identify the authenticity of hybrid progenies, the results of different primers were inconsistent. Han et al. [13] thought that different SSR primers had different identification efficiency for homozygous dominant markers and heterozygous dominant markers. Theoretically, only one homozygous dominant marker is required to identify all hybrid offspring, while for heterozygous dominant markers, at least five parent-specific markers are required to identify 97% of true hybrids. Yin et al. [38] found the first type of AA × BB, AB × CD, and AB × CC marker, and there is no same allele locus between parents. Theoretically, only one SSR primer is needed to identify all true hybrids, and the identification rate is high. For the second type of AA × AB and AB × BC type marker, because there is one identical allele locus between parents, the identification ability is poor, which needs further verification. In this study, eight pairs of SSR markers with high polymorphism were used to identify the authenticity of 65 mango F1 hybrids. A total of 62 true hybrids were identified, and the hybrid rate was 95.38%. The results showed that the fluorescent SSR markers could be applied for the identification of true and false mango hybrids. The identification process was simple, fast, and low cost, and the results were stable and reliable. It is a useful supplement to molecular biology identification technology of mango hybrids, which could be widely used in mango germplasm innovation, hybrid breeding, and other fields in the future.
Mango breeding is a long-term activity, complicated by a heterozygous genome, juvenility, low fruit set and retention rates, long evaluation periods, polyembryony, and outcrossing behavior. These factors make genetic improvement through conventional parental selection and breeding slow and unpredictable. It is difficult to obtain genetic materials and the fruit has a long growth cycle, which makes mango genetic research far behind other fruit crops. The results of previous studies showed that genetic composition of ‘Jinhuang’ and ‘Renong No.1′ was quite different, the progenies were widely separated, and strong heterosis was observed in hybrids for most of the traits examined. The quantitative traits such as average fruit weight, total soluble solids, and descriptive traits such as peel color, pulp color, and fruit flavor of hybrid offspring showed rich diversity [39], which could be used as excellent gene resources for subsequent breeding. The number of hybrid population constructed in this study was small, so it is necessary to expand the population number to lay a foundation for construction of mango molecular marker linkage map and subsequent genetic analysis. The adoption of molecular markers and genomics-based breeding strategies will likely improve predictability and breeding efficiency.
5. Conclusions
In the present study, we used eight SSR markers to test the authenticity of hybrids derived from crossing combinations of ‘Jinhuang’ and ‘Renong No.1′, clarified the inheritance of parental alleles, and developed fingerprints for hybrid populations, which could facilitate the breeding of mango new cultivars. The results showed that SSR fluorescent labeling technology could quickly and effectively identify the authenticity of mango hybrid progenies, and could be applied to the identification of true and false hybrids, which would lay a foundation for the follow-up mango cross breeding and molecular marker-assisted breeding.
Author Contributions
Conceptualization, H.W., M.Q. and S.W.; methodology, H.W. and X.L.; data curation, X.L., B.Z. and W.X.; writing—original draft preparation, X.L., X.M. and H.W.; writing—review and editing, X.L., S.W., M.Q. and H.W.; funding acquisition, H.W. and M.Q. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Hainan Province Key Research and Development Plan (grant number: ZDYF2022XDNY255), National Key Research and Development Plan of China (grant number: 2018YFD1000504, 2019YFD1000504), Guangdong Provincial Special Fund for Modern Agriculture Industry Technology Innovation Teams (grant number: 2022KJ108).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Deshpande, A.; Anamika, K.; Jha, V.; Chidley, H.; Oak, P.; Kadoo, N.Y.; Pujari, K.; Giri, A.; Gupta, V. Transcriptional transitions in Alphonso mango (Mangifera indica L.) during fruit development and ripening explain its distinct aroma and shelf life characteristics. Reports 2017, 7, 8711. [Google Scholar] [CrossRef]
