Abstract
Late spring frosts can become one of the limiting factors for the expansion of cultivation area towards a harsher climate for the almond [Prunus amygdalus Batsch syn P. dulcis (Mill.) D.A. Webb] crop as spring frost can damage up to 90% of the harvest. In order to identify key genes favoring cold tolerance in almonds, branches from three late-blooming genotypes: ‘Guara’, ‘Soleta’ and ‘Belona’ were exposed at −4 °C during 24 h in a constant climate chamber. Phenotype analysis showed that ‘Guara’ and ‘Soleta’ had a greater acclimation capacity to cold than ‘Belona’. The qRT-PCR BioMark System technology was used to monitor the relative expression of 30 candidate genes with a potential relation to cold response, which are either involved in the ICE-CBF-COR pathway or the independent CBF pathway, and also genes not yet characterized or with unknown function in almond genome. Differences in the gene expression profiles were found among the three studied genotypes and the three time-points of cold exposure (0, 2 and 24 h). BBX20 and CLO genes behaved as differentiator genes between tolerant and susceptible genotypes in cold stress response in almond pistils. In addition, the differences of expression among the tolerant genotypes suggested the intervention of different mechanisms responding to cold stress in almonds.
1. Introduction
Plants develop different mechanisms to cope with abiotic stresses, including cold stress by freezing temperatures. These mechanisms can be categorized into three different strategies. The first one is the stress escape strategy. Plants are not capable of surviving to freeze periods during the active growth phase. They have to adapt their phenological cycle to favorable periods [1], for instance by delaying the blooming period to reduce the risk of injury in floral organs. The other two strategies are stress avoidance and stress tolerance. Under low temperatures, plants, including fruit-tree species, with a stress avoidance strategy aim at delaying the frost injuries in sensitive tissues by performing processes including supercooling, which prevent ice crystal formation in their cells by decreasing the cell water content, allowing it to reach a lower freezing point [1,2,3,4,5]. Following a cold-stress tolerance strategy implies changes at physiological, biochemical and morphological levels which lead to resist the action of stress, these metabolic changes include mechanisms to avoid plasma membrane degradation and preservation of its lipid composition, synthesis of osmoprotectants like soluble sugars, sugar alcohols and cryoprotective proteins to maintain an osmotic adjustment, and increase in antioxidative-enzyme activity to control the cell homeostasis. An intricate gene network to avoid cell dehydration under freezing temperatures regulates all these processes.
During spring, temperatures may drop below zero Celsius degrees for many hours. Thus, newly opened almond flowers can undergo frost stress. In almond, the most sensitive floral organs to freezing temperatures are the base of style, the internal wall of the ovary and the base of the ovule, whose tissue brownish is the main frost-damage symptom [6]. Therefore, this browning pistil-tissue may be considered as a phenotypic marker for the evaluation of frost injuries that include symptoms in flowers [7]. Frost temperatures negatively affect the development, productivity as well as distribution of horticulture crops [8]. Particularly, almond [Prunus amygdalus Batsch syn. P. dulcis (Mill.) D.A. Webb], with a low chilling requirement for blooming, presents a high risk of suffering spring-frost damages due to its early flowering [9,10]. For this reason, almond production has been restricted to areas with a low risk of frosts. The geographical expansion of almond to land with a higher altitude of Mediterranean areas (e.g., Central Spain) allows sustainable cultivation of otherwise unused plots, but it can significantly decrease the yield, due to the incidence of spring frosts being more frequent in these regions [10]. Furthermore, warmer late-winter temperatures induced by climate change produce a precocity in flowing and bud breaking times [11]. Therefore, it is necessary to introduce late-blooming almond genotypes to avoid the spring-frost period, but also evaluate the genetic resistance to cold stress during flowering and fruit development processes to obtain almond selections adapted to freezing temperatures.
This study aims at unraveling the molecular response to frost stress in order to identify key genes related to a cold-adaptive mechanism in almond, analyzing three different genotypes: ‘Guara’, ‘Soleta’ and ‘Belona’. Our hypothesis is that the molecular cold response should match the phenotypic response. Therefore, marker-assisted selection could potentially be applied in new cold-tolerant-breeding programs using the genes identified in this research as markers for cold stress tolerance.
2. Materials and Methods
2.1. Plant Material and Cold Treatment
Branches at phenological stages ‘E’–‘F’ (stamens visible-open flowers) [12] were harvested from the field (41°43′33.8″ N, 0°49′01.1″ W) at 9 a.m. in three late-blooming genotypes: ‘Guara’, ‘Belona’ and ‘Soleta’, whose flowering is in early-March and use is extended in the almond orchards of the Mediterranean Basin. Cut branches were introduced in a constant climate chamber (model KMF 115, BINDER GmbH, Tuttlingen, Germany). The temperature was previously set at −4 °C. Pistil samples were harvested at 0, 2 and 24 h (three biological replicates for each time point) and flash-frozen in liquid nitrogen before storage at −80 °C for subsequent RNA extraction.
2.2. Phenotype Analysis of the Cold Damages
A total of 100 pistils of the treated flowers were taken at each time point (0, 2 and 24 h) for each genotype. Browning pistil-tissue (necrotic pistil) was counted and evaluated as a cold-damage symptom [13]. Cold damage was measured as the percentage of necrotic pistils.
2.3. RNA Isolation and cDNA Synthesis
Total RNA was extracted from 0.35 g of pistil tissue for each time point and each biological replicate (each biological replicate was considered as group of randomly taken pistils from the different branches of the same tree) using the CTAB method as described by Meisel et al. [14] with some modifications [15,16,17]. Extracted RNA was quantified using a NanoDrop ND-1000 UV-vis spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Contaminated genomic DNA in RNA samples was removed using DNase I [TURBO DNA-Free™ (Invitrogen, Thermo Fisher Scientific, Carlsbad, CA, USA)] following the manufacturer’s instructions. RNA integrity was verified by 1% agarose gel electrophoresis. RNA (2500 ng) was reverse-transcribed with the SuperScript™ III First-Strand Synthesis System for RT-PCR (Invitrogen, Thermo Fisher Scientific, Carlsbad, CA, USA) in a total volume of 21 µL according to manufacturer’s instructions.
2.4. High-Throughput qRT-PCR
High-throughput qRT-PCR analysis was performed by the Centre for Research in Agricultural Genomics (CRAG—Barcelona, Spain) using the BioMark 48:48 Dynamic Array integrated fluidic circuits (Fluidigm, https://www.fluidigm.com; date last accessed 12 July 2021). Specific primers corresponding to 30 early-cold-responsive genes, as well as the two genes used as reference [a Translocation Elongation Factor 2 gene (TEF2), accession number TC3544; and an RNA Polymerase subunit II (RPII), accession number TC1717], were designed based on their nucleotide sequences present in the assembled and annotated peach genome (Prunus persica genome v2.0; https://www.rosaceae.org/; date last accessed 20 January 2021) (Table S1) using Primer3 [18,19]. Since the primer design was performed before the release of the almond genome, the presence of all primer sequences was checked against the P. dulcis genome cv. ‘Texas’ [20] with NCBI Megablast after the primers design. Briefly, each cDNA sample was pre-amplified with a pool of primers specific to the target genes by using the following program: 95 °C for 10 min, then 14 cycles of 95 °C for 15 s and 60 °C for 4 min. The pre-amplified products were diluted 1:5 in 10 mM Tris-HCl, pH 8; 0.1 mM EDTA and analyzed by qRT-PCR using the following conditions: 95 °C for 10 min, then 35 cycles of 95 °C for 15 s and 60 °C for 30 s. The specificity of the PCR products was then confirmed by analyzing melting curves, following the final PCR cycle. Only primers that produced a linear amplification and qRT-PCR products with a single-peak melting curve were used for further analysis.
