Abstract
B-box (BBX) genes, as zinc finger transcription factors (TFs), play essential roles in regulating plant growth and development. In this study, we identified 23 BBX genes in the pomegranate (Punica granatum L.) genome. These genes were classified into five groups based on the distribution of conserved domains and phylogenetic relationships. Each PgBBX group exhibited similar molecular weights, theoretical isoelectric points (pI), gene structures, and conserved motif distributions compared with BBX members in Arabidopsis and Chinese white pear in corresponding groups. Syntenic analysis revealed segmental duplications of eight PgBBX gene pairs within the pomegranate genome. Additionally, twenty-seven and thirty-one orthologous BBX pairs were identified between PgBBX and AtBBX, and PgBBX and PbBBX, respectively. Promoter analysis revealed the presence of five types of cis-acting elements responding to light, phytohormones, stress, developmental signaling, and potential transcription factors (TFs). GO enrichment analysis confirmed that most PgBBX genes function as TF involved in plant growth and development. RNA-seq data indicated that PgBBX5 was primarily expressed in leaves and flowers, with increased expression in different fruit tissues during ripening. Moreover, PgBBX5 showed a high degree of sequence similarity with anthocyanin-related homologs, including AtBBX24, PhBBX24, FaBBX24, MdCOL4, and PyBBX24. During the ripening of ‘Tunisia’ fruits, PgBBX5 expression was positively correlated with the dynamic changes in anthocyanin content and the expression of key anthocyanin biosynthetic and transport genes. Furthermore, subcellular localization suggested that PgBBX5 encodes a nuclear-localized protein. This study provides a comprehensive characterization of the PgBBX family, offering valuable insights into the mechanisms underlying anthocyanin accumulation during pomegranate fruit ripening.
1. Introduction
Transcription factors (TFs) are key regulatory proteins in gene expression, essential for plant development, growth, and adaptation to environmental stress. They function by binding to cis-acting elements within promoter regions. B-box (BBX) proteins, as members of the zinc finger protein family, are characterized by the presence of single- or double-conserved B-box domains in their N-terminus and a conserved C-terminal CCT (CONSTANS, CO-like, and TOC1) domain in several members [1]. Initially, many BBX genes in Arabidopsis were designated as CO-Like (COL) due to CO (CONSTANS), which was the first BBX gene to be identified in Arabidopsis [2]. B-box domains are hypothesized to mediate DNA binding, protein–protein interactions, and transcriptional regulation, while CCT domains are primarily involved in transcriptional regulation and nuclear localization of BBX proteins [3]. Based on the conserved domain distribution, the BBX family has been classified into five groups [2]. Groups I and II possess two B-box domains and a CCT domain, group III contains a single B-box domain and a CCT domain, group IV members have two B-box domains, and group V sequences contain only a single B-box domain [1].
BBX TFs are integral to light signaling networks and play regulatory roles in seedling photomorphogenesis, seed germination, photoperiodic regulation of flowering, shade avoidance, and responses to biotic and abiotic stresses [1,2]. They are also implicated in the regulation of multiple plant secondary metabolite pathways. For instance, SlBBX20 in tomato [4], LbaBBX2 and LbaBBX4 in wolfberry [5], and CaBBX20 in pepper [6] modulate carotenoid biosynthesis. In sweet potato, the overexpression of IbBBX29 increases flavonoid contents in leaves and storage roots [7], while GmBBX4 in soybean exhibits a negative regulatory role in isoflavonoid biosynthesis [8]. In Artemisia annua, AaBBX22 positively activates the promoter activity of AaADS, AaCYP71AV1, AaDBR2, and AaALDH1 in the artemisinin biosynthetic pathway, thereby increasing artemisinin content [9]. Conversely, AaBBX21, another positive regulator of artemisinin biosynthesis, indirectly activates the transcription of artemisinin biosynthesis-related genes [10]. Many BBX genes are reported to regulate anthocyanin metabolism pathways, such as AtBBX20, AtBBX21, AtBBX22, AtBBX23, AtBBX24, AtBBX25, and AtBBX32 in Arabidopsis [11,12,13]; MdBBX1, MdCOL11, MdBBX20, MdBBX21, MdBBX22, MdCOL4, and MdBBX37 in apple [14,15,16,17,18,19,20]; PpBBX16, PpBBX18, PpBBX21, PpBBX24 and PyBBX24 in pear [21,22,23,24]; PavBBX6 and PavBBX9 in sweet cherry [25]; FaBBX22 and FaBBX24 in strawberry [26,27]; RcBBX26 in Rubus chingii [28]; VvBBX44 in grape [29]; PtrBBX23 in poplar [30]; SlBBX20 in tomato [31]; MaBBX20 and MaBBX51 in grape hyacinth [32]; OsBBX14 in rice [33]; FtBBX22 in Tartary buckwheat [34]; and LvBBX24 in lily [35]. Among these genes, MdBBX22 [36], FaBBX24 [27], and PtrBBX23 [30] are also involved in the regulation of proanthocyanidin content.
