Identification of F1 Hybrid Progenies in Mango Based on Fluorescent SSR Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Extraction
2.3. The Choice of SSR
2.4. PCR Amplication and SSR Analysis
2.5. Data Analysis
3. Results
3.1. Genetic Identification of Mango Using SSR Markers
3.2. Segregation of SSR Loci
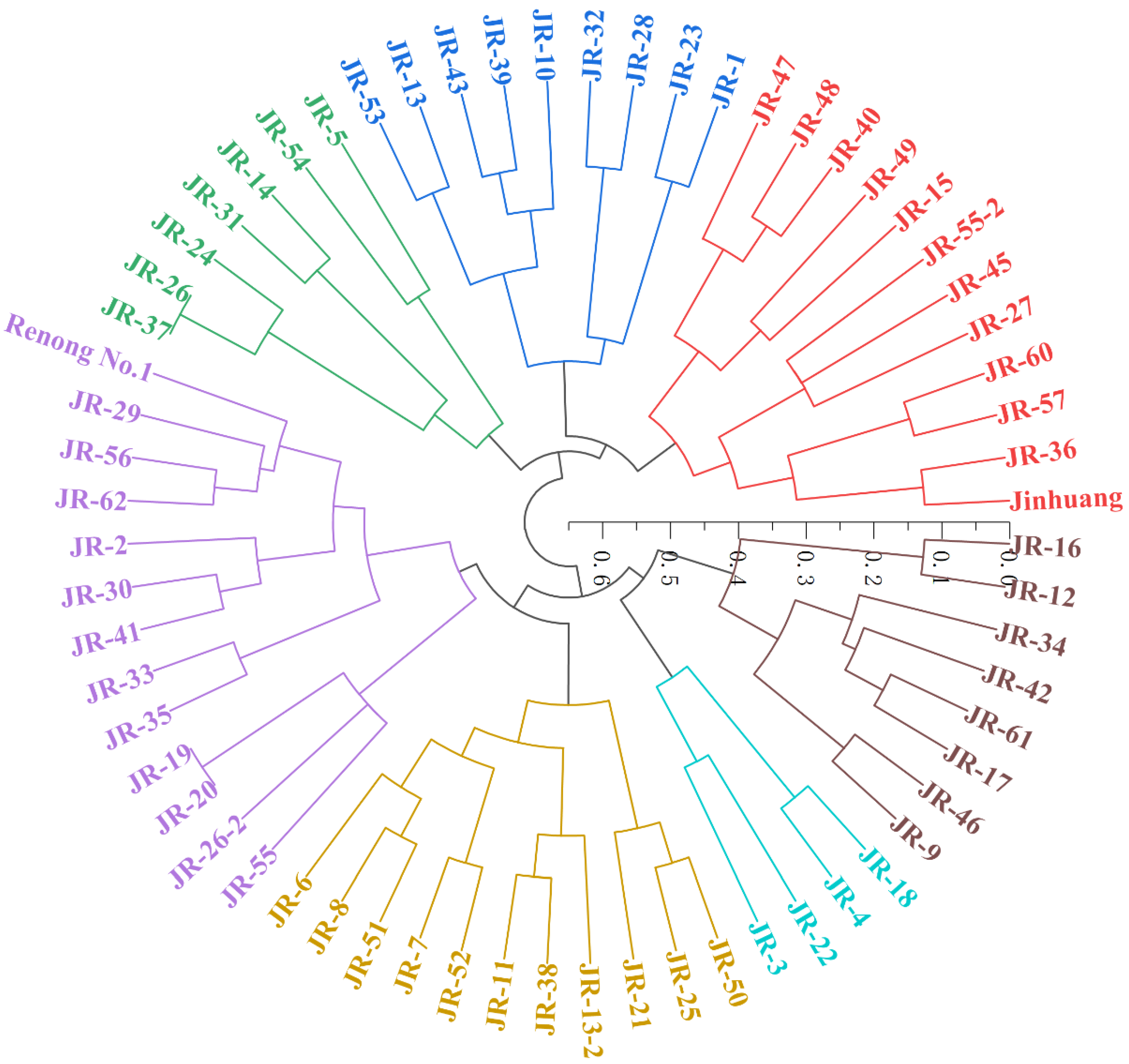
3.3. Dendrogram Analysis of Parents and Progenies
3.4. Development of Hybrids DNA Fingerprinting
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Deshpande, A.; Anamika, K.; Jha, V.; Chidley, H.; Oak, P.; Kadoo, N.Y.; Pujari, K.; Giri, A.; Gupta, V. Transcriptional transitions in Alphonso mango (Mangifera indica L.) during fruit development and ripening explain its distinct aroma and shelf life characteristics. Reports 2017, 7, 8711. [Google Scholar] [CrossRef]
- Bompard, J. Taxonomy and systematics. In The Mango. Botany, Production and Use, 2nd ed.; Litz, R.E., Ed.; CABI Publishing: Wallingford, UK, 2009; pp. 19–41. [Google Scholar]
- Chen, Q. Perspectives on the mango industry in mainland China. Acta Hortic. 2013, 992, 25–36. [Google Scholar] [CrossRef]
- Wang, S.; Wu, H.; Ma, W.; Zhan, R.; Yao, Q.; Sun, G. Observating and evaluating on 16 Mango (Mangifera indica L.) varieties introduced from abroad. Chin. J. Trop. Crops. 2010, 21, 1655–1660. (In Chinese) [Google Scholar]
- Huang, J.; Wu, H.; Wang, S.; Liang, G.; Wang, W.; Ma, W. Effect of low temperature on nucleolus and chromosomes’ behavior of mango (Mangifera indica L.) PMC during meiosis. Acta Hortic. Sin. 2008, 35, 1101–1108. (In Chinese) [Google Scholar]
- Wang, S.; Zhan, R.; Zhao, J.; Guo, X.; Wu, H.; Yao, Q. Countermeasures for industrialization development of late ripening mango in Jinshajiang dry-hot valley. Chin. J. Trop. Agric. 2010, 30, 36–39. (In Chinese) [Google Scholar]
- Ma, X.; Wang, J.; Su, M.; Liu, B.; Du, B.; Zhang, Y.; He, L.; Wang, S.; Wu, H. The link between mineral elements variation and internal flesh breakdown of ‘keitt’ mango in a steep slope mountain area, Southwest China. Horticulture 2022, 8, 533. [Google Scholar] [CrossRef]
- Campbell, R. (Ed.) A guide to Mangos in Florida; Fairchild Tropical Garden: Miami, FL, USA, 1992. [Google Scholar]
- Human, C.; Rheeder, S.; Sippel, A. New cultivars and hybrid selections from the mango breeding program of the Agricultural Research Council in South Africa. Acta Hortic. 2009, 820, 119–126. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Z.; Gu, X.; Mao, S.; Li, X. Genetic diversity of pepper (Capsicum spp.) germplasm resources in China reflects selection for cultivar types and spatial distribution. J. Integr. Agric. 2016, 15, 1991–2001. [Google Scholar] [CrossRef]
