Abstract
Touch stimulus responses are common in plants. Some flowering plants sense the arrival of their pollinators and secrete nectar or release pollen sacs, facilitating successful pollination. Molecular mechanisms for mechanical stimulus responses in plants are well characterized in Arabidopsis leaves, but not in non-model plants or other organs such as flowers. Here, we performed RNA-seq analysis of touched flower buds of Dianthus hybrida, a major ornamental plant. Upon touch treatment, 931 and 132 genes were upregulated and downregulated, respectively. GO enrichment analysis revealed that genes encoding serine/threonine protein kinases were significantly abundant among the upregulated genes, which is consistent with previous studies that demonstrated the pivotal role of protein phosphorylation in the touch stimulus response of Arabidopsis leaves. In comparison with the gene expression profile of touched Arabidopsis leaves, the same families but different homologs of the representative touch-induced genes encoding protein kinases were upregulated, showing that phosphorelay signaling was the common mechanism for touch stimulus response in flowers and leaves, but the players of the phosphorelay signaling were different. These results will contribute to further studies on the mechanical stimulus responses of ornamental flowers and the utilization of this mechanism for breeding programs.
1. Introduction
Plants continuously encounter and can sense environmental mechanical stimuli such as wind, rainfall, sounds, and touch to control their growth and development [1,2]. Mechanical stimuli contribute to the successful pollination of some flowers. For example, in the legume Desmodium setigerum, flower color and shape are changed by pollination and bee visits [3]. In the case of Oenothera drummondii, flowers produce nectar that is sweeter than usual after being treated with the playback sound of a flying bee [4]. The male flower of the dioecious Catasetum releases pollen sacs when pollinators touch the center of the flower, helping to effectively pollinate this species [5]. Although flowers are thought to sense mechanical stimuli, their response to them is less documented.
Molecular mechanisms of touch stimulus responses in plants have been studied exclusively in Arabidopsis thaliana. Many signaling molecules, including potential secondary messengers and plant hormones, have been implicated in touch-induced responses. First, a very rapid change in cytosolic free Ca2+ concentration occurs after the stimulation of plant cells [6,7,8,9,10,11,12]. An increase in Ca2+ concentrations is sensed by Ca2+-binding proteins that regulate downstream molecular processes [8,9,10,11,12] such as the phosphorylation of proteins, resulting in the modulation of protein conformation [13,14,15]. In Arabidopsis, 24 touch-responsive phosphopeptides, including kinases, phosphatases, cytoskeleton proteins, membrane proteins, and ion transporters, were identified after initiation of touch treatments [16]. A non-phosphorylated isoform of touch-regulated phosphoprotein 1 (TREPH1) exhibited insensitive phenotypes to touch treatments and loss of the induction of touch-induced gene expression [16].
Touch-induced gene expression occurred 10 to 40 min after the initiation of touch treatments, while the increase in Ca2+ concentrations and Ca2+-independent protein phosphorylation occurred shortly after treatment. The representative touch-induced genes, TCHs, were originally isolated as responsible genes after stimuli, such as touch, wind, rain, wounding, and darkness [1]. TCH1 encodes an Arabidopsis calmodulin, CAM2, whereas TCH2 and TCH3 encode calmodulin-like (CML) proteins, CML24 and CML12, respectively [1,17,18,19], which is consistent with the above description that Ca2+-dependent pathways are involved in touch stimulation. Touch-induced genes were also identified throughout the genome, and a representative gene, CML39 was isolated [20]. TCH4 encodes xyloglucan endotransglucosylase/hydrolase [21].
The phytohormone jasmonate (JA) is also an important molecule involved in touch-induced responses in plants. An Arabidopsis mutant defective in allene oxide synthase (aos) and JA did not show touch-induced morphological changes [22]. In contrast, the expression of touch-induced genes was detected in the mutant, indicating that this was not sufficient to show touch-induced morphological changes, and that the expression of these genes was independent of the JA signaling pathway or upstream of the pathway [22].