With the progress in plant whole-genome sequencing, a varying number of BBX genes have been identified across diverse plant species. Specifically, 32 BBX members were identified in Arabidopsis [2]; 17 in sugar beet [37]; 18 in Chinese chestnut [38]; 19 in pineapple [39]; 22 in woodland strawberry [40]; 23 in pepper [41] and blueberry [42]; 24 in grapevine [43]; 25 in Chinese white pear [44]; 26 in cucumber [45]; 27 in Moso bamboo [46], A. annua [9], and Salvia miltiorrhiza [47]; 28 in wolfberry [5] and Tartary buckwheat [48]; 30 in rice [49]; 31 in tomato [50]; 32 in R. chingii [28]; 34 in sweet potato wild ancestor Ipomoea trifida [51]; 36 in maize [52]; 49 in tobacco [53]; 51 in cultivated strawberry [54]; 57 in soybean [55]; 64 in apple [56]; and 77 in cultivated peanut [57]. However, details regarding the BBX genes in pomegranate remain limited.
Pomegranate (Punica granatum L.), a perennial deciduous fruit tree native to Central Asia, is experiencing significant growth in the global market, with estimates projecting an increase from USD 235.94 million in 2021 to USD 338.6 million by 2030 [58]. Pomegranates are rich in phenolic compounds, which exhibit a range of health-beneficial properties, including antioxidant, antimicrobial, antidiabetic, and anticancer activities [58]. Anthocyanins, a key category of phenolic compounds, accumulate in the seed coat and pericarp, contributing to the diverse colors of pomegranate fruits, which range from white to purple-red [59]. The core structural genes encoding enzymes involved in anthocyanins biosynthesis and transport have been well characterized in pomegranate, including chalcone synthase (CHS), flavonoid 3’-hydroxylase (F3’H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), UDP-glucose flavonoid-3-O-glucosyl transferase (UFGT), and glutathione S-transferase (GST) [59,60,61]. Functional studies have also been conducted on several well-known anthocyanin-related TFs. For instance, PgMYB1, similar to MdMYB1 and CmMYB6, directly bound to the promoter of PgGSTF6, thereby activating its expression and promoting anthocyanin accumulation [61]. PgMYB5-like interacted with PgbHLH to activate PgF3H, PgF3’H, and PgF3’5’H, leading to the accumulation of dihydroflavonols, which is not directly correlated with anthocyanin production [62]. PgWD40, which encoded a WD40-repeat protein, interacted with grape VlMYB-A1 and is responsible for producing over-pigmented cotyledons with a purple color in the Arabidopsis ttg1 mutant [63]. Additionally, the overexpression of PgbZIP16, a member of the basic leucine zipper (bZIP) TF, significantly upregulated the expression of UF3GT, ANS, and DFR in Arabidopsis, thereby increasing anthocyanin accumulation [64]. However, the specific contributions of BBX genes to the anthocyanin metabolism remain to be elucidated in pomegranate.
In this study, we conducted a genome-wide identification of 23 BBX genes in pomegranate. We investigated their evolutionary relationships, gene structure, conserved motifs, synteny relationships, promoter elements, and GO annotation. Expression patterns and sequence alignment reveal that PgBBX5, a homolog of anthocyanin-related BBX TFs, may play a role in anthocyanin accumulation during fruit ripening. Our findings enhance the understanding of the potential roles of the PgBBX gene family members, particularly in the fruit ripening in pomegranate.
2. Materials and Methods
2.1. Plant Materials
Six-year-old ‘Tunisia’ pomegranate trees were cultivated in the greenhouse at the horticultural experimental station of Huaibei Normal University, Huaibei, China. For this study, healthy and uniform fruits were randomly selected at 15, 30, 60, 90, and 135 days after pollination (DAPs), with three replicates of 15 fruits at each stage. The fruit pericarps were carefully peeled using stainless steel knives from the outermost layer to ensure consistent tissue collection. This standardized method was applied uniformly across all sampling time points. The collected pericarp samples were immediately frozen in liquid nitrogen and stored at −80 °C until further analysis of anthocyanin content and gene expression.