- Zhou, R.; Wu, Z.; Jiang, F.; Liang, M. Comparison of gSSR and EST-SSR markers for analyzing genetic variability among tomato cultivars (Solanum lycopersicum L.). Genet. Mol. Res. 2015, 14, 13184–13194. [Google Scholar] [CrossRef]
- Zhong, H.; Zhang, F.; Zhou, X.; Pan, M.; Wu, X. Genome-wide identification of sequence variations and SSR marker development in the Munake Grape cultivar. Front. Ecol. Evol. 2021, 9, 190. [Google Scholar] [CrossRef]
- Han, G.; Xiang, S.; Wang, W.; Wei, X.; He, B.; Li, X.; Liang, G. Identification and genetic diversity of hybrid progenies from Shatian Pummelo by SSR. Sci. Agric. Sin. 2010, 43, 4678–4686. (In Chinese) [Google Scholar]
- Sun, Q.; Ma, S.; Liu, W.; Jiang, N.; Fang, J.; Yang, X.; Xiang, X. Identification and analysis on true or false hybrids from hybridization populations of Litchi (Litchi chinensis Sonn.) using EST-SSR markers. Agric. Biotec. 2015, 4, 38–43,46. [Google Scholar]
- Duval, M.; Bunel, J.; Sitbon, C.; Risterucci, A. Development of microsatellite markers for mango (Mangifera indica L.). Mol. Ecol. Resour. 2010, 5, 824–826. [Google Scholar] [CrossRef]
- Didiana, G.; Sanjuana, H.; Maurilio, G.; Noe, B.E.; Maurilio, S.; Netzahualcoyot, M. Genetic analysis of mango landraces from Mexico based on molecular markers. Plant Genet. Resour. C 2009, 7, 244–251. [Google Scholar]
- Ravishankar, K.; Dinesh, M.; Nischita, P.; Sandya, B. Development and characterization of microsatellite markers in mango (Mangifera indica) using next-generation sequencing technology and their transferability across species. Mol. Breed. 2015, 35, 93. [Google Scholar] [CrossRef]
- Viruel, M.; Escribano, P.; Barbieri, M.; Ferri, M.; Hormaza, J. Fingerprinting, embryo type and geographic differentiation in mango (Mangifera indica L., Anacardiaceae) with microsatellites. Mol. Breed. 2005, 15, 383–393. [Google Scholar] [CrossRef]
- Zhu, J.; Guo, Y.; Liu, Z.; Kun, L.; Yang, X.; Shi, G.; Niu, Z.; Li, C.; Guo, X. Identification of the F1 hybrids of grape using SSR molecular markers. J. Shenyang Agric. Univ. 2016, 47, 148–152. (In Chinese) [Google Scholar]
- Wang, G.; Shen, Y.; Jia, H.; Jiao, Y.; Chai, C.; Bao, J.; Sun, D.; Jia, H.; Gao, Z. Construction of a crossing population between two Chinese bayberry cultivars ‘Biqi’ and ‘Dongkui’ and hybrid identification by polymorphic SSRs. J. Fruit Sci. 2015, 32, 555–560. (In Chinese) [Google Scholar]
- Cheng, W.; Luo, X.; Hong, J.; Wang, S.; Huang, X.; Du, L.; Jiang, L.; Hong, Z.; Chen, G. Using the fluorescent labeled SSR marker to identify hybrid purity of Watermelon. China Fruit Veg. 2020, 40, 36–41. (In Chinese) [Google Scholar]
- Saxena, R.; Saxena, K.; Varshney, R. Application of SSR markers for molecular characterization of hybrid parents and purity assessment of ICPH 2438 hybrid of pigeonpea [Cajanus cajan (L.) Millspaugh]. Mol. Breed. 2010, 26, 371–380. [Google Scholar] [CrossRef]