Although these characteristics have been well documented in Arabidopsis leaves, touch-induced gene expression in flowers remains mostly unknown. Here, we investigated the gene expression profiles after exposure to touch stimulus of the flowers of an ornamental plant, Dianthus hybrida, which is an interspecific hybrid between D. chinensis and D. barbatus. The genus Dianthus, a member of the family Caryophyllaceae [23], is one of the major ornamental flowers of commercial importance, with production similar to that of Chrysanthemum and Rosa. The Dianthus is commonly known as “carnation,” which refers to Dianthus caryophyllus and several intra/interspecific hybrids. Dianthus flowers vary widely in colors and shapes [24,25,26,27]. Molecular understanding of physiological mechanism underlying environmental stimulus effects on flower morphology will facilitate to develop new varieties. The genome project of D. caryophyllus is almost complete [28], allowing us to perform transcriptome analyses. In this report, we aimed to list the differentially expressed genes after touch stimulation in D. hybrida flowers and indicate directions for future research that will serve as a basis for further development and utilization of this response in breeding programs.
2. Materials and Methods
2.1. Plant Materials and RNA Extraction
D. hybrida cultivar “Telstar Scarlet” plants were grown in plastic pots (10.5 cm diameter and 9.5 cm height) in a wind- and vibration-free PVC box placed in a growth chamber under the following conditions; 14 h light/10 h dark, temperature of 28 °C and relative humidity of 70% (Figure 1A). Photosynthetic photon flux density (PPFD) was 315 μmol m−2 s−1. Three untouched flower buds were sampled as controls, and another three buds were touched by hand for 40 s with hands and sampled 40 min later, following a previous study on Arabidopsis leaves [20]. All biological replicates were sampled from different plants. Total RNA was extracted using the RNeasy Plant Mini Kit (QIAGEN, Germantown, MD, USA) following the manufacturer’s instructions.
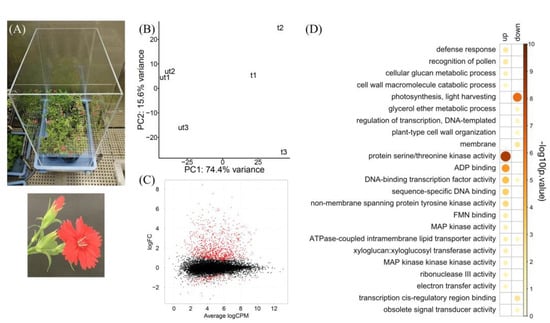
Figure 1.
RNA-seq summary of touched D. hybrida flower buds. (A) D. hybrida plants grown in a wind- and vibration-proofed box. The bottom panel shows a flower and a bud. (B) PCA plot of three replicates each of touched (t) and untouched (ut) samples using uniquely mapped reads. (C) MA plot of expressed genes (TPM > 2.5 at least in all three replicates of either touched or untouched sample). Red dots indicate DEGs (FDR < 0.001). (D) GO enrichment analysis of DEGs.
2.2. RNA-seq Analysis
Total RNA sequencing was conducted by Rhelixa (Tokyo, Japan). Strand-specific RNA-seq libraries were prepared using the NEBNext Ultra II Directional mRNA-seq Kit for Illumina (New England Biolabs, Hitchin, Hertfordshire, UK). The libraries were bulked with others and sequenced using the NovaSeq6000 (Illumina, San Diego, CA, USA) with paired-end 150-bp reads.
Due to unavailability of chromosome-level genome assembly of D. hybrida, the genome scaffold of the closely related species Dianthus caryophyllus (carnation), the highest-quality genome sequence within the genus, was used as a reference. Scaffold, gene annotation, and Arabidopsis homolog data for carnation were downloaded from the Carnation DB [28] (http://carnation.kazusa.or.jp, accessed on 7 December 2021). RNA-seq reads were mapped to carnation scaffolds using the STAR aligner ver. 2.7.3a [29] with options “--outFilterMultimapNmax 1 --quantMode GeneCounts”. Differentially expressed genes (DEGs) were called with glmLRT of R package edgeR ver. 3.26.8 [30], and transcripts per kilobase million (TPM) values were computed after trimmed mean of M values (TMM) normalization. GO enrichment analysis was performed using the R package clusterProfiler ver 3.12.0 [31] and visualized using the R package corrplot ver 0.92 [32]. To compare the transcriptome profile of D. hybrida flower buds with that of Arabidopsis leaves, microarray data from a previous study [20] was used.