The dynamic array raw data resulting from the RNA expression analysis was examined with the Fluidigm Real-Time PCR Analysis software using a quality threshold of 0.65 and an automatic selection method of the Ct threshold. CTs were obtained with Fluidigm Real-Time PCR Analysis Software version 4.1.3 (Fluidigm, https://www.fluidigm.com; date last accessed 12 July 2021). The Fold of Change (FC) was calculated with the relative expression values of the genes, referenced to the expression of the housekeeping genes (TEF2 and RPII). The relative gene expression was normalized to ‘Guara’ at 0 h.
2.5. Statistical Analysis
Statistical analyses were performed with the statistical software R package (3.5.0) through R Studio software environment (1.2.5042) [21]. Phenotype data were subjected to ANOVA to test for significant differences among genotypes and among time points. Statistical significance was assessed with Tukey’s honest significant difference (HSD) test (p ≤ 0.05). For gene expression statistical analysis, data distribution of the did not follow a normal distribution. Therefore, a Log-transformation of the data was carried out because the distribution of the data did not follow a normal distribution. After calculating the log2 of FC, data were subjected to a three-way ANOVA (p ≤ 0.05) following Tukey’s HSD test (p < 0.05) for comparison among hours and among genotypes for each gene. Expression values of the genes with significant differences were clustered through the Spearman correlation method (p < 0.05) and represented as a heatmap. Principal Component Analysis (PCA) was performed to explore the data structure, the relationships among genotypes and the variables time point and injury percentage, as well as correlations among gene expression profiles.
3. Results and Discussion
3.1. Necrotic Pistil as a Cold-Sensitiveness Phenotypic Trait
Low and freezing temperatures can generate injuries by ice formation in extracellular spaces in tissues of sensitive plants [22]. After analyzing cold damage detected in the pistil tissue of all cultivars (Figure 1) as a percentage of necrotic pistils [13], statistical differences were not found among genotypes at 0 h (Table 1). At 2 h of cold experiment, ‘Guara’ presented a necrotic-pistil percentage of 2%, whereas ‘Soleta’ and ‘Belona’ did not show any necrotic pistil. (Table 1). However, damages detected at 2 h did not predict damages at 24 h. Significant differences were observed at 24 h of exposure at −4 °C. At that time point, ‘Guara’ and ‘Soleta’ stood out for presenting the lowest rate of necrotic pistils, while ‘Belona’ showed a rate of 62% of necrotic pistils (Table 1). So far, few papers related to the physiological response to cold stress have been published in these almond genotypes. Masip et al. [23] subjected different almond cultivars such as ‘Marinada’, ‘Vayro’, ‘Belona’, ‘Guara’, ‘Mardia’, ‘Soleta’, ‘Penta’ and ‘Tardona’ to freezing temperatures. Despite the fact that work was conducted at phenological stage ‘H’ and not at stages ‘E’–‘F’, ‘Belona’ was also highlighted as the genotype most sensitive to frost, as in our study.
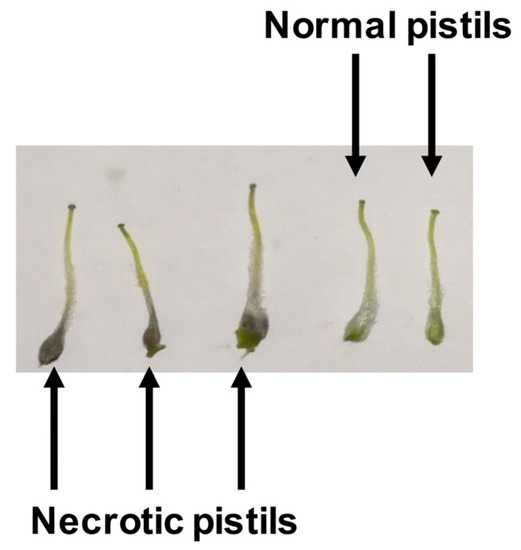
Figure 1.
Detail of frost injuries in almond pistils (necrotic pistils) in comparison to normal almond pistils.

Table 1.
Percentage of necrotic pistils throughout the cold experiment. Similar letter values indicate no significant difference (p ≤ 0.05) following Tukey’s honest significant difference (HSD) test among genotypes for each hour (in capital letters), and among hours for each genotype (in lowercase). (h = hours).
3.2. Changes in Gene Expression as a Response to Cold Stress
During cold stress exposure, the first molecular event triggered is stress perception by cell membrane receptors that in turn activates secondary messengers. Phosphorylation cascades are triggered, resulting in a downstream activation of regulatory genes which in turn modulate the expression of effector genes of cold stress tolerance [24,25,26]. As a result, plants can experience different biochemical changes such as accumulation of osmoprotectants, soluble sugars and other cryoprotective components to prevent dehydration of cell (tolerance strategy), as well as physiological and morphological changes including seasonal adjustments and unsaturated fatty acids accumulation in the cell membrane to maintain cellular homeostasis (resistance strategy). All these changes allow cold acclimation of plants [8,27].
In this study, the expression patterns of 30 candidate genes with a potential relation to cold response were analyzed, founding differences among the three studied genotypes and time of cold exposure (Figure 2). These include genes encoding several kinase proteins, transcription factors (TFs) such as B-Box Zinc finger proteins and central-cold regulators, and effector genes related to dehydration-protective functions. In addition, other functional and uncharacterized proteins were studied. The gene expression profile indicates a higher level of gene expression in ‘Soleta’ throughout the cold exposure compared to the other genotypes (Figure 2).
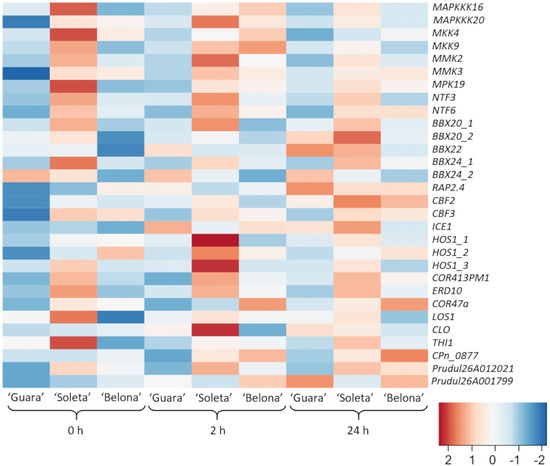
Figure 2.
Expression profile of the 30 studied genes in ‘Guara’, ‘Soleta’ and ‘Belona’ almond genotypes analyzed for each time point of the experiment (0, 2 and 24 h). Color scale represents the row Z-score, where blue indicates underexpression level and red indicates overexpression level.
Principal Component Analysis (PCA) was performed with the gene expression of the 30 genes and the percentage of necrotic pistils at three-time points (0 h, 2 h, and 24 h) in the studied genotypes to determine the level of interaction between the gene expression profile and the cold response for genotypes throughout the experiment (Figure 3). When all data were compared, 62.89% of the total variance was explained by two components (PC1: 48.97%; PC2: 13.92%). ‘Guara’, ‘Soleta’ and ‘Belona’ were distributed in three different groups (Figure 3a). Three PCAs were done at each time point of the cold experiment individually for detecting when each gene could play a role in the response to cold. Therefore, in the 0 h-PCA, 76.29% of the total variance was explained (PC1: 59.48%; PC2: 16.81%); at 2 h, the total variance was explained at 76.34% by two components (PC1:54.78%; PC2: 21.56%); and, finally at 24 h, 77.18% of the total variance was explained by two components (PC1: 50.69%; PC2: 26.49%).