2.2. BBX Family Gene Identification in Pomegranate
Genome sequences and annotations for the ‘Tunisia’ pomegranate (Punica granatum L., assembly ASM765513v2) were sourced from NCBI Genome datasets (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_007655135.1/, accessed on 5 May 2025) [65]. Genetic information for Chinese white pear (Pyrus bretschneideri) was obtained from the PearMODB (https://pearomics.njau.edu.cn/, accessed on 5 May 2025) [66]. A total of 32 BBX gene family members from Arabidopsis thaliana were collected from TAIR (https://www.arabidopsis.org, accessed on 5 May 2025) and employed as queries for a local BLASTP v 2.14.0 search against the pomegranate protein database to identify putative PgBBX genes (E-value ≤ 1 × 10−5) at the genome-wide level. Hidden Markov model (HMM) profiles specific to the B-box-type zinc finger domain (PF00643) were retrieved from the InterPro database (https://www.ebi.ac.uk/interpro/entry/pfam, accessed on 20 February 2025) and applied to search for PgBBX candidates using HMMER v 3.1 (E-value ≤ 1 × 10−5). After removing redundant sequences, the identified candidates were further validated using the SMART tool (http://smart.embl-heidelberg.de/, accessed on 20 February 2025) to confirm the presence of conserved B-BOX domains. The number of amino acids, molecular weight, and theoretical isoelectric points (pI) of the PgBBX genes were evaluated using the ExPasy ProtParam tool (http://web.expasy.org/protparam/, accessed on 20 February 2025). The subcellular localization of the PgBBX protein was predicted using the BUSCA web server (http://busca.biocomp.unibo.it/, accessed on 20 February 2025).
2.3. Phylogenetic Analysis and Group Classification
A total of 32 AtBBX, 25 PbBBX, and 23 PgBBX proteins were aligned using Clustal Omega (https://www.ebi.ac.uk/jdispatcher/msa/clustalo, accessed on 20 February 2025), and a phylogenetic tree was constructed using MEGA X software with the neighbor-joining method and 1000 bootstrap replicates. The 80 BBX proteins were classified into five groups based on the presence of B-BOX and CCT domains, and their evolutionary relationships with Arabidopsis BBX members.
2.4. Gene Structure and Conserved Motif Analysis
The gene structure information for 80 BBX genes from Arabidopsis, Chinese white pear, and pomegranate was extracted individually from their respective genomic annotation files. The MEME Suite v 5.5.7 (http://meme-suite.org/tools/meme, accessed on 20 February 2025) was used to identify the composition and distribution of conserved motifs within the 80 BBX proteins. The parameters for the motif search were configured as follows: allowing any number of repetitions, a maximum of ten motifs, and setting the motif length to range from 6 to 200 amino acids.
2.5. Chromosomal Mapping and Collinearity Analysis of BBX Genes
The chromosomal positions of the PgBBX genes were plotted according to the pomegranate genomic annotation file. To investigate collinear relationships, we analyzed the PgBBX genes in the pomegranate genome and conducted comparisons with BBX genes from Arabidopsis and Chinese white pear using the One-step MCScanX—Super Fast tool in TBtools-II v 2.119 with default parameters [67]. The collinear results were visualized using the Advanced Circos and Dual Systeny Plot tools in TBtools-II. Genes from different groups were highlighted with corresponding lines to facilitate the identification of collinear gene pairs.
2.6. Cis-Acting Elements Analysis of PgBBX Genes
The promoter sequences of all PgBBX genes were extracted from genomic DNA sequences, specifically the 1500 nucleotides (nt) upstream of the transcriptional initiation codon (ATG), using TBtools-II. The putative cis-acting elements with these promoters were identified using the PlantCARE program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 20 February 2025). After excluding common, unnamed, and redundant elements, the remaining elements were classified into five main types based on their annotation. The results were visualized as a heatmap using TBtools-II [67].
2.7. Gene Ontology (GO) Annotation and Enrichment Analysis of PgBBX Proteins
GO annotation of pomegranate proteins was conducted using eggNOG-mapper v 2.1.12 (http://eggnog-mapper.embl.de/, accessed on 20 February 2025), an online tool for assigning orthologs and functional predictions based on the eggNOG database [68]. GO terms enrichment analysis for the PgBBX protein was performed to identify significantly enriched molecular functions, cellular components, and biological processes. The results were visualized using TBtools-II [67].
2.8. Expression Pattern Analysis of PgBBX Genes in Pomegranate
The RNA-seq dataset, which includes the root, leaf, and flower, and three fruit tissues (inner seed coat, outer seed coat, and pericarp) at 50, 95, and 140 days after pollination (DAPs) (PRJNA360679) [69], was used to analyze the expression patterns of PgBBX genes. Additionally, the transcriptome dataset of inner and outer seed coats in ‘Dabenzi’ and ‘Tunisia’ cultivars at 50, 95, and 140 DAPs (PRJNA548841) was used to confirm the expression characteristics of PgBBX genes during fruit ripening [70]. The raw data were downloaded from the NCBI SRA database (https://www.ncbi.nlm.nih.gov/sra, accessed on 20 February 2025) and processed according to previously reported methods [71]. The expression heatmap was visualized using TBtools-II [67]. Correlation analysis of gene expression levels, based on TPM values, between PgBBX5 and PgCHS, PgF3’H, PgDFR, PgANS, PgUFGT, and PgGSTF6 was performed using the corr.test function from the psych package in RStudio v 4.2.1.