- Shi, F.; Wang, Q.; Zhao, M.; Wang, J.; Guan, L. Identification of interspecific hybrids derived from Fragaria × ananassa× F. viridis by morphological features and SSR markers. J. Fruit Sci. 2017, 34, 175–185. (In Chinese) [Google Scholar]
- Dillon, N.; Bally, I.; Hucks, L.; Wright, C.; Innes, D.; Dietzgen, R. Implementation of SSR markers in mango breeding in Australia. Acta Hortic. 2013, 992, 259–267. [Google Scholar] [CrossRef]
- Ravishankar, K.; Bommisetty, P.; Bajpai, A.; Srivastava, N.; Mani, B.; Vasugi, C.; Rajan, S.; Dinesh, M. Genetic diversity and population structure analysis of mango (Mangifera indica) cultivars assessed by microsatellite markers. Trees 2015, 29, 775–783. [Google Scholar] [CrossRef]
- Srivastav, M.; Singh, S.; Prakash, J.; Singh, R.; Singh, N. New hyper-variable SSRs for diversity analysis in mango (Mangifera indica L.). Indian J. Genet. Plant Breed. 2021, 81, 119–126. [Google Scholar] [CrossRef]
- Surapaneni, M.; Vemireddy, L.; Hameedunnisa, B.; Reddy, B.; Anwar, S.; Siddiq, E. Genetic variability and DNA fingerprinting of elite mango genotypes of india using microsatellite markers. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2018, 89, 1251–1258. [Google Scholar] [CrossRef]
- Sajana, S.; Thomas, P.; Ravishankar, K.; Nandeesha, P.; Kurian, R.; Sane, A.; Bindu, H. Marker-assisted confirmation of maternal or zygotic origin of embryogenic cultures and regenerated plantlets of two polyembryonic mango cultivars using SSR markers. J. Hortic. Sci. Biotech. 2020, 96, 201–208. [Google Scholar]
- Luo, C.; Wu, H.; Yao, Q.; Wang, S.; Xu, W. Development of EST-SSR and TRAP markers from transcriptome sequencing data of the mango. Genet. Mol. Res. 2015, 14, 7914–7919. [Google Scholar] [CrossRef]
- Yeh, F.; Yang, R.; Boyle, T. POPGENE version 1.31. Microsoft Windows-Based Freeware for Population Genetic Analysis; University Alberta: Edmonton, AB, Canada, 1999. [Google Scholar]
- Shinsuke, Y.; Fumiko, H.; Masato, M.; Yuko, O.-M.; Kenji, N.; Naoya, U.; Tatsushi, O.; Moriyuki, S.; Toshiya, Y. Genetic diversity and relatedness of mango cultivars assessed by SSR markers. Breed. Sci. 2019, 69, 332–344. [Google Scholar]
- Wu, H.; Jia, H.; Ma, X.; Wang, S.; Yao, Q.; Xu, W.; Zhou, Y.; Gao, Z.; Zhan, R. Transcriptome and proteomic analysis of mango (Mangifera indica Linn.) fruits. J. Proteom. 2014, 105, 19–30. [Google Scholar] [CrossRef]
- Han, Z.; Bi, Z.; Lu, F.; Gao, P.; Zhang, P.; Li, J.; Zhang, J. Analysis of the relative relationship of different hawthorn cultivars by apple SSR primers. Hebei J. For. Orchard. Res. 2014, 29, 165–168. (In Chinese) [Google Scholar]