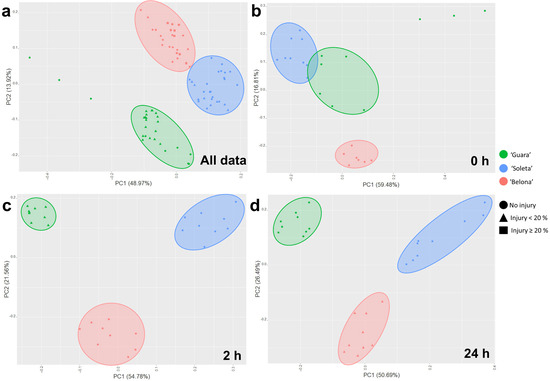
Figure 3.
Principal Component Analysis (PCA) showing the genotype distribution based on gene expression and injury percentage, as well as the gene expression influence on the cold response in ‘Guara’, ‘Soleta’ and ‘Belona’ cultivars (a) for all data, (b) at 0 h, (c) at 2 h and (d) at 24 h of cold treatment.
At 0 h, ‘Soleta’ and ‘Belona’ replicates showed an opposite distribution on the PCA in two different groups (Figure 3b). ‘Guara’ did not present a clear distribution pattern (Figure 3b). The genes whose expression showed the highest influence in the performance of ‘Soleta’ at 0 h were MAPKKK16, MKK9, MPK19, NTF3 and BBX20_1. On one hand, Prudul26A001799, HOS1_2 and RAP2.4 were the genes with the most influence in the performance of ‘Belona’ (Figure 3b). After 2 h, the replicates for each genotype were distributed in three differenced groups (Figure 3c,d). In both, 2 and 24 h, the expression of many of the studied genes influenced ‘Soleta’ behavior (Figure 3c,d). However, the performance of ‘Guara’ was hardly influenced by the analyzed genes (Figure 3c,d) at both time points.
It is noteworthy that when the samples of ‘Soleta’ and ‘Belona’ were collected, at the 0 h time-point, the ambient temperature was 0.74 °C, while for ‘Guara’ was 5.85 °C. This lowered temperature registered from ‘Soleta’ and ‘Belona’ could explain the overexpression in these genotypes at 0 h shown by several stress-responsive genes in the experiment (Figure 2). These two genotypes could have activated their cold-response signaling mechanisms earlier, while ‘Guara’ had not already perceived the stress stimulus. This behavior in ‘Guara’ was also observed at 2 and 24 h. We did not observe many differentially expressed genes in ‘Guara’ either (i) because the response pathway requires fewer genes or was never triggered; (ii) that analyzed genes are not involved in ‘Guara’ response; or (iii) the cold-stress response occurs after 24 h, thus not being detectable in our experimental setup.
3.2.1. ICE-CBF-COR Pathway
CBFs (C-repeat-Binding Factors) belong to AP2/ERF family TFs (CBF1, CBF2 and CBF3), which are bound to promoters cold-responsive genes such as genes coding for COR (Cold-Regulated) proteins [28]. The CBF-mediated signaling pathway, named ICE-CBF-COR, is well described in the literature and plays a major role in the cold response. This pathway is a complex network whose central regulator is ICE (inducer of CBF expression) that controls the induction of CBFs. ICE1 (Inducer of CBF expression 1), a MYC basic helix-loop-helix (bHLh) TF, binds to MYC elements in the promoter region of CBF3 inducing their expression [29]. In addition, other components, including different protein kinases and transcription factors such as HOS1 (High expression of osmotically responsive 1) and LOS1 (Low expression of osmotically responsive 1), act as activators or repressors of the principal components of this intricate pathway [29,30,31,32,33,34,35].
MAPK (mitogen-activated protein kinase) signaling cascades are induced by the transmission of the stress-stimuli sensed in plasma membrane receptors. Kinases are phosphorylated as the signal is transmitted. One of these protein kinases is MAPKKK16 which is promoted by freezing temperatures. The phosphorylation of this protein activates the MAPKs cascade which leads to degradation of ICE1 [34,36,37]. Wu et al. [37] found that phosphorylation of MAPKKK16 initiated MAPKs cascade during cold stress in Denbrobium officinale. As a consequence, other kinase cascades such as MKK4/5 and MPK3/6 are activated [34,36,38]. It is known that the MPK3/6 cascade is also activated by MKK9, which acts as a negative regulator by CBF inhibition via the ethylene signaling pathway of freezing tolerance [32]. As a result, MPK3/6 interacts with ICE1 promoting its degradation and then, reducing its transcriptional activity [34,36]. However, another kinase cascade, MEKK1-MMK2-MPK4, acts as an inhibitor of the ICE1 phosphorylation via the MPK3/6 cascade [34,36,39]. In our study, there were not strong (over 2-FC) changes in the relative expression values of genes code for MAPKKK16, MKK4, MKK9 (except to ‘Belona’ at 2 h), MMK2 and MMK3 (Figure 4a–e). These results suggest that ICE1 expression is not impaired by this MAPK signaling pathway. However, in the three genotypes, ICE1 did not show changes in its relative expression levels, being stable throughout the experiment (Figure 4i). It is possible that ICE1 expression could be controlled by other components such as HOS1. The three isoforms of HOS1 analyzed in our almond genotypes showed different expression levels and similar patterns within each other (Figure 4f–h). Two different mRNA of HOS1 were described for P. persica LOC18776389, called HOS1_1 and HOS1_2. Within this analysis, we added a third sequence previously described in almond, KT202287.1 [13], which was renamed as HOS1_3 (Table S1). After the almond genome sequence was published [20], it was revealed that these three sequences were found on the same locus in almond and we observed similar changes of expression according to our three probes (Figure 4f–h). No significant changes of expression for any of the three mRNA variants were observed in ‘Guara’ and ‘Belona’. An early response peak of expression was observed in ‘Soleta’ for the three probes in HOS1_1 and HOS1_2 at 2 h (Figure 4f,g). These results are surprising since it is known that HOS1 negatively affects CBF3 expression via ICE1 ubiquitination and degradation [29,30]. We have seen that the decrease of MAPK expression might result in lower phosphorylation of ICE1, which HOS1 will not be able to degrade anyway. However, CBF3 gene expression was switched on in almond pistils upon cold. CBF3 was significantly overexpressed from 2 h in ‘Guara’, while in ‘Soleta’ and ‘Belona’; it was relatively stable from 0 h with significantly higher values in both cultivars than in ‘Guara’ (Figure 4j). These results suggest that other genes would be more influential in the positive regulation CBF3 [40,41,42]. The CBF2 was statistically overexpressed upon cold stress with the higher magnitude changes observed in this study, presenting the highest relative expression level at 24 h of cold exposure in the three genotypes. In ‘Guara’ and ‘Soleta’, CBF2 expression gradually increased, while the expression increased only between 2 and 24 h in ‘Belona’ (Figure 4k). The CBF2 expression results are in agreement with previous results in almond, where authors demonstrated that CBF2 plays a role in cold acclimation by ABA induction [40]. Novillo et al. [43] reported that CBF2 negatively regulates CBF3 in Arabidopsis, as well as the induction of CBF3 is earlier than CBF2 in response to cold. However, in our data, while CBF2 expression increased, we only observed a slight decrease in ‘Soleta’ and even an increase in ‘Guara’ (Figure 4j,k). Therefore, it would be possible that CBF3 was not repressed by CBF2 because its expression might be controlled by other signaling pathways and genes [44,45]. Our results highlight that almond-cold response is a complex regulatory network integrated by many interconnected signaling pathways, which show crosstalk among regulated genes signaling pathways.