3. Results and Discussion
To explore the gene expression profiles in the flower buds of D. hybrida in response to touch stimulus, we performed transcriptome analysis of the flower buds 40 min after touch treatment. On average, 22.8 million reads were obtained for each sample, and 71.5% of the total reads were uniquely mapped to carnation scaffolds (Table S1). Transcripts from 15,258 genes were detected (TPM ≥ 2.5, in all three replicates in either touched or untouched conditions). Principal component analysis based on the uniquely mapped read count showed that touched and untouched samples were clearly separated by the PC1 value with a 74.4% contribution rate (Figure 1B), suggesting touch-induced transcriptional changes in the flower buds. Genes with the highest loading values along the PC2 axis were a CEP1 homolog and three UDP-glycosyltransferase genes, which are involved in pollen development [33], indicating that the PC2 represented inflorescence developmental stage. Gene expression analysis showed that 961 and 132 genes were up- or downregulated by touch treatment, respectively (FDR < 0.001, edgeR; Figure 1C; Supplementary Excel File). GO enrichment analysis revealed that genes with GO “protein serine/threonine kinase activity” were significantly upregulated in response to touch stimuli (Figure 1D), which was consistent with previous studies that demonstrated the importance of protein phosphorylation in Arabidopsis touch-delayed bolting [16,34]. Other GO terms related to phosphorelay signaling, such as “non-membrane spanning protein tyrosine kinase activity”, “MAP kinase activity”, and “MAP kinase kinase kinase activity” were also enriched in touch-induced upregulated genes. In addition, GO “xyloglucan:xyloglucosyl transferase activity,” a representative mechanosensitive response [21], was detected. Although related GO terms were not enriched significantly, calmodulin genes, such as a CAM5 homolog, and JA biosynthetic (LOX1, 4, OPR3, and AOC3 homologs) and signaling genes (JAZ1 and NINJA homologs) were upregulated. On the other hand, genes with GO “photosynthesis” were significantly downregulated upon touch stimulus, which resembled responses to other biotic stresses including bacteria, insects, and wounding [35].
To explore whether touch stimulus responses are generally conserved between flowers and leaves, we compared the RNA-seq data of D. hybrida flower buds with the microarray data of Arabidopsis leaves from a previous study [20]. All DEGs commonly detected in both experiments showed the same expression changes (Figure 2A,B), except for one clock-related gene, FKF1, which appeared to reflect the difference in sampling time. TCH4, which encodes xyloglucan endotransglucosylase [21], and its two Dianthus homologs were commonly upregulated. For calmodulin, CML49, but not CAM5, was upregulated in touched Arabidopsis leaves, while their homologs, Dca25393 and Dca20630, respectively, showed the opposite expression patterns in the flower buds of D. hybrida, which implies that different organs utilize different calmodulin family members as touch signaling components. GO enrichment analysis revealed that genes for “protein serine/threonine kinase activity” were enriched in commonly detected and flower bud-specific DEGs, but not in leaf-specific ones (Figure 2C). In fact, 32 out of 44 genes encoding serine/threonine kinases were detected as DEGs only in flower buds, while no kinase gene was differentially expressed specifically in leaves. These kinases might be key players in a flower-specific touch stimulus response. Although it must be considered that these DEGs specific to one condition might reflect the difference between the two species and/or growth condition, these results suggest that an organ-specific molecular mechanism for touch stimulus response might exist, which could be the basis of organ function, as in the case of flowers listening to bees [4].
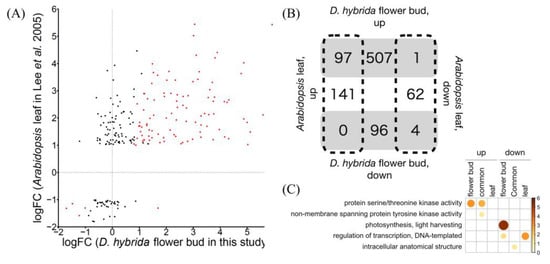
Figure 2.
Comparison of transcriptome profile between D. hybrida flower buds in this study and Arabidopsis leaves [20]. (A) Pairwise gene expression comparison, red dots indicating DEGs in this study (FDR < 0.001). (B) Venn diagram of up- and downregulated genes. (C) GO enrichment analysis of common, D. hybrida flower bud-specific, and Arabidopsis leaf-specific DEGs.