2.9. Protein Sequence Alignment and Structure Analysis of PgBBX5 with Its Homologs
A phylogenetic tree, including PgBBX group IV members and PgBBX5 with its homologs, was generated using the neighbor-joining (NJ) method in MEGA X software. Multiple protein sequence alignment and sequence similarity analysis of PgBBX5 with its homologs were conducted using Clustal Omega (https://www.ebi.ac.uk/jdispatcher/msa/clustalo, accessed on 20 February 2025). The protein secondary structures were predicted using the NPS@ web server, which detected four conformational states: alpha helix, extended strand, beta turn, and random coil (https://npsa.lyon.inserm.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html, accessed on 20 February 2025). The tertiary structural models were built using the SWISS-MODEL protein structure homology-modeling server (https://swissmodel.expasy.org/, accessed on 20 February 2025).
2.10. Anthocyanin Content Assay
After the pomegranate fruits were photographed for pericarp color comparison, anthocyanins were extracted using a 1% (v/v) HCl–methanol solution, and the anthocyanin content was quantified according to the previous report [72].
2.11. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR) Analysis
Total RNA was extracted from triplicate biological samples using a plant RNA extraction kit (V1.5, Biofit, Chengdu, China). Genomic DNA contamination was eliminated, and complementary DNA (cDNA) was synthesized using a one-step gDNA removal and cDNA synthesis supermix kit (Transgen Biotech, Beijing, China). The RT-qPCR reactions were prepared with ChamQ universal SYBR qPCR master mix (Vazyme, Nanjing, China) and analyzed using an ABI-QuantStudio6 Flex real-time PCR system. Gene expression was normalized using the PgActin gene as an internal reference. Relative gene expression levels were calculated using the 2−ΔΔCt method [73]. The primers used for RT-qPCR are listed in Table S1.
2.12. Subcellular Localization
For the subcellular localization assay, the CDS of PgBBX5 (without the stop codon) was cloned into the pCAMBIA1302-35S:GFP vector to create the 35S:PgBBX5-GFP vector. The empty vector (35S:GFP) was used as the control. Both the recombination and empty vectors were transformed into Agrobacterium tumefaciens strain GV3101 (pSoup-p19). These were then introduced into 4-week-old tobacco (Nicotiana benthamiana) leaves, which also contained an mCherry-tagged nuclear protein as a marker. After 48-72 h, the GFP and mCherry signals were observed using a confocal laser scanning microscope (LSM-880, Zeiss, Germany).
2.13. Statistical Analysis
Data came from at least three biological replications and were analyzed using SPSS software (version 22.0, IBMCorp., Armonk, N.Y., USA) for statistical analysis. Statistically significant differences were assessed using one-way ANOVA, followed by a post hoc Tukey test at a significance level of p < 0.05.
3. Results
3.1. BBX Gene Family Members Identification and Phylogenetic Relationship Analysis in Pomegranate
We identified a total of 23 PgBBX genes in the pomegranate genome by utilizing BLAST and HMM analyses, followed by conserved domain validation and redundancy elimination (Table 1). These genes were named as PgBBX1 to PgBBX23, corresponding to their chromosomal locations. Each PgBBX member contained at least one B-BOX domain, which comprises approximately 40 residues with the consensus sequence C-X2-C-X8-C-X2-D-X4-C-X2-C-D-X3-H-X8-H. Several members also possessed an additional conserved CCT domain with the sequence R-X5-R-Y-X-E-K-X3-R-X3-K-X2-R-Y-X2-R-K-X2-A-X2-R-X-R-X-K-G-R-F-X-K (Figure 1A). Based on the distribution of conserved domains and phylogenetic relationships with BBX genes from Arabidopsis and Chinese white pear, the 23 PgBBX genes were categorized into five groups (Figure 1B). Group I and II members each possessed three conserved domains: B-BOX1, B-BOX2, and CCT domains. Members of group III typically comprised B-BOX1 and CCT domains. Group IV members featured two B-BOX domains but lacked the CCT domain. Group V members exclusively contained the B-BOX1 domain. Specifically, group IV had the highest number of members, with eight PgBBX genes, followed by groups I and II with five genes each, group V with three genes, and group III with two genes. We also compared the identified PgBBX genes with those