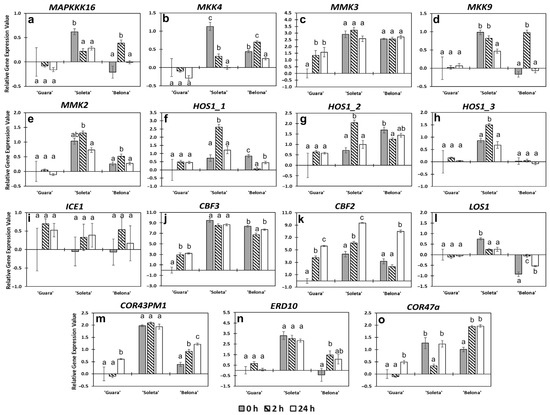
Figure 4.
Relative gene expression profiles of the studied genes involved in the ICE-CBF-COR pathway. (a) MAPPPK16, (b) MKK4, (c) MMK3, (d) MKK9, (e) MMK2, (f) HOS1_1, (g) HOS1_2, (h) HOS1_3, (i) ICE1, (j) CBF3, (k) CBF2, (l) LOS1, (m) COR43PM1, (n) ERD10, (o) COR47a. Error bars represent the standard error among replicates. Letters indicate statistical differences following Tukey’s honest significant difference (HSD) test (p < 0.05) among hours for each genotype.
COR (cold-responsive) genes are activated by CBFs via binding to the promoters of these COR genes. Adaptation and tolerance to low temperatures are facilitated by genes that play an important role in the regulation of processes involved in osmoprotective functions to maintain cellular membrane stability. In this study, the relative expression of three COR genes with cryoprotective functions was analyzed. These genes comprise a Cold regulated 413 plasma membrane 1 (COR413PM1) and two dehydrins: An Early response to dehydration 10 (ERD10) and the gene coding for a COR47a protein. COR413PM1 is a multispanning transmembrane protein belonging to the COR protein family [46] associated with cold acclimation [31,47,48]. Cold induction of COR413PM1 has been reported previously. Fu et al. [47] found this gene overexpressed under cold stress in the tolerant genotype of Elymus nutans in comparison with the sensitive. In addition, a proteomic study revealed COR413PM1 confers cold tolerance by intervening in biochemical changes related to changes in lipid-plasma-membrane composition, as well as changes related to carbohydrate metabolism by sugar accumulation [48]. Transgenic tobacco plants expressing SikCOR413PM1 were more tolerant to cold than wild-type plants [31]. In our almond genotypes, the expression of this gene also showed differences. In ‘Guara’, COR413PM1 was found significantly overexpressed at 24 h of cold treatment, while in ‘Soleta’ COR413PM1 remained constitutively highly expressed from 0 to 24 h of cold stress treatment. Finally, ‘Belona’ showed an increase in gene expression throughout the experiment, presenting its maximum value at 24 h (Figure 4m). These findings agree with the previously published studies above mentioned. Furthermore, Guo et al. [31] reported that SikCOR413PM1 regulates the activity of other cold-responsive genes such as ERD10. In our experiment, the dehydrin ERD10 was also found significantly overexpressed in ‘Soleta’ compared with ‘Guara’ and ‘Belona’ (Figure 4n). ERD10 works as a chaperone [49]. Previously, ERD10 has been described as abscisic acid (ABA)-dependent in Brassica napus [50], and necessary for the expression of the CBF regulon and downstream genes in Arabidopsis [49]. CBFs and ERD10 genes showed lower expression in ‘Guara’ compared to the other two genotypes (Figure 4j,k,n). Thus, it could be evidenced that ABA-dependent pathways might have been activated in ‘Belona’ and ‘Soleta’ through a constitutive presence of ERD10 expression, but not in ‘Guara’. Surprisingly, the COR47a expression level reached its higher values in the sensitive genotype ‘Belona’ at 2 and 24 h than in the tolerant ‘Soleta’ (Figure 4o). ‘Guara’ showed a significant change of COR47a expression at 24 h, but with much lower values than the other genotypes, COR413PM1 and ERD10 genes (Figure 4m–o). COR47a belongs to the Group II LEA protein family, is induced by ABA and water stress by drought and freezing temperatures [51]. Previously published studies have demonstrated that the expression of COR47a is controlled by different proteins including ICE1, HOS15 and MYB96, among others which are recognized by CBFs in the promoter region of this dehydrine [29,44,45]. Guo et al. [52] demonstrated the key role played by LOS1 in cold response. LOS1 is coding for a translation elongation factor 2, which controls the CBF-transcript accumulation. The los1-1 mutation in Arabidopsis plants induces an over-accumulation of CBFs; however, the lack of protein synthesis by the mutation impairs the COR activation downstream of CBFs transcription [52]. In our data, LOS1 was inhibited in ‘Guara’ and ‘Belona’ to a lesser extent at 0 h compared to ‘Soleta’. In ‘Soleta’, its relative expression values slightly decreased from 2 h of cold treatment (Figure 4l). Then, in these almond genotypes, the lack of LOS1 induction under cold exposure might explain the high accumulation observed for CBF2 and CBF3. Nevertheless, other signaling response pathways might regulate this transcript accumulation and its post-transcriptional functions inducing COR genes. Then, in our results, COR47a expression might be regulated by CBF2 and CBF3, which were found highly overexpressed in ‘Soleta’ and ‘Belona’. Under this assumption, further analysis would be necessary for almond genotypes to determine which genes are involved in the induction of CBFs and how the downstream COR induction is produced.
3.2.2. CBF-Independent Responses
As mentioned above, kinase proteins are one of the first links in that chain of transmission. They are essential in the control of signaling cascade activity in various stresses [53]. In this study, besides the ones involved in the CBF cascade, two genes belonging to the MAPK family: MAPKKK20 and MPK19; and two Nicotiana tabacum FUS3-related kinase genes (NTF3 and NTF6) were analyzed. ‘Guara’, ‘Soleta’ and ‘Belona’ presented different gene expression patterns for these kinases, except for the gene encoding a MAPKKK20 whose relative expression was maintained stable upon cold stress (Figure 5a, Table S2). Meanwhile, a mapkkk20 mutant of Arabidopsis shows an enhancement of tolerance to salinity [54], Kim et al. [55] determined that MAPKKK20 overexpression plays a role in cold response by osmotic adjustment in Arabidopsis from 1 h of cold exposure, decreasing at 12 h. A similar tendency was found in the MAPKKK20 expression pattern in our three almond genotypes, although no significant changes in their expression levels were found (Figure 5a). During the cold exposure, ‘Guara’ showed no significant differences in the expression values of the genes coding for MPK19 (Figure 5b). However, this gene was overexpressed significantly in ‘Soleta’ and ‘Belona’ compared to ‘Guara’, with different expression patterns. Meanwhile, MPK19 showed its highest expression levels at 0 h, this gene was maintained overexpressed until 24 h in ‘Soleta’. In ‘Belona’, MPK19 reached the highest value at 2 h, maintaining it also at 24 h (Figure 5b). According to Zhou et al. [56], MPK19 is involved in the ABA-activated signaling pathway by interaction with PP2C16. In addition, this kinase was found downregulated in radish submitted to salinity stress [57]. The different MPK19 expression levels found in our almond genotypes, without a relation to the cold-tolerance phenotypes shown (Table 1), could be due to crosstalk among different signaling pathways.
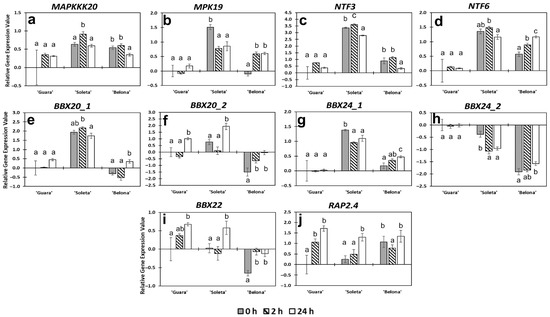
Figure 5.