- Dillon, N.; Innes, D.J.; Bally, I.; Wright, C.; Devitt, L.; Dietzgen, R. Expressed sequence tag-simple sequence repeat (EST-SSR) marker resources for diversity analysis of mango (Mangifera indica L.). Diversity 2014, 6, 72–87. [Google Scholar] [CrossRef]
- Yao, Q.; Lei, X.; Huang, Z.; Zhu, M.; Wang, S.; Huang, L.; Ma, X.; Zhan, R.; Wu, H. Construction of the resistance to anthracnose for crossed mango population and identification by SSR molecular markers. J. Fruit Sci. 2010, 27, 265–269. (In Chinese) [Google Scholar]
- Gao, Y.; Wang, K.; Tian, L.; Cao, Y.; Liu, F. Identification of apple cultivars by TP-M13-SSR technique. J. Fruit Sci. 2010, 27, 833–837. (In Chinese) [Google Scholar]
- Lei, Y.; Duan, J.; Huang, F.; Kang, Y.; Luo, Y.; Chen, Y.; Ding, D.; Yao, L.; Dong, L.; Li, S. Identification and genetic diversity of Tea F1 hybrids based on SSR markers. Plant. Genet. Resour. 2021, 22, 748–757. [Google Scholar]
- Yin, B.; Zhang, Y.; Sun, F. Identification and genetic diversity of apple F1 hybrids based on SSR markers. J. Hebei Agric. Univ. 2019, 42, 46–51. (In Chinese) [Google Scholar]
- Wu, H.; Li, X.; Xu, W.; Zheng, B.; Ma, X.; Su, M.; Liang, Q.; Yao, Q.; Wang, S. Genetic analysis of fruit traits in F1 progenies of ‘Chinhuang’ב Renong 1’ mango. Acta Hortic. Sin. 2022, 49, 416–426. (In Chinese) [Google Scholar]
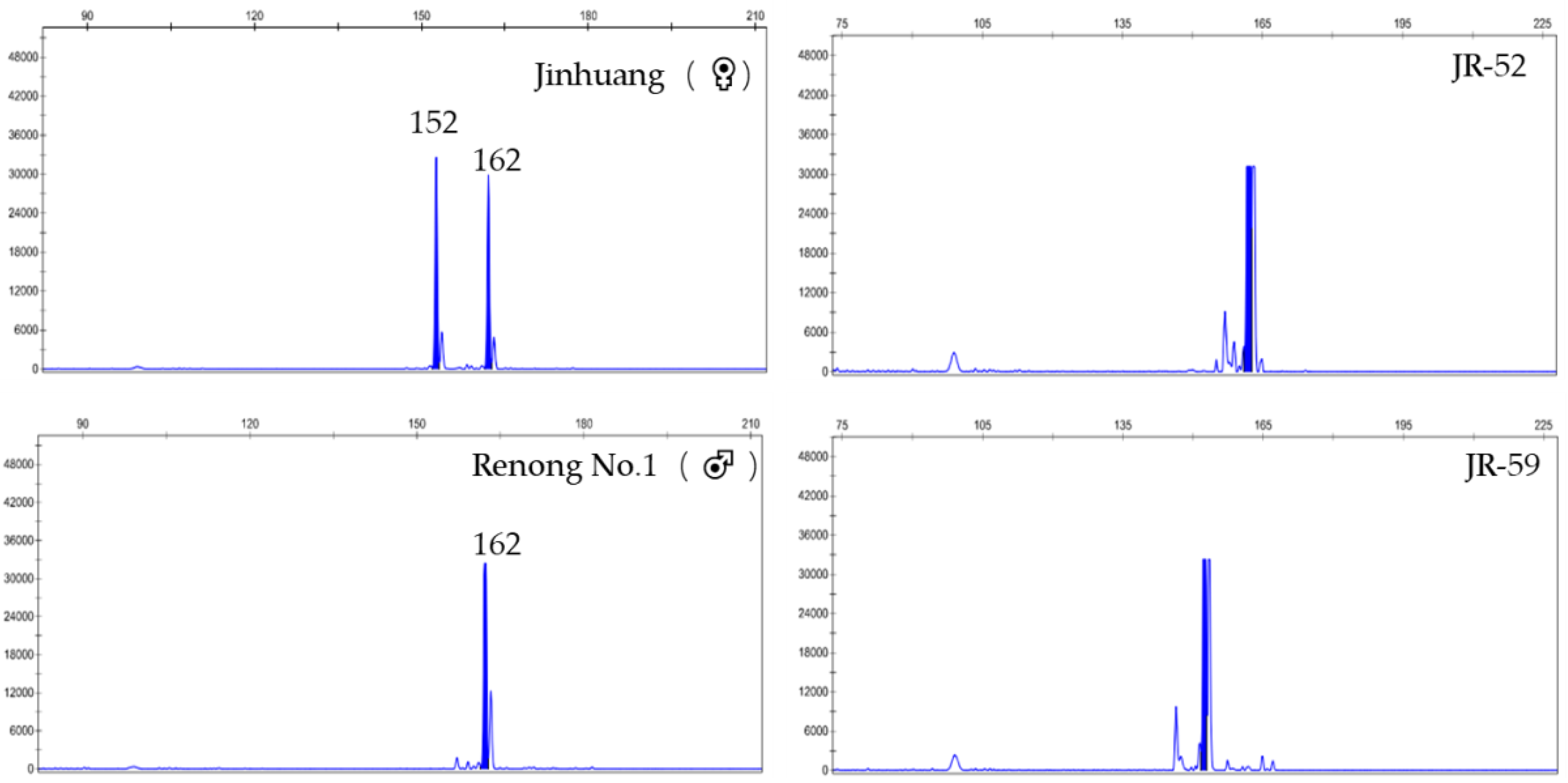
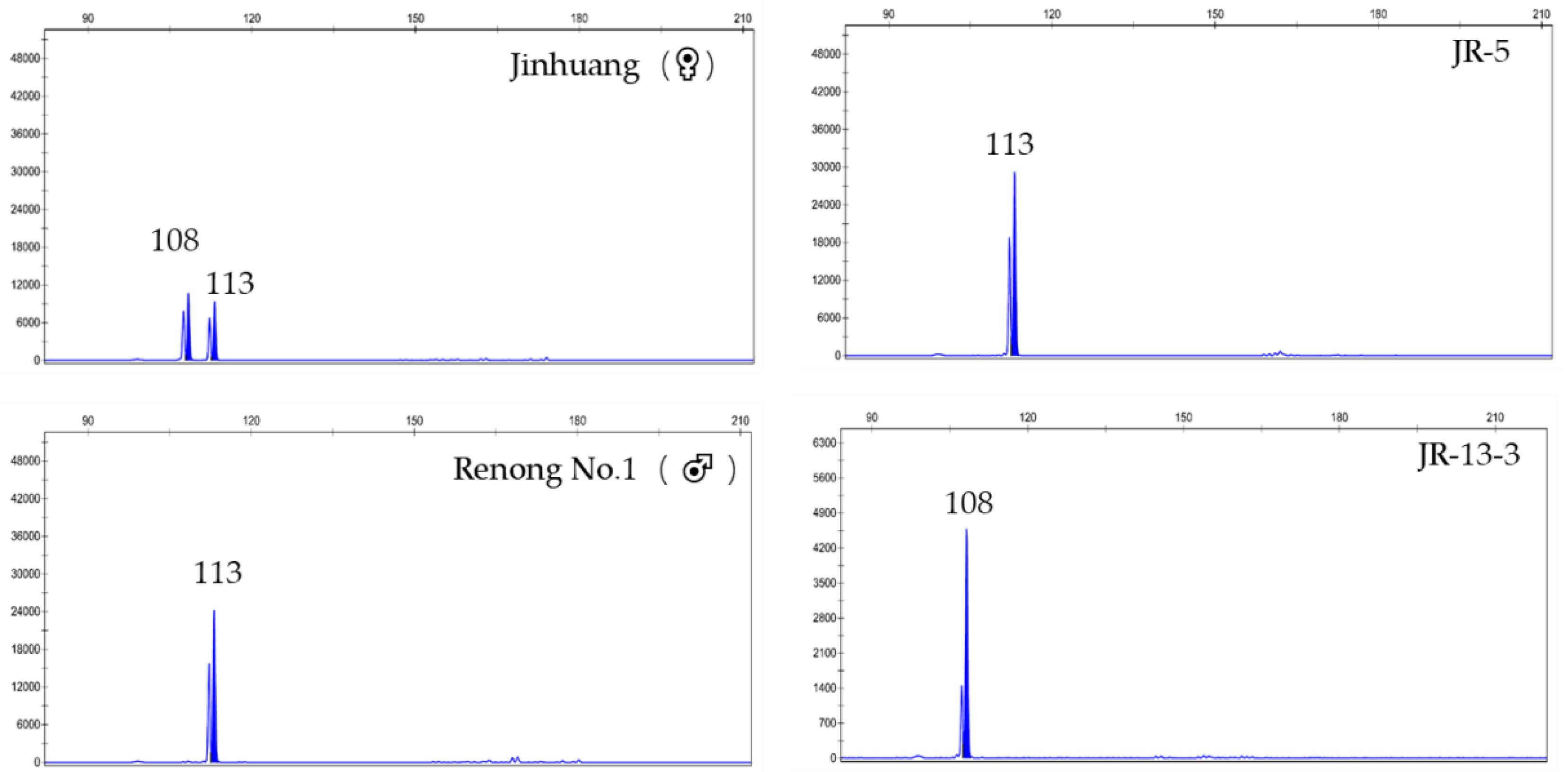
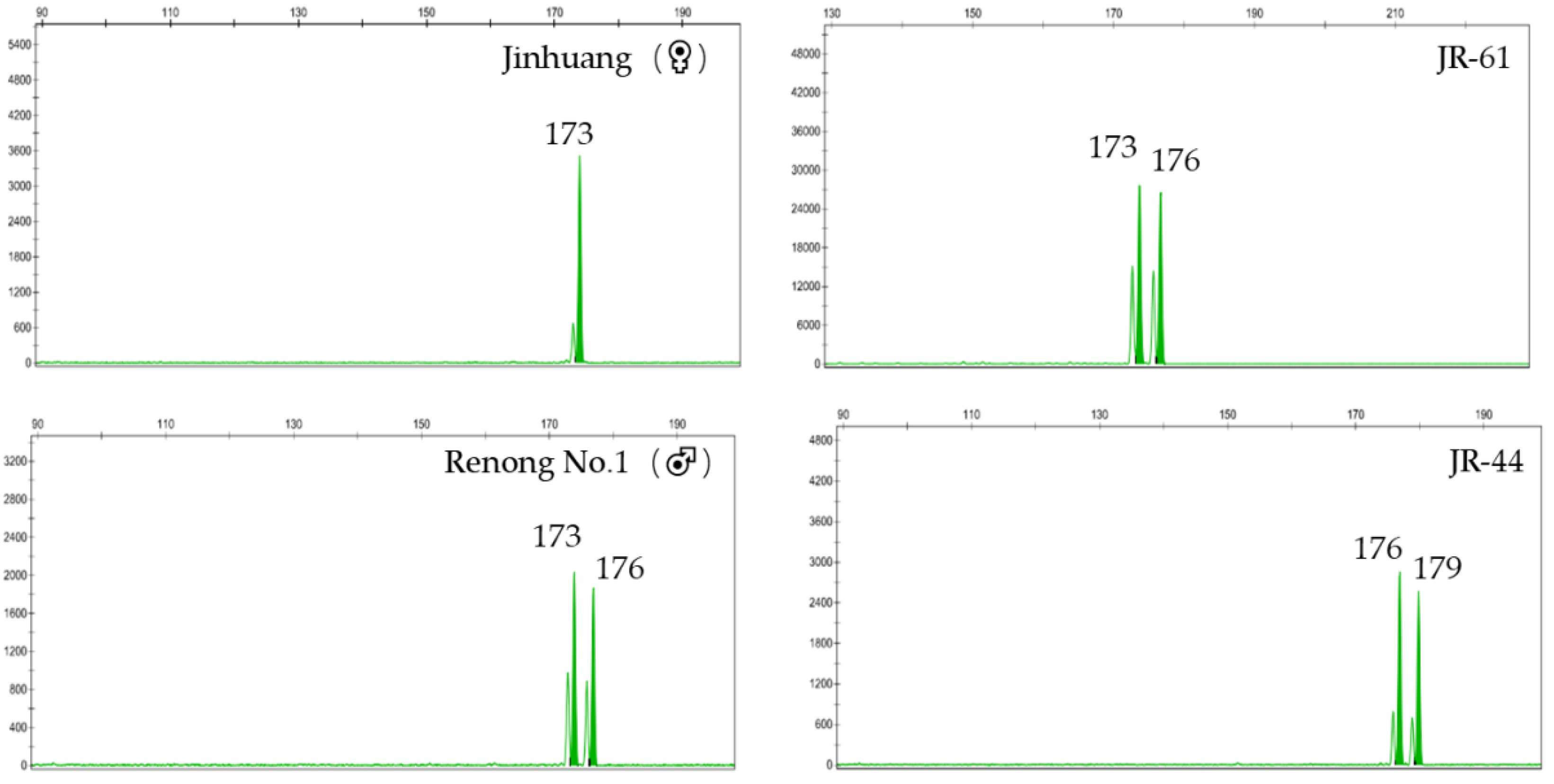
| SSR Marker | Repeat Motif | Primer Sequence (5′-3′) | 5′Fluorescent Labeling | Expected Product (bp) |
|---|---|---|---|---|
| ES35 | AGA(6) | F: TCCATTTTTGACAAATCTCCACTR: TAGCCTTGAATTCCTCAAACAAA | NED | 156 |
| ES39 | TGT(5) | F: GGTTGTTATCGTCGTTATTGTGGR: CGGAAAATGTTCGACTTGTAACT | FAM | 143 |
| ES55 | ACCTC(4) | F: AAATCAAGCTACAACTGCAACAAR: AGCTTGGCTATGTAAAAATGGGT | FAM | 100 |
| ES63 | TCC(5) | F: TCACTTCCACTAAAATCACCACCR: AGTGAAACTGAATCAGAAGAGCG | HEX | 159 |
| ES78 | ACA(6) | F: TTTGATTAATCTGCTGCAACAACR: ACCGGTAAAACAAGCTTACAACA | NED | 137 |
| ES83 | GCT(6) | F: TCCAAATGTTGCAGCCTATTTACR: ATTATGTCCAGATTCACCATTCG | PET | 145 |
| ES98 | TCTG(6) | F: AATTTTCTCATCTGCCTCTGTTGR: CGAGAAAACGATAAACGAAGAGA | PET | 142 |
| ES172 | CAT(7) | F: TATCATCAGAACTACCGCCATTTR: CTCTTTTCTTCGCTGTGTTTTGT | FAM | 146 |
| JR | ES35 | ES39 | ES55 | ES63 | ES78 | ES83 | ES98 | ES172 |
|---|---|---|---|---|---|---|---|---|
| J (♀) | 172/172 | 152/162 | 108/113 | 173/173 | 157/157 | 160/163 | 160/170 | 161/161 |
| R (♂) | 172/175 | 162/162 | 113/113 | 173/176 | 147/157 | 157/163 | 160/160 | 152/161 |
| JR-1 | 172/172 | 162/162 | 113/113 | 173/176 | 157/157 | 157/163 | 160/170 | 161/161 |
| JR-2 | 172/175 | 162/162 | 108/113 | 173/176 | 157/157 | 160/163 | 160/160 | 161/161 |