4. Conclusions
Touch stimuli induced upregulation of genes encoding protein kinase, xyloglucan endotransglucosylase, calmodulin, and JA biosynthetic and signaling components and downregulation of photosynthetic genes in D. hybrida flower buds. This response was consistent with that of Arabidopsis leaves, in which protein phosphorylation events occur immediately after mechanical stimulation, followed by the increase in Ca2+ and JA concentrations [16,20,22], implying that molecular mechanisms for the touch stimulus response are generally conserved among plant organs and among plant species. In contrast, a large proportion of protein kinase genes was induced only in flower buds, suggesting an organ-specific response to tactile stimulation. Our results will contribute to further studies on physiological mechanisms underlying touch stimulus response of ornamental flowers and utilization of this response in future breeding programs.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/horticulturae8100918/s1, Table S1: mapping rate of RNA-seq reads to carnation scaffold sequences, File S1: statistical summary of differentially expressed genes.
Author Contributions
H.S. and Y.K. designed the research; A.S. and Y.K. performed the experiments; R.N. analyzed the data; R.N. and Y.K. wrote the manuscript with contributions from all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Grant for Strategic Issue Research Promotion 03-04R020207 from Fukui Prefectural University to H.S. and Y.K., and JSPS KAKENHI grant number 21K20585 to R.N. and 22H05071 and 21KK0128 to Y.K.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The RNA-seq data reported in this study have been deposited in the DDBJ database (accession No. DRA014742).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Braam, J.; Davis, R.W. Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell 1990, 60, 357–364. [Google Scholar] [CrossRef]
- Braam, J. In touch: Plant responses to mechanical stimuli. New Phytol. 2005, 165, 373–389. [Google Scholar] [CrossRef] [PubMed]
- Willmer, P.; Stanley, D.A.; Steijven, K.; Matthews, I.M.; Nuttman, C.V. Bidirectional flower color and shape changes allow a second opportunity for pollination. Curr. Biol. 2009, 19, 919–923. [Google Scholar] [CrossRef]
- Veits, M.; Khait, I.; Obolski, U.; Zinger, E.; Boonman, A.; Goldshtein, A.; Saban, K.; Seltzer, R.; Ben-Dor, U.; Estlein, P.; et al. Flowers respond to pollinator sound within minutes by increasing nectar sugar concentration. Ecol. Lett. 2019, 22, 1483–1492. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, C.C.; Bales, J.W.; Palmer-Fortune, J.E.; Nicholson, R.G. Darwin’s bee-trap: The kinetics of Catasetum, a new world orchid. Plant Signal. Behav. 2008, 3, 19–23. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Knight, M.R.; Campbell, A.K.; Smith, S.M.; Trewavas, A.J. Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 1991, 352, 524–526. [Google Scholar] [CrossRef]
- Zhu, X.; Feng, Y.; Liang, G.; Liu, N.; Zhu, J.K. Aequorin-based luminescence imaging reveals stimulus- and tissue-specific Ca2+ dynamics in Arabidopsis Plants. Mol. Plant 2013, 6, 444–455. [Google Scholar] [CrossRef]
- Harper, J.F.; Breton, G.; Harmon, A. Decoding Ca2+ Signals through plant protein kinases. Annu. Rev. Plant Biol. 2004, 55, 263–288. [Google Scholar] [CrossRef]
- Kim, M.C.; Chung, W.S.; Yun, D.J.; Cho, M.J. Calcium and calmodulin-mediated regulation of gene expression in plants. Mol. Plant 2009, 2, 13–21. [Google Scholar] [CrossRef]
- Luan, S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009, 14, 37–42. [Google Scholar] [CrossRef]
- DeFalco, T.A.; Bender, K.W.; Snedden, W.A. Breaking the code: Ca2+ sensors in plant signalling. Biochem. J. 2009, 425, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Kudla, J.; Batistič, O.; Hashimoto, K. Calcium signals: The lead currency of plant information processing. Plant Cell 2010, 22, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Hunter, T. Protein kinases and phosphatases: The yin and yang of protein phosphorylation and signaling. Cell 1995, 80, 225–236. [Google Scholar] [CrossRef]