Relative gene expression profiles of the studied genes involved in CBF-independent response to cold. (a) MAPPPK20, (b) MPK19, (c) NTF3, (d) NTF6, (e) BBX20_1, (f) BBX20_2, (g) BBX24_1, (h) BBX24_2, (i) BBX22, (j) RAP2.4. Error bars represent the standard error among replicates. Letters indicate statistical differences following Tukey’s honest significant difference (HSD) test (p < 0.05) among hours for each genotype.
When the relative expression of NTF3 and NTF6 were studied, no changes were found among the three-time points for each genotype (Figure 5c,d). However, differences were observed among genotypes for each time point (Table S2). ‘Soleta’ showed the highest relative gene expression level for both genes under freezing temperatures. In ‘Guara’, NTF3 and NTF6 presented the lowest expression, and ‘Belona’ showed an intermediate level and a gradual non-significant increase in the NTF6 expression level until reaching a similar gene expression value than ‘Soleta’ at 24 h of cold stress (Figure 4d). NTF3, also known as NtMPK8 [58], codes for a kinase in tobacco. It was speculated that it could possess a specific function because of its divergent sequence compared to other kinases [59]. Its ortholog, OsMPK8, could be regulated by drought and other abiotic stresses [60], suggesting that it may play an important role in response to various stresses [61]. Our results could suggest a possible role of NTF3 in the response to cold stress since the first hours of exposition in ‘Soleta’ almond pistils. However, further analysis of phosphorylation activity of this kinase could help elucidate actual intervention in cold response in almond. On the other hand, the NTF6 gene, which is activated by H2O2 presence, is overexpressed in Anabasis aphylla under cold stress [62]. Therefore, the changes in NTF6 found in our almond genotypes could evidence response to cold stress mediated by the interaction with oxidative stress signaling pathways caused by low temperatures in the ‘Soleta’ and ‘Belona’ genotypes.
Some B-Box zinc finger (BBX) proteins have shown to play key roles in response to cold stress in several species [63,64]. Differences in the expression of the genes encoding three B-Box zinc finger proteins (BBX20, BBX22 and BBX24) were found in our study (Figure 5e–i). In almond, the gene encoding BBX20 presents two different paralogs: Prudul26A027241, called BBX20_1, and Prudul26A030197, called BBX20_2 (Table S1). BBX20_1 was found overexpressed in ‘Soleta’ from 0 h against both ‘Guara’ and ‘Belona’. However, we observed no significant changes over time (Figure 5e). By contrast, we found an increase in the expression of BBX20_2 in the three genotypes during the cold stress experiment (Figure 5f). ‘Guara’ and ‘Soleta’ presented significant overexpression in BBX20_2 at 24 h (log2FC of 1.018 and 1.934, respectively) compared to both 0 h (log2FC of 0 and 0.770, respectively) and 2 h (log2FC of −0.332 and 0.103, respectively). This TF was only overexpressed in ‘Belona’ in comparison to 0 h (Figure 5f). Its expression was always significantly lower than the two other genotypes. BBX24 also has two paralogues in almond: Prudul26A012331, called BBX24_1, and Prudul26A022757, called BBX24_2 (Table S1). BBX24_1 was found overexpressed in ‘Soleta’ from 0 h against both ‘Guara’ and ‘Belona’. However, any significant changes over time were not found (Figure 5g). We neither observed change over time for BBX24_2, but it was more significantly expressed in ‘Guara’ than ‘Soleta’ (Figure 5h). The differential expression shown by BBX20 and BBX24 paralogs in the three almond genotypes suggested that different isoforms might play a role in ‘Guara’ and ‘Soleta’, but they are almost always underexpressed in ‘Belona’ compared to the other genotypes. Except for BBX20_2, none of them responded to freezing temperatures. We examined another BBX, the BBX22 gene, but it did not present significant changes between cultivar or over time (Figure 5i). BBX20, BBX22 and BBX24 are implied with both positive and negative roles in the control of processes including photomorphogenesis [65], hormonal signaling [64,66], anthocyanin biosynthesis [67], and different abiotic stress responses as drought, cold and salinity in different plant species. Our results might suggest that the activity of BBX20_1 BBX24_1 and BBX24_2 could be not related directly to cold stress response, but the differences between genotype, and correlation of expression with tolerance might be related to innate protection mechanism and could indicate a crucial role enhancing the cold-tolerance response in almond pistils. The expression of BBX20_2 increase upon cold stress, the role of BBX20 in cold response have been reported by Fang et al. [67], which demonstrated that an accumulation of anthocyanin was produced by the interaction of MdBBX20 with HY5 under UV-B radiation and low temperatures.
The AP2/ERF (APETALA/Ethylene-Responsive Factor) family TF, RAP2.4 (Related to APETALA2-4), is implicated in key processes regulating drought and salt responses [68]. Changes in expression of this gene were found under low temperatures in our almond genotypes. Therefore, RAP2.4 showed a gradual increase of expression in the tolerant genotypes, ‘Guara’ and ‘Soleta’ (Figure 5j), but its expression was stable and high in ‘Belona’ (Figure 5j). Our results indicate that RAP2.4 responded not only to drought and to salt, but also could contribute to cold stress; however, its potential implication in the tolerance mechanism is extremely unclear.
3.2.3. Other Functional and Uncharacterized Proteins
In this study, two other stress-related genes were also analyzed: a Clotho/gametophytic factor 1 (CLO) and a gene encoding a thiamine thiazole synthase 1 (THI1). When expression changes in CLO were determined, a significant overexpression was found in ‘Soleta’ compared to the other genotypes at 2 h of cold exposure (Figure 6a). In this genotype, the expression significantly increased between 0 h and 2 h from a log2FC of 0.072 to 1.095. At 24 h, CLO expression decreased in ‘Soleta’ (log2FC of 0.460) showing a value similar to ‘Guara’ (lof2FC of 0.549) (Figure 6a). CLO codes for an elongation factor related to floral organ growth in Arabidopsis [69]. CLO has been reported to participate in other aspects of plant development as cellular fate [70] and the generation of gametophytic cells [71]. The results found in this study suggested that ‘Soleta’ would have triggered the induction of different floral growth processes at 2 h of cold stress treatment, which could protect the floral tissue under low temperatures. Therefore, ‘Soleta’ could present a possible tolerant response, not found in ‘Belona’. A thiamine thiazole synthase 1 (THI1) was analyzed in our cold stress. There is no expression change with cold stress, although we observed contrasts between genotypes. THI1 is overexpressed in ‘Soleta’ compared to ‘Belona’ at 0 h, but the expression is similar after 2 h (Figure 6b). Although its role in other abiotic stress responses as drought has been reported previously as being involved in anion channels activation and stomatal closure through Ca2+-dependent proteins in Arabidopsis [72], our data suggest no role of this protein in almond pistils under cold stress.
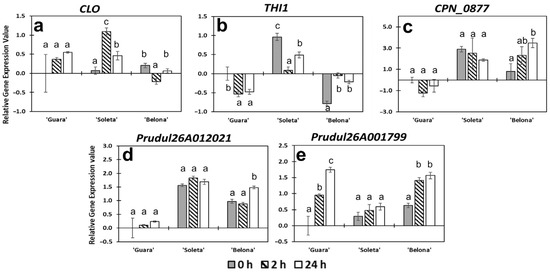
Figure 6.
Relative gene expression profiles of genes encoding for other functional and uncharacterized proteins. (a) CLO, (b) THI1, (c) CPn_0877, (d) Prudul26A012021, (e) Prudul26A001799. Error bars represent the standard error among replicates. Letters indicate statistical differences following Tukey’s honest significant difference (HSD) test (p < 0.05) among hours for each genotype.