| JR-3 | 172/172 | 162/162 | 108/113 | 173/176 | 147/157 | 160/163 | 160/170 | 161/161 |
| JR-4 | 172/172 | 162/162 | 113/113 | 173/176 | 147/157 | 157/160 | 160/160 | 152/161 |
| JR-5 | 172/172 | 162/162 | 113/113 | 173/173 | 157/157 | 157/160 | 160/160 | 161/161 |
| JR-6 | 172/172 | 162/162 | 108/113 | 173/173 | 157/157 | 160/163 | 160/160 | 152/161 |
| JR-7 | 172/172 | 162/162 | 108/113 | 173/173 | 157/157 | 157/163 | 160/170 | 152/161 |
| JR-8 | 172/172 | 162/162 | 113/113 | 173/173 | 157/157 | 163/163 | 160/160 | 152/161 |
| JR-9 | 172/172 | 152/162 | 113/113 | 173/176 | 157/157 | 157/163 | 160/160 | 152/161 |
| JR-10 | 172/175 | 152/162 | 108/113 | 173/176 | 157/157 | 157/160 | 160/170 | 161/161 |
| JR-11 | 172/172 | 162/162 | 108/113 | 173/176 | 157/157 | 160/163 | 160/160 | 152/161 |
| JR-12 | 172/172 | 152/162 | 113/113 | 173/176 | 157/157 | 157/160 | 160/160 | 161/161 |
| JR-13 | 172/175 | 162/162 | 108/113 | 173/176 | 157/157 | 160/163 | 160/170 | 161/161 |
| JR-13-2 | 172/172 | 162/162 | 108/113 | 173/176 | 157/157 | 157/160 | 160/160 | 152/161 |
| JR-13-3 | 172/172 | 162/162 | 108/108 | 173/176 | 140/157 | 163/163 | 170/170 | 152/161 |
| JR-14 | 172/175 | 162/162 | 113/113 | 173/176 | 147/157 | 163/163 | 160/170 | 161/161 |
| JR-15 | 172/175 | 162/162 | 108/113 | 173/173 | 147/157 | 160/163 | 160/170 | 161/161 |
| JR-16 | 172/172 | 152/162 | 108/113 | 173/176 | 157/157 | 157/160 | 160/160 | 161/161 |
| JR-17 | 172/172 | 152/162 | 113/113 | 173/176 | 147/157 | 163/163 | 160/160 | 152/161 |
| JR-18 | 172/172 | 162/162 | 113/113 | 173/176 | 147/157 | 157/160 | 160/170 | 152/161 |
| JR-19 | 172/175 | 152/162 | 108/113 | 173/173 | 147/157 | 160/163 | 160/160 | 152/161 |
| JR-20 | 172/175 | 152/162 | 108/113 | 173/173 | 147/157 | 160/163 | 160/160 | 152/161 |
| JR-21 | 172/172 | 162/162 | 108/113 | 173/173 | 147/157 | 160/163 | 160/160 | 161/161 |
| JR-22 | 172/172 | 162/162 | 108/113 | 173/176 | 147/157 | 157/163 | 160/170 | 152/161 |
| JR-23 | 172/172 | 152/162 | 113/113 | 173/176 | 157/157 | 157/163 | 160/170 | 161/161 |
| JR-24 | 172/175 | 162/162 | 113/113 | 173/173 | 147/157 | 157/163 | 160/160 | 161/161 |
| JR-25 | 172/172 | 162/162 | 108/113 | 173/173 | 157/157 | 160/163 | 160/160 | 161/161 |
| JR-26 | 172/175 | 152/162 | 113/113 | 173/173 | 147/157 | 157/163 | 160/160 | 161/161 |
| JR-26-2 | 172/172 | 152/162 | 108/113 | 173/176 | 147/157 | 160/163 | 160/160 | 152/161 |
| JR-27 | 172/172 | 152/162 | 113/113 | 173/173 | 147/157 | 157/160 | 160/170 | 161/161 |
| JR-28 | 172/175 | 152/162 | 108/113 | 173/176 | 157/157 | 157/163 | 160/170 | 152/161 |
| JR-29 | 172/175 | 162/162 | 108/113 | 173/176 | 147/157 | 157/163 | 160/160 | 161/161 |