- Bompard, J. Taxonomy and systematics. In The Mango. Botany, Production and Use, 2nd ed.; Litz, R.E., Ed.; CABI Publishing: Wallingford, UK, 2009; pp. 19–41. [Google Scholar]
- Chen, Q. Perspectives on the mango industry in mainland China. Acta Hortic. 2013, 992, 25–36. [Google Scholar] [CrossRef]
- Wang, S.; Wu, H.; Ma, W.; Zhan, R.; Yao, Q.; Sun, G. Observating and evaluating on 16 Mango (Mangifera indica L.) varieties introduced from abroad. Chin. J. Trop. Crops. 2010, 21, 1655–1660. (In Chinese) [Google Scholar]
- Huang, J.; Wu, H.; Wang, S.; Liang, G.; Wang, W.; Ma, W. Effect of low temperature on nucleolus and chromosomes’ behavior of mango (Mangifera indica L.) PMC during meiosis. Acta Hortic. Sin. 2008, 35, 1101–1108. (In Chinese) [Google Scholar]
- Wang, S.; Zhan, R.; Zhao, J.; Guo, X.; Wu, H.; Yao, Q. Countermeasures for industrialization development of late ripening mango in Jinshajiang dry-hot valley. Chin. J. Trop. Agric. 2010, 30, 36–39. (In Chinese) [Google Scholar]
- Ma, X.; Wang, J.; Su, M.; Liu, B.; Du, B.; Zhang, Y.; He, L.; Wang, S.; Wu, H. The link between mineral elements variation and internal flesh breakdown of ‘keitt’ mango in a steep slope mountain area, Southwest China. Horticulture 2022, 8, 533. [Google Scholar] [CrossRef]
- Campbell, R. (Ed.) A guide to Mangos in Florida; Fairchild Tropical Garden: Miami, FL, USA, 1992. [Google Scholar]
- Human, C.; Rheeder, S.; Sippel, A. New cultivars and hybrid selections from the mango breeding program of the Agricultural Research Council in South Africa. Acta Hortic. 2009, 820, 119–126. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Z.; Gu, X.; Mao, S.; Li, X. Genetic diversity of pepper (Capsicum spp.) germplasm resources in China reflects selection for cultivar types and spatial distribution. J. Integr. Agric. 2016, 15, 1991–2001. [Google Scholar] [CrossRef]
- Zhou, R.; Wu, Z.; Jiang, F.; Liang, M. Comparison of gSSR and EST-SSR markers for analyzing genetic variability among tomato cultivars (Solanum lycopersicum L.). Genet. Mol. Res. 2015, 14, 13184–13194. [Google Scholar] [CrossRef]
- Zhong, H.; Zhang, F.; Zhou, X.; Pan, M.; Wu, X. Genome-wide identification of sequence variations and SSR marker development in the Munake Grape cultivar. Front. Ecol. Evol. 2021, 9, 190. [Google Scholar] [CrossRef]
- Han, G.; Xiang, S.; Wang, W.; Wei, X.; He, B.; Li, X.; Liang, G. Identification and genetic diversity of hybrid progenies from Shatian Pummelo by SSR. Sci. Agric. Sin. 2010, 43, 4678–4686. (In Chinese) [Google Scholar]
- Sun, Q.; Ma, S.; Liu, W.; Jiang, N.; Fang, J.; Yang, X.; Xiang, X. Identification and analysis on true or false hybrids from hybridization populations of Litchi (Litchi chinensis Sonn.) using EST-SSR markers. Agric. Biotec. 2015, 4, 38–43,46. [Google Scholar]
- Duval, M.; Bunel, J.; Sitbon, C.; Risterucci, A. Development of microsatellite markers for mango (Mangifera indica L.). Mol. Ecol. Resour. 2010, 5, 824–826. [Google Scholar] [CrossRef]
- Didiana, G.; Sanjuana, H.; Maurilio, G.; Noe, B.E.; Maurilio, S.; Netzahualcoyot, M. Genetic analysis of mango landraces from Mexico based on molecular markers. Plant Genet. Resour. C 2009, 7, 244–251. [Google Scholar]
- Ravishankar, K.; Dinesh, M.; Nischita, P.; Sandya, B. Development and characterization of microsatellite markers in mango (Mangifera indica) using next-generation sequencing technology and their transferability across species. Mol. Breed. 2015, 35, 93. [Google Scholar] [CrossRef]
- Viruel, M.; Escribano, P.; Barbieri, M.; Ferri, M.; Hormaza, J. Fingerprinting, embryo type and geographic differentiation in mango (Mangifera indica L., Anacardiaceae) with microsatellites. Mol. Breed. 2005, 15, 383–393. [Google Scholar] [CrossRef]
- Zhu, J.; Guo, Y.; Liu, Z.; Kun, L.; Yang, X.; Shi, G.; Niu, Z.; Li, C.; Guo, X. Identification of the F1 hybrids of grape using SSR molecular markers. J. Shenyang Agric. Univ. 2016, 47, 148–152. (In Chinese) [Google Scholar]
- Wang, G.; Shen, Y.; Jia, H.; Jiao, Y.; Chai, C.; Bao, J.; Sun, D.; Jia, H.; Gao, Z. Construction of a crossing population between two Chinese bayberry cultivars ‘Biqi’ and ‘Dongkui’ and hybrid identification by polymorphic SSRs. J. Fruit Sci. 2015, 32, 555–560. (In Chinese) [Google Scholar]