The gene expression of three uncharacterized proteins was also studied. CPn_0877 is an uncharacterized protein homologous to AT5G01300 from Arabidopsis, which belongs to the PEBP (Phosphatidylethanolamine-binding protein) family protein. The main functions of this family protein are related to reproductive processes such as flowering differentiation [73]. In our study, CPn_0877 gene expression was inhibited in ‘Guara’ compared to the other genotypes. Expression in ‘Belona’, rose throughout the cold treatment, reaching similarly high values to ‘Soleta’ at 2 h and its high value at 24 h (Figure 6c). This protein might be a potential role in cold response in almond. Similar results were reported in a previous transcriptome analysis done in Thellungiella salsuginea under cold stress, finding a putative phosphatidylethanolamine-binding protein gene overexpressed [74].
The other two uncharacterized proteins have been described in the almond genome. The first one corresponded to the protein coded by the gene Prudul26A012021. This gene was overexpressed in ‘Soleta’ and ‘Belona’ in comparison with the transcript level found in ‘Guara’ (Figure 6d). Prudul26A001799 gene was found significantly overexpressed in ‘Guara’ and ‘Belona’ genotypes from 2 h of cold stress exposure, but not in ‘Soleta’ (Figure 6e). These two proteins are homologous to AT5G55530 and AT2G33320, respectively, which are described as Calcium-dependent lipid-binding (CaLB domain) proteins. De Silva et al. [75] evidenced that an AtCLB protein acts as a negative regulator in abiotic stress. In addition, in a microarray analysis in rice, a C2 CaLB domain-containing protein was found downregulated under salt and drought stresses [76]. However, in a recent study, this protein was found induced by a cold effect in Chlamydomonas reinhardtii [77]. Agreeing with these pieces of evidence, Prudul26A012021 and Prudul26A001799 might encode a CaLB domain protein with a potential role in almond cold stress response.
4. Conclusions
Our study showed that cold response in almond pistils is mediated through the CBF pathway, but also through alternative pathways that can be differentially activated among genotypes. This response is substantially different between the two tolerant genotypes, ‘Guara’ and ‘Soleta’, with many genes overexpressed in ‘Soleta’ upon cold stress while expression is generally stable in ‘Guara’ (Figure 4, Figure 5 and Figure 6). However, these two genotypes were capable of behaving as tolerant in the phenotypic analysis (Table 1). Our study suggests that there is not a unique cold-response strategy in almond. While ‘Soleta’ followed a tolerant strategy upon cold stress, ‘Guara’ presented an intrinsic tolerance. Finally, ‘Belona’ did not develop cold tolerance compared to the other tolerant genotypes under cold stress conditions, probably due to lack of expression of certain candidate genes, including BBX20_2 and CLO (Figure 5f and Figure 6a). In addition, based on our data, it could be considered that in ‘Belona’, even if the stress response genes were overexpressed, this would not be enough to produce the synthesis of proteins with cryoprotective functions. Studies at the proteomic level would be necessary to validate this hypothesis. Based on our results, two genes (BBX20_2 and CLO) described in our three genotypes represent promising targets for use as cold-tolerant biomarkers in almond breeding programs.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/horticulturae7120527/s1, Table S1: Primer list and amplicon sizes for the thirty studied genes and the two reference genes in pistils of the three almond genotypes by qRT-PCR; Table S2: ANOVA results from relative gene expression during the cold experiment for the studied genotypes. Same letter values indicate a no significant difference (p ≤ 0.05) following Tuckey’s post hoc test among genotypes for each time-point of treatment.
Author Contributions
Conceptualization, J.G. and M.J.R.-C.; methodology, B.B., J.I.Á.-A., J.G. and M.J.R.-C.; validation, B.B., J.I.Á.-A., J.G. and M.J.R.-C.; formal analysis, B.B., J.I.Á.-A. and J.G.; investigation, B.B. and J.I.Á.-A.; resources, M.J.R.-C.; data curation, B.B., J.I.Á.-A., Á.F.i.M. and J.G.; writing—original draft preparation, B.B. and J.I.Á.-A.; writing—review and editing, B.B., J.G. and M.J.R.-C.; visualization, B.B.; supervision, J.G. and M.J.R.-C.; project administration, M.J.R.-C.; funding acquisition, M.J.R.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This research is part of the I+D+I project no. RTI2018-094210-R100 funded by MCIN/AEI/10.13039/501100011033, as well as by Gobierno de Aragón—European Social Fund, European Union (Grupo Consolidado A12).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Varela, S.A. Aspectos básicos de la fisiología en respuesta al estrés y el clima como condicionante del mismo en las plantas. INTA EEA Bariloche. Comun. Técnica 2010, 78, 1–23. [Google Scholar]
- Janská, A.; Maršík, P.; Zelenková, S.; Ovesná, J. Cold stress and acclimation—What is important for metabolic adjustment? Plant Biol. 2010, 12, 395–405. [Google Scholar] [CrossRef]
- Rodrigo, J. Spring frosts in deciduous fruit trees. Morphological damage and flower hardiness. Sci. Hortic. (Amsterdam) 2000, 85, 155–173. [Google Scholar] [CrossRef]
- Wisniewski, M.; Gusta, L.; Neuner, G. Adaptive mechanisms of freeze avoidance in plants: A brief update. Environ. Exp. Bot. 2014, 99, 133–140. [Google Scholar] [CrossRef]
- Lodolini, E.M.; Alfei, B.; Santinelli, A.; Cioccolanti, T.; Polverigiani, S.; Neri, D. Frost tolerance of 24 olive cultivars and subsequent vegetative re-sprouting as indication of recovery ability. Sci. Hortic. (Amsterdam) 2016, 211, 152–157. [Google Scholar] [CrossRef]
- Urbina, V. Daños por heladas en frutales. Sintomatología y evaluación. In Valoració de Danys Climatoògics i Incendis; Centre de Formació i Estudis Agrorurals, Generalitat de Catalunya—Departament d’Agricultura, Alimentació I Acció Rural: Reus, Spain, 2007; pp. 1–19. [Google Scholar]
- Yu, D.J.; Lee, H.J. Evaluation of freezing injury in temperate fruit trees. Hortic. Environ. Biotechnol. 2020, 61, 787–794. [Google Scholar] [CrossRef]
- Bañuelos, M.; Moreno, L.; Winzerling, J.; Orozco, J.; Gardea, A. Winter metabolism in deciduous trees: Mechanisms, genes and associated proteins. Rev. Fitotec. Mex. 2008, 31, 295–308. [Google Scholar]
- Alonso Segura, J.M.; Socias i Company, R.; Kodad, O. Late-blooming in almond: A controversial objective. Sci. Hortic. (Amsterdam) 2017, 224, 61–67. [Google Scholar] [CrossRef]
- Kodad, O.; Socias i Company, R. Daños diferenciales por heladas en flores y frutos y criterios de selección para la tolerancia a heladas en el almendro. ITEA Inf. Tec. Econ. Agrar. 2005, 101, 349–363. [Google Scholar]
- Lorite, I.J.; Cabezas-Luque, J.M.; Arquero, O.; Gabaldón-Leal, C.; Santos, C.; Rodríguez, A.; Ruiz-Ramos, M.; Lovera, M. The role of phenology in the climate change impacts and adaptation strategies for tree crops: A case study on almond orchards in Southern Europe. Agric. For. Meteorol. 2020, 294, 108142. [Google Scholar] [CrossRef]