| JR-30 | 172/175 | 162/162 | 108/113 | 173/176 | 157/157 | 157/163 | 160/160 | 161/161 |
| JR-31 | 172/175 | 162/162 | 113/113 | 173/176 | 147/157 | 157/163 | 160/170 | 161/161 |
| JR-32 | 172/175 | 152/162 | 113/113 | 173/176 | 157/157 | 157/163 | 160/170 | 152/161 |
| JR-33 | 172/175 | 152/162 | 108/113 | 173/176 | 157/157 | 163/163 | 160/160 | 152/161 |
| JR-34 | 172/175 | 152/162 | 113/113 | 173/176 | 147/157 | 163/163 | 160/160 | 152/161 |
| JR-35 | 172/175 | 162/162 | 108/113 | 173/176 | 157/157 | 163/163 | 160/160 | 152/161 |
| JR-36 | 172/172 | 152/162 | 113/113 | 173/173 | 157/157 | 160/163 | 160/170 | 161/161 |
| JR-37 | 172/175 | 152/162 | 113/113 | 173/173 | 147/157 | 157/163 | 160/160 | 161/161 |
| JR-38 | 172/172 | 162/162 | 108/113 | 173/176 | 157/157 | 160/163 | 160/170 | 152/161 |
| JR-39 | 172/175 | 152/162 | 108/113 | 173/176 | 157/157 | 160/163 | 160/170 | 161/161 |
| JR-40 | 172/172 | 162/162 | 108/113 | 173/173 | 147/157 | 163/163 | 160/170 | 161/161 |
| JR-41 | 172/175 | 162/162 | 108/113 | 173/173 | 157/157 | 157/163 | 160/160 | 161/161 |
| JR-42 | 172/172 | 152/162 | 113/113 | 173/176 | 147/157 | 157/163 | 160/160 | 152/161 |
| JR-43 | 172/175 | 152/162 | 108/113 | 173/173 | 157/157 | 160/163 | 160/170 | 161/161 |
| JR-44 | 172/172 | 152/162 | 113/113 | 176/179 | 147/157 | 157/163 | 160/160 | 161/161 |
| JR-45 | 172/172 | 152/162 | 113/113 | 173/173 | 147/157 | 160/163 | 160/170 | 152/161 |
| JR-46 | 172/172 | 152/162 | 113/113 | 173/176 | 157/157 | 160/163 | 160/160 | 152/161 |
| JR-47 | 172/172 | 162/162 | 108/113 | 173/173 | 157/157 | 163/163 | 160/170 | 161/161 |
| JR-48 | 172/172 | 162/162 | 113/113 | 173/173 | 147/157 | 163/163 | 160/170 | 161/161 |
| JR-49 | 172/175 | 152/162 | 108/113 | 173/173 | 147/157 | 163/163 | 160/170 | 161/161 |
| JR-50 | 172/172 | 152/162 | 108/113 | 173/173 | 157/157 | 160/163 | 160/160 | 161/161 |
| JR-51 | 172/172 | 162/162 | 113/113 | 173/173 | 157/157 | 160/163 | 160/160 | 152/161 |
| JR-52 | 172/172 | 162/162 | 108/113 | 173/173 | 157/157 | 157/163 | 160/160 | 152/161 |
| JR-53 | 172/175 | 162/162 | 113/113 | 173/176 | 157/157 | 160/163 | 160/170 | 161/161 |
| JR-54 | 172/175 | 162/162 | 113/113 | 173/173 | 157/157 | 157/160 | 160/170 | 161/161 |
| JR-55 | 172/175 | 152/162 | 108/113 | 173/176 | 147/157 | 160/163 | 160/160 | 161/161 |
| JR-55-2 | 172/172 | 152/162 | 113/113 | 173/176 | 147/157 | 160/163 | 160/170 | 161/161 |
| JR-56 | 172/175 | 162/162 | 108/113 | 173/176 | 147/157 | 157/163 | 160/160 | 152/161 |
| JR-57 | 172/172 | 152/162 | 108/113 | 173/173 | 157/157 | 163/163 | 160/170 | 152/161 |
| JR-59 | 172/172 | 152/152 | 108/113 | 173/173 | 147/157 | 157/160 | 160/170 | 161/161 |