- Soderling, T.R. The Ca2+–calmodulin-dependent protein kinase cascade. Trends Biochem. Sci. 1999, 24, 232–236. [Google Scholar] [CrossRef]
- Clapham, D.E. Calcium signaling. Cell 2007, 131, 1047–1058. [Google Scholar] [CrossRef]
- Wang, K.; Yang, Z.; Qing, D.; Ren, F.; Liu, S.; Zheng, Q.; Liu, J.; Zhang, W.; Dai, C.; Wu, M.; et al. Quantitative and functional posttranslational modification proteomics reveals that TREPH1 plays a role in plant touch-delayed bolting. Proc. Natl. Acad. Sci. USA 2018, 115, E10265–E10274. [Google Scholar] [CrossRef]
- Sistrunk, M.L.; Antosiewicz, D.M.; Purugganan, M.M.; Braam, J. Arabidopsis TCH3 encodes a novel Ca2+ binding protein and shows environmentally induced and tissue-specific regulation. Plant Cell 1994, 6, 1553–1565. [Google Scholar] [CrossRef]
- Khan, A.R.; Johnson, K.A.; Braam, J.; James, M.N.G. Comparative modeling of the three-dimensional structure of the calmodulin-related TCH2 protein from Arabidopsis. Proteins 1997, 27, 144–153. [Google Scholar] [CrossRef]
- McCormack, E.; Braam, J. Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol. 2003, 159, 585–598. [Google Scholar] [CrossRef]
- Lee, D.; Polisensky, D.H.; Braam, J. Genome-wide identification of touch- and darkness-regulated Arabidopsis Genes: A focus on calmodulin-like and XTH genes. New Phytol. 2005, 165, 429–444. [Google Scholar] [CrossRef]
- Rose, J.K.C.; Braam, J.; Fry, S.C.; Nishitani, K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002, 43, 1421–1435. [Google Scholar] [CrossRef] [PubMed]
- Chehab, E.W.; Yao, C.; Henderson, Z.; Kim, S.; Braam, J. Arabidopsis touch-induced morphogenesis is jasmonate mediated and protects against pests. Curr. Biol. 2012, 22, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Harbaugh, D.T.; Nepokroeff, M.; Rabeler, R.K.; McNeill, J.; Zimmer, E.A.; Wagner, W.L. A new lineage-based tribal classification of the family Caryophyllaceae. Int. J. Plant Sci. 2010, 171, 185–198. [Google Scholar] [CrossRef]
- Onozaki, T.; Mato, M.; Shibata, M.; Ikeda, H. Differences in flower color and pigment composition among white carnation (Dianthus caryophyllus L.) Cultivars. Sci. Hortic. 1999, 82, 103–111. [Google Scholar] [CrossRef]
- Okamura, M.; Yasuno, N.; Ohtsuka, M.; Tanaka, A.; Shikazono, N.; Hase, Y. Wide variety of flower-color and -shape mutants regenerated from leaf cultures irradiated with ion beams. Nucl. Instrum. Methods Phys. Res. B 2003, 206, 574–578. [Google Scholar] [CrossRef]
- Okamura, M.; Umemoto, N.; Onishi, N. Breeding glittering carnations by an efficient mutagenesis system. Plant Biotechnol. 2012, 29, 209–214. [Google Scholar] [CrossRef]
- Okamura, M.; Nakayama, M.; Umemoto, N.; Cano, E.A.; Hase, Y.; Nishizaki, Y.; Sasaki, N.; Ozeki, Y. Crossbreeding of a metallic color carnation and diversification of the peculiar coloration by ion-beam irradiation. Euphytica 2013, 191, 45–56. [Google Scholar] [CrossRef]
- Yagi, M.; Kosugi, S.; Hirakawa, H.; Ohmiya, A.; Tanase, K.; Harada, T.; Kishimoto, K.; Nakayama, M.; Ichimura, K.; Onozaki, T.; et al. Sequence analysis of the genome of carnation (Dianthus caryophyllus L.). DNA Res. 2014, 21, 231–241. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Simko, V.R. Package “Corrplot”: Visualization of a Correlation Matrix (Version 0.92). Available online: https://github.com/taiyun/corrplot (accessed on 1 September 2022).
- Zhang, D.; Liu, D.; Lv, X.; Wang, Y.; Xun, Z.; Liu, Z.; Li, F.; Lu, H. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 2014, 26, 2939–2961. [Google Scholar] [CrossRef] [PubMed]
- Darwish, E.; Ghosh, R.; Ontiveros-Cisneros, A.; Tran, H.C.; Petersson, M.; de Milde, L.; Broda, M.; Goossens, A.; van Moerkercke, A.; Khan, K.; et al. Touch signaling and thigmomorphogenesis are regulated by complementary CAMTA3- and JA-dependent pathways. Sci. Adv. 2022, 8, eabm2091. [Google Scholar] [CrossRef]
- Bilgin, D.D.; Zavala, J.A.; Zhu, J.; Clough, S.J.; Ort, D.R.; Delucia, E.H. Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ. 2010, 33, 1597–1613. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).