- Felipe, J. Almendro. Estados fenológicos. Inf. Tec. Econ. Agrar. 1977, 27, 8–9. [Google Scholar]
- Alisoltani, A.; Shiran, B.; Fallahi, H.; Ebrahimie, E. Gene regulatory network in almond (Prunus dulcis Mill.) in response to frost stress. Tree Genet. Genomes 2015, 11, 1–15. [Google Scholar] [CrossRef]
- Meisel, L.; Fonseca, B.; González, S.; Baeza-Yates, R.; Cambiazo, V.; Campos, R.; González, M.; Orellana, A.; Retamales, J.; Silva, H. A Rapid and Efficient Method for Purifying High Quality Total RNA from Peaches (Prunus persica) for Functional Genomics Analyses. Biol. Res. 2005, 38, 83–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Salzman, R.A.; Fujita, T.; Zhu-Salzman, K.; Hasegawa, P.M.; Bressan, R.A. An Improved RNA Isolation Method for Plant Tissues Containing High Levels of Phenolic Compounds or Carbohydrates. Plant Mol. Biol. Rep. 1999, 17, 11–17. [Google Scholar] [CrossRef]
- Zeng, Y.; Yang, T. RNA Isolation From Highly Viscous Samples Rich in Polyphenols and Polysaccharides. Plant Mol. Biol. Rep. 2002, 20, 417. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Alioto, T.; Alexiou, K.G.; Bardil, A.; Barteri, F.; Castanera, R.; Cruz, F.; Dhingra, A.; Duval, H.; Martí, Á.F.I.; Frias, L.; et al. Transposons played a major role in the diversification between the closely related almond and peach genomes: Results from the almond genome sequence. Plant J. 2019, 101, 455–472. [Google Scholar] [CrossRef] [Green Version]
- Team, R. RStudio: Integrated Development Environment for R; RStudio, PBC: Boston, MA, USA, 2020. [Google Scholar]
- Zuther, E.; Lee, Y.P.; Erban, A.; Kopka, J.; Hincha, D.K. Natural variation in freezing tolerance and cold acclimation response in Arabidopsis thaliana and related species. In Survival Strategies in Extreme Cold and Desiccation. Advances in Experimental Medicine and Biology; Iwaya-Inoue, M., Sakurai, M., Uemura, M., Eds.; Springer: Singapore, 2018; Volume 1081, pp. 81–98. ISBN 9789811312441. [Google Scholar]
- Masip, D.; Torguet, L.; Batlle, I.; Alegre, S.; Miarnau, X. Almond fruit tolerance to frost temperatures in new Spanish cultivars. Acta Hortic. 2018, 1219, 67–72. [Google Scholar] [CrossRef]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef]
- Roychoudhury, A.; Paul, S.; Basu, S. Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 2013, 32, 985–1006. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional Regulatory Networks in Cellular Responses and Tolerance to Dehydration and Cold Stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuñez, M.M.; Hernández, P.F.V.; Rivas, M.R.; López, M.A.V.; Becerra, A.A.; Suárez, S.L.; Cárdenas, F.R. Resistencia y Tolerancia a Estrés Abiótico: Mecanismos Sofisticados de Adaptación de las Plantas ante Distintas Condiciones de Estrés. Available online: https://www.researchgate.net/profile/Pedro-Vera-Hernandez-2/publication/324482888_Resistencia_y_tolerancia_a_estres_abiotico_Mecanismos_sofisticados_de_adaptacion_de_las_plantas_ante_distintas_condiciones_de_estres/links/5ad62b86458515c60f55f5ff/Resistenc (accessed on 14 April 2021).
- Medina, J.; Catalá, R.; Salinas, J. The CBFs: Three arabidopsis transcription factors to cold acclimate. Plant Sci. 2011, 180, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.; Hong, X.; Agarwal, M.; Zhu, J.-K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [Green Version]
- Dong, C.H.; Agarwal, M.; Zhang, Y.; Xie, Q.; Zhu, J.K. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 2006, 103, 8281–8286. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Zhang, L.; Dong, G.; Xu, Z.; Li, G.; Liu, N.; Wang, A.; Zhu, J. A novel cold-regulated protein isolated from Saussurea involucrata confers cold and drought tolerance in transgenic tobacco (Nicotiana tabacum). Plant Sci. 2019, 289, 110246. [Google Scholar] [CrossRef]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, Y.; Wang, Y.; Liu, H.; Lei, L.; Yang, H.; Liu, G.; Ren, D. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 2008, 283, 26996–27006. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Wang, P.; Si, T.; Hsu, C.C.; Wang, L.; Zayed, O.; Yu, Z.; Zhu, Y.; Dong, J.; Tao, W.A.; et al. MAP Kinase Cascades Regulate the Cold Response by Modulating ICE1 Protein Stability. Dev. Cell 2017, 43, 618–629. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Wang, C.; Zhu, F.; Li, Y. Mild osmotic stress promotes 4-methoxy indolyl-3-methyl glucosinolate biosynthesis mediated by the MKK9–MPK3/MPK6 cascade in Arabidopsis. Plant Cell Rep. 2017, 36, 543–555. [Google Scholar] [CrossRef]
- Li, H.; Ding, Y.; Shi, Y.; Zhang, X.; Zhang, S.; Gong, Z.; Yang, S. MPK3- and MPK6-Mediated ICE1 Phosphorylation Negatively Regulates ICE1 Stability and Freezing Tolerance in Arabidopsis. Dev. Cell 2017, 43, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.G.; Jiang, W.; Chen, S.L.; Mantri, N.; Tao, Z.M.; Jiang, C.X. Insights from the cold transcriptome and metabolome of Dendrobium officinale: Global reprogramming of metabolic and gene regulation networks during cold acclimation. Front. Plant Sci. 2016, 7, 1653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, K.; Raina, S.K.; Sultan, S.M. Arabidopsis MAPK signaling pathways and their cross talks in abiotic stress response. J. Plant Biochem. Biotechnol. 2020, 29, 700–714. [Google Scholar] [CrossRef]
- Wisniewski, M.; Nassuth, A.; Arora, R. Cold hardiness in trees: A mini-review. Front. Plant Sci. 2018, 9, 1394. [Google Scholar] [CrossRef] [PubMed]
- Barros, P.M.; Gonçalves, N.; Saibo, N.J.M.; Oliveira, M.M. Functional characterization of two almond C-repeat-binding factors involved in cold response. Tree Physiol. 2012, 32, 1113–1128. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Zhang, L.; Zhang, T.; Zhang, H.; Xu, S.; An, L. Transcriptional regulation network of cold-responsive genes in higher plants. Plant Sci. 2005, 169, 987–995. [Google Scholar] [CrossRef]
- Zhang, J.Z.; Creelman, R.A.; Zhu, J.K. From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol. 2004, 135, 615–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.G.; Seo, P.J. The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J. 2015, 82, 962–977. [Google Scholar] [CrossRef]
- Park, J.; Lim, C.J.; Shen, M.; Park, H.J.; Cha, J.Y.; Iniesto, E.; Rubio, V.; Mengiste, T.; Zhu, J.K.; Bressan, R.A.; et al. Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. USA 2018, 115, E5400–E5409. [Google Scholar] [CrossRef] [Green Version]
- Breton, G.; Danyluk, J.; Frenette Charron, J.B.; Sarhan, F. Expression profiling and bioinformatic analyses of a novel stress-regulated multispanning transmembrane protein family from cereals and Arabidopsis. Plant Physiol. 2003, 132, 64–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.J.; Geng, J.C.; Miao, Y.J.; Xu, Y.M.; Hu, T.M.; Yang, P.Z. Transcriptomic analyses reveal genotype- and organ-specific molecular responses to cold stress in Elymus nutans. Biol. Plant. 2018, 62, 671–683. [Google Scholar] [CrossRef]