| JR-60 | 172/172 | 152/162 | 108/113 | 173/173 | 157/157 | 157/160 | 160/170 | 152/161 |
| JR-61 | 172/172 | 152/162 | 113/113 | 173/176 | 147/157 | 163/163 | 160/160 | 161/161 |
| JR-62 | 172/175 | 162/162 | 108/113 | 173/176 | 157/157 | 157/163 | 160/160 | 152/161 |
| Locus | ES35 | ES39 | ES55 | ES63 | ES78 | ES83 | ES98 | ES172 |
|---|---|---|---|---|---|---|---|---|
| Jinhuang♀ | 172/172 | 152/162 | 108/113 | 173/173 | 157/157 | 160/163 | 160/170 | 161/161 |
| a:a | a:b | a:b | a:a | a:a | a:b | a:b | a:a | |
| Renong No.1♂ | 172/175 | 162/162 | 113/113 | 173/176 | 147/157 | 157/163 | 160/160 | 152/161 |
| a:b | b:b | b:b | a:b | b:a | c:b | a:a | b:a | |
| Theoretical Genotypes | aa:ab | ab:bb | ab:bb | aa:ab | aa:ab | ab:ac:bb:bc | aa:ab | aa:ab |
| Segregation | 1:1 | 1:1 | 1:1 | 1:1 | 1:1 | 1:1:1:1 | 1:1 | 1:1 |
| Observed | 38:27 | 30:34 | 36:28 | 28:36 | 35:29 | 22:11:13:19 | 35:29 | 36:29 |
| He | 0.34 | 0.36 | 0.41 | 0.41 | 0.34 | 0.62 | 0.35 | 0.35 |
| HO | 0.44 | 0.47 | 0.56 | 0.56 | 0.44 | 0.81 | 0.45 | 0.45 |
| Chi-square | 4.81 | 5.76 | 9.48 | 9.48 | 4.81 | 12.59 | 5.27 | 5.27 |
| p value | 0.028 | 0.016 | 0.002 | 0.002 | 0.028 | 0.005 | 0.021 | 0.021 |
| JR | ES35 | ES39 | ES55 | ES63 | ES78 | ES83 | ES98 | ES172 |
|---|---|---|---|---|---|---|---|---|
| J (♀) | 11 | 12 | 12 | 11 | 22 | 23 | 12 | 22 |
| R (♂) | 12 | 22 | 22 | 12 | 12 | 13 | 11 | 12 |
| JR-1 | 11 | 22 | 22 | 12 | 22 | 13 | 12 | 22 |
| JR-2 | 12 | 22 | 12 | 12 | 22 | 23 | 11 | 22 |
| JR-3 | 11 | 22 | 12 | 12 | 12 | 23 | 12 | 22 |
| JR-4 | 11 | 22 | 22 | 12 | 12 | 12 | 11 | 12 |
| JR-5 | 11 | 22 | 22 | 11 | 22 | 12 | 11 | 22 |
| JR-6 | 11 | 22 | 12 | 11 | 22 | 23 | 11 | 12 |
| JR-7 | 11 | 22 | 12 | 11 | 22 | 13 | 12 | 12 |
| JR-8 | 11 | 22 | 22 | 11 | 22 | 33 | 11 | 12 |
| JR-9 | 11 | 12 | 22 | 12 | 22 | 13 | 11 | 12 |
| JR-10 | 12 | 12 | 12 | 12 | 22 | 12 | 12 | 22 |
| JR-11 | 11 | 22 | 12 | 12 | 22 | 23 | 11 | 12 |
| JR-12 | 11 | 12 | 22 | 12 | 22 | 12 | 11 | 22 |
| JR-13 | 12 | 22 | 12 | 12 | 22 | 23 | 12 | 22 |
| JR-13-2 | 11 | 22 | 12 | 12 | 22 | 12 | 11 | 12 |
| JR-14 | 12 | 22 | 22 | 12 | 12 | 33 | 12 | 22 |
| JR-15 | 12 | 22 | 12 | 11 | 12 | 23 | 12 | 22 |
| JR-16 | 11 | 12 | 12 | 12 | 22 | 12 | 11 | 22 |
| JR-17 | 11 | 12 | 22 | 12 | 12 | 33 | 11 | 12 |
| JR-18 | 11 | 22 | 22 | 12 | 12 | 12 | 12 | 12 |
| JR-19 | 12 | 12 | 12 | 11 | 12 | 23 | 11 | 12 |
| JR-20 | 12 | 12 | 12 | 11 | 12 | 23 | 11 | 12 |
| JR-21 | 11 | 22 | 12 | 11 | 12 | 23 | 11 | 22 |
| JR-22 | 11 | 22 | 12 | 12 | 12 | 13 | 12 | 12 |
| JR-23 | 11 | 12 | 22 | 12 | 22 | 13 | 12 | 22 |
| JR-24 | 12 | 22 | 22 | 11 | 12 | 13 | 11 | 22 |
| JR-25 | 11 | 22 | 12 | 11 | 22 | 23 | 11 | 22 |
| JR-26 | 12 | 12 | 22 | 11 | 12 | 13 | 11 | 22 |