- Cheng, W.; Luo, X.; Hong, J.; Wang, S.; Huang, X.; Du, L.; Jiang, L.; Hong, Z.; Chen, G. Using the fluorescent labeled SSR marker to identify hybrid purity of Watermelon. China Fruit Veg. 2020, 40, 36–41. (In Chinese) [Google Scholar]
- Saxena, R.; Saxena, K.; Varshney, R. Application of SSR markers for molecular characterization of hybrid parents and purity assessment of ICPH 2438 hybrid of pigeonpea [Cajanus cajan (L.) Millspaugh]. Mol. Breed. 2010, 26, 371–380. [Google Scholar] [CrossRef]
- Shi, F.; Wang, Q.; Zhao, M.; Wang, J.; Guan, L. Identification of interspecific hybrids derived from Fragaria × ananassa× F. viridis by morphological features and SSR markers. J. Fruit Sci. 2017, 34, 175–185. (In Chinese) [Google Scholar]
- Dillon, N.; Bally, I.; Hucks, L.; Wright, C.; Innes, D.; Dietzgen, R. Implementation of SSR markers in mango breeding in Australia. Acta Hortic. 2013, 992, 259–267. [Google Scholar] [CrossRef]
- Ravishankar, K.; Bommisetty, P.; Bajpai, A.; Srivastava, N.; Mani, B.; Vasugi, C.; Rajan, S.; Dinesh, M. Genetic diversity and population structure analysis of mango (Mangifera indica) cultivars assessed by microsatellite markers. Trees 2015, 29, 775–783. [Google Scholar] [CrossRef]
- Srivastav, M.; Singh, S.; Prakash, J.; Singh, R.; Singh, N. New hyper-variable SSRs for diversity analysis in mango (Mangifera indica L.). Indian J. Genet. Plant Breed. 2021, 81, 119–126. [Google Scholar] [CrossRef]
- Surapaneni, M.; Vemireddy, L.; Hameedunnisa, B.; Reddy, B.; Anwar, S.; Siddiq, E. Genetic variability and DNA fingerprinting of elite mango genotypes of india using microsatellite markers. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2018, 89, 1251–1258. [Google Scholar] [CrossRef]
- Sajana, S.; Thomas, P.; Ravishankar, K.; Nandeesha, P.; Kurian, R.; Sane, A.; Bindu, H. Marker-assisted confirmation of maternal or zygotic origin of embryogenic cultures and regenerated plantlets of two polyembryonic mango cultivars using SSR markers. J. Hortic. Sci. Biotech. 2020, 96, 201–208. [Google Scholar]
- Luo, C.; Wu, H.; Yao, Q.; Wang, S.; Xu, W. Development of EST-SSR and TRAP markers from transcriptome sequencing data of the mango. Genet. Mol. Res. 2015, 14, 7914–7919. [Google Scholar] [CrossRef]
- Yeh, F.; Yang, R.; Boyle, T. POPGENE version 1.31. Microsoft Windows-Based Freeware for Population Genetic Analysis; University Alberta: Edmonton, AB, Canada, 1999. [Google Scholar]
- Shinsuke, Y.; Fumiko, H.; Masato, M.; Yuko, O.-M.; Kenji, N.; Naoya, U.; Tatsushi, O.; Moriyuki, S.; Toshiya, Y. Genetic diversity and relatedness of mango cultivars assessed by SSR markers. Breed. Sci. 2019, 69, 332–344. [Google Scholar]
- Wu, H.; Jia, H.; Ma, X.; Wang, S.; Yao, Q.; Xu, W.; Zhou, Y.; Gao, Z.; Zhan, R. Transcriptome and proteomic analysis of mango (Mangifera indica Linn.) fruits. J. Proteom. 2014, 105, 19–30. [Google Scholar] [CrossRef]
- Han, Z.; Bi, Z.; Lu, F.; Gao, P.; Zhang, P.; Li, J.; Zhang, J. Analysis of the relative relationship of different hawthorn cultivars by apple SSR primers. Hebei J. For. Orchard. Res. 2014, 29, 165–168. (In Chinese) [Google Scholar]
- Dillon, N.; Innes, D.J.; Bally, I.; Wright, C.; Devitt, L.; Dietzgen, R. Expressed sequence tag-simple sequence repeat (EST-SSR) marker resources for diversity analysis of mango (Mangifera indica L.). Diversity 2014, 6, 72–87. [Google Scholar] [CrossRef]
- Yao, Q.; Lei, X.; Huang, Z.; Zhu, M.; Wang, S.; Huang, L.; Ma, X.; Zhan, R.; Wu, H. Construction of the resistance to anthracnose for crossed mango population and identification by SSR molecular markers. J. Fruit Sci. 2010, 27, 265–269. (In Chinese) [Google Scholar]
- Gao, Y.; Wang, K.; Tian, L.; Cao, Y.; Liu, F. Identification of apple cultivars by TP-M13-SSR technique. J. Fruit Sci. 2010, 27, 833–837. (In Chinese) [Google Scholar]
- Lei, Y.; Duan, J.; Huang, F.; Kang, Y.; Luo, Y.; Chen, Y.; Ding, D.; Yao, L.; Dong, L.; Li, S. Identification and genetic diversity of Tea F1 hybrids based on SSR markers. Plant. Genet. Resour. 2021, 22, 748–757. [Google Scholar]
- Yin, B.; Zhang, Y.; Sun, F. Identification and genetic diversity of apple F1 hybrids based on SSR markers. J. Hebei Agric. Univ. 2019, 42, 46–51. (In Chinese) [Google Scholar]
- Wu, H.; Li, X.; Xu, W.; Zheng, B.; Ma, X.; Su, M.; Liang, Q.; Yao, Q.; Wang, S. Genetic analysis of fruit traits in F1 progenies of ‘Chinhuang’ב Renong 1’ mango. Acta Hortic. Sin. 2022, 49, 416–426. (In Chinese) [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).