- Su, C.; Chen, K.; Ding, Q.; Mou, Y.; Yang, R.; Zhao, M.; Ma, B.; Xu, Z.; Ma, Y.; Pan, Y.; et al. Proteomic analysis of the function of a novel cold-regulated multispanning transmembrane protein COR413-PM1 in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.Y.; Nam, K.H. Physiological roles of ERD10 in abiotic stresses and seed germination of Arabidopsis. Plant Cell Rep. 2010, 29, 203–209. [Google Scholar] [CrossRef]
- Deng, Z.; Pang, Y.; Kong, W.; Chen, Z.; Wang, X.; Liu, X.; Pi, Y.; Sun, X.; Tang, K. A novel ABA-dependent dehydrin ERD10 gene from Brassica napus. DNA Seq. J. DNA Seq. Mapp. 2005, 16, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Artus, N.N.; Thomashow, M.F. cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol. Biol. 1992, 18, 13–21. [Google Scholar] [CrossRef]
- Guo, Y.; Xiong, L.; Ishitani, M.; Zhu, J.-K. An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proc. Natl. Acad. Sci. USA 2002, 99, 7786–7791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moustafa, K.; AbuQamar, S.; Jarrar, M.; Al-Rajab, A.J.; Trémouillaux-Guiller, J. MAPK cascades and major abiotic stresses. Plant Cell Rep. 2014, 33, 1217–1225. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Xiang, C. Bin The genetic locus At1g73660 encodes a putative MAPKKK and negatively regulates salt tolerance in Arabidopsis. Plant Mol. Biol. 2008, 67, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Woo, D.H.; Kim, S.H.; Lee, S.Y.; Park, H.Y.; Seok, H.Y.; Chung, W.S.; Moon, Y.H. Arabidopsis MKKK20 is involved in osmotic stress response via regulation of MPK6 activity. Plant Cell Rep. 2012, 31, 217–224. [Google Scholar] [CrossRef]
- Zhou, A.; Ma, H.; Liu, E.; Jiang, T.; Feng, S.; Gong, S.; Wang, J. Transcriptome sequencing of Dianthus spiculifolius and analysis of the genes involved in responses to combined cold and drought stress. Int. J. Mol. Sci. 2017, 18, 849. [Google Scholar] [CrossRef]
- Sun, X.; Xu, L.; Wang, Y.; Luo, X.; Zhu, X.; Kinuthia, K.B.; Nie, S.; Feng, H.; Li, C.; Liu, L. Transcriptome-based gene expression profiling identifies differentially expressed genes critical for salt stress response in radish (Raphanus sativus L.). Plant Cell Rep. 2016, 35, 329–346. [Google Scholar] [CrossRef]
- Wilson, C.; Eller, N.; Gartner, A.; Vicente, O.; Heberle-Bors, E. Isolation and characterization of a tobacco cDNA clone encoding a putative MAP kinase. Plant Mol. Biol. 1993, 23, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Anglmayer, R.; Vicente, O.; Heberlebors, E. Molecular cloning, functional expression in Escherichia coli, and characterization of multiple mitogen-activated-protein kinases From tobacco. Eur. J. Biochem. 1995, 233, 249–257. [Google Scholar] [CrossRef]
- Reyna, N.S.; Yang, Y. Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol. Plant-Microbe Interact. 2006, 19, 530–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Cheng, T.; Wang, G.; Yan, Y.; Xia, Q. Cloning and evolutionary analysis of tobacco MAPK gene family. Mol. Biol. Rep. 2013, 40, 1407–1415. [Google Scholar] [CrossRef]
- Wang, T.; Ye, C.; Wang, M.; Chu, G. Identification of cold-stress responsive proteins in Anabasis aphylla seedlings via the iTRAQ proteomics technique. J. Plant Interact. 2017, 12, 505–519. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–471. [Google Scholar] [CrossRef]
- Shalmani, A.; Jing, X.Q.; Shi, Y.; Muhammad, I.; Zhou, M.R.; Wei, X.Y.; Chen, Q.Q.; Li, W.Q.; Liu, W.T.; Chen, K.M. Characterization of B-BOX gene family and their expression profiles under hormonal, abiotic and metal stresses in Poaceae plants. BMC Genom. 2019, 20, 27. [Google Scholar] [CrossRef]
- Wei, C.-Q.; Chien, C.-W.; Ai, L.-F.; Zhao, J.; Zhang, Z.; Li, K.; Burlingame, A.; Sun, Y.; Wang, Z.-Y. The Arabidopsis B-BOX Protein BZS1/BBX20 Interacts with HY5 and Mediates Strigolactone Regulation of Photomorphogenesis Chuang-Qi. J. Genet. Genom. 2016, 43, 555–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.Y.; Sun, Y.; Cao, D.M.; Bai, M.Y.; Luo, X.M.; Yang, H.J.; Wei, C.Q.; Zhu, S.W.; Sun, Y.; Chong, K.; et al. BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol. Plant 2012, 5, 591–600. [Google Scholar] [CrossRef] [Green Version]
- Fang, H.; Dong, Y.; Yue, X.; Hu, J.; Jiang, S.; Xu, H.; Wang, Y.; Su, M.; Zhang, J.; Zhang, Z.; et al. The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 2019, 42, 2090–2104. [Google Scholar] [CrossRef]
- Lin, R.C.; Park, H.J.; Wang, H.Y. Role of Arabidopsis RAP2.4 in regulating lightand ethylene-mediated developmental processes and drought stress tolerance. Mol. Plant 2008, 1, 42–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yagi, N.; Takeda, S.; Matsumoto, N.; Okada, K. VAJ/GFA1/CLO is involved in the directional control of floral Organ growth. Plant Cell Physiol. 2009, 50, 515–527. [Google Scholar] [CrossRef] [Green Version]
- Berger, F.; Twell, D. Germline specification and function in plants. Annu. Rev. Plant Biol. 2011, 62, 461–484. [Google Scholar] [CrossRef]
- Moll, C.; Von Lyncker, L.; Zimmermann, S.; Kägi, C.; Baumann, N.; Twell, D.; Grossniklaus, U.; Groß-Hardt, R. CLO/GFA1 and ATO are novel regulators of gametic cell fate in plants. Plant J. 2008, 56, 913–921. [Google Scholar] [CrossRef]
- Li, C.L.; Wang, M.; Wu, X.M.; Chen, D.H.; Lv, H.J.; Shen, J.L.; Qiao, Z.; Zhang, W. THI1, a Thiamine Thiazole Synthase, Interacts with Ca2+ -Dependent Protein Kinase CPK33 and Modulates the S-Type Anion Channels and Stomatal Closure in Arabidopsis. Plant Physiol. 2016, 170, 1090–1104. [Google Scholar] [CrossRef] [Green Version]
- Khosa, J.; Bellinazzo, F.; Goldstein, R.K.; Macknight, R.; Immink, R.G.H. PEBP proteins: The conductors of dual reproduction in plants with vegetative storage organs. J. Exp. Bot. 2021, 72, 2845–2856. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Q.; Cui, F.; Hou, L.; Zhao, S.; Xia, H.; Qiu, J.; Li, T.; Zhang, Y.; Wang, X.; et al. Genome-wide analysis of gene expression provides new insights into cold responses in Thellungiella salsuginea. Front. Plant Sci. 2017, 8, 713. [Google Scholar] [CrossRef] [Green Version]
- de Silva, K.; Laska, B.; Brown, C.; Sederoff, H.W.; Khodakovskaya, M. Arabidopsis thaliana calcium-dependent lipid-binding protein (AtCLB): A novel repressor of abiotic stress response. J. Exp. Bot. 2011, 62, 2679–2689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campo, S.; Baldrich, P.; Messeguer, J.; Lalanne, E.; Coca, M.; San Segundo, B. Overexpression of a Calcium-Dependent Protein Kinase Confers Salt and Drought Tolerance in Rice by Preventing Membrane Lipid Peroxidation. Plant Physiol. 2014, 165, 688–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Peng, H.; Tan, S.; Zhou, J.; Fang, Z.; Hu, Z.; Gao, L.; Li, T.; Zhang, W.; Chen, L. Effects of early cold stress on gene expression in Chlamydomonas reinhardtii. Genomics 2020, 112, 1128–1138. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).