| JR-26-2 | 11 | 12 | 12 | 12 | 12 | 23 | 11 | 12 |
| JR-27 | 11 | 12 | 22 | 11 | 12 | 12 | 12 | 22 |
| JR-28 | 12 | 12 | 12 | 12 | 22 | 13 | 12 | 12 |
| JR-29 | 12 | 22 | 12 | 12 | 12 | 13 | 11 | 22 |
| JR-30 | 12 | 22 | 12 | 12 | 22 | 13 | 11 | 22 |
| JR-31 | 12 | 22 | 22 | 12 | 12 | 13 | 12 | 22 |
| JR-32 | 12 | 12 | 22 | 12 | 22 | 13 | 12 | 12 |
| JR-33 | 12 | 12 | 12 | 12 | 22 | 33 | 11 | 12 |
| JR-34 | 12 | 12 | 22 | 12 | 12 | 33 | 11 | 12 |
| JR-35 | 12 | 22 | 12 | 12 | 22 | 33 | 11 | 12 |
| JR-36 | 11 | 12 | 22 | 11 | 22 | 23 | 12 | 22 |
| JR-37 | 12 | 12 | 22 | 11 | 12 | 13 | 11 | 22 |
| JR-38 | 11 | 22 | 12 | 12 | 22 | 23 | 12 | 12 |
| JR-39 | 12 | 12 | 12 | 12 | 22 | 23 | 12 | 22 |
| JR-40 | 11 | 22 | 12 | 11 | 12 | 33 | 12 | 22 |
| JR-41 | 12 | 22 | 12 | 11 | 22 | 13 | 11 | 22 |
| JR-42 | 11 | 12 | 22 | 12 | 12 | 13 | 11 | 12 |
| JR-43 | 12 | 12 | 12 | 11 | 22 | 23 | 12 | 22 |
| JR-45 | 11 | 12 | 22 | 11 | 12 | 23 | 12 | 12 |
| JR-46 | 11 | 12 | 22 | 12 | 22 | 23 | 11 | 12 |
| JR-47 | 11 | 22 | 12 | 11 | 22 | 33 | 12 | 22 |
| JR-48 | 11 | 22 | 22 | 11 | 12 | 33 | 12 | 22 |
| JR-49 | 12 | 12 | 12 | 11 | 12 | 33 | 12 | 22 |
| JR-50 | 11 | 12 | 12 | 11 | 22 | 23 | 11 | 22 |
| JR-51 | 11 | 22 | 22 | 11 | 22 | 23 | 11 | 12 |
| JR-52 | 11 | 22 | 12 | 11 | 22 | 13 | 11 | 12 |
| JR-53 | 12 | 22 | 22 | 12 | 22 | 23 | 12 | 22 |
| JR-54 | 12 | 22 | 22 | 11 | 22 | 12 | 12 | 22 |
| JR-55 | 12 | 12 | 12 | 12 | 12 | 23 | 11 | 22 |
| JR-55-2 | 11 | 12 | 22 | 12 | 12 | 23 | 12 | 22 |
| JR-56 | 12 | 22 | 12 | 12 | 12 | 13 | 11 | 12 |
| JR-57 | 11 | 12 | 12 | 11 | 22 | 33 | 12 | 12 |
| JR-60 | 11 | 12 | 12 | 11 | 22 | 12 | 12 | 12 |
| JR-61 | 11 | 12 | 22 | 12 | 12 | 33 | 11 | 22 |
| JR-62 | 12 | 22 | 12 | 12 | 22 | 13 | 11 | 12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Zheng, B.; Xu, W.; Ma, X.; Wang, S.; Qian, M.; Wu, H. Identification of F1 Hybrid Progenies in Mango Based on Fluorescent SSR Markers. Horticulturae 2022, 8, 1122. https://doi.org/10.3390/horticulturae8121122
Li X, Zheng B, Xu W, Ma X, Wang S, Qian M, Wu H. Identification of F1 Hybrid Progenies in Mango Based on Fluorescent SSR Markers. Horticulturae. 2022; 8(12):1122. https://doi.org/10.3390/horticulturae8121122
Chicago/Turabian StyleLi, Xing, Bin Zheng, Wentian Xu, Xiaowei Ma, Songbiao Wang, Minjie Qian, and Hongxia Wu. 2022. "Identification of F1 Hybrid Progenies in Mango Based on Fluorescent SSR Markers" Horticulturae 8, no. 12: 1122. https://doi.org/10.3390/horticulturae8121122
APA StyleLi, X., Zheng, B., Xu, W., Ma, X., Wang, S., Qian, M., & Wu, H. (2022). Identification of F1 Hybrid Progenies in Mango Based on Fluorescent SSR Markers. Horticulturae, 8(12), 1122. https://doi.org/10.3390/horticulturae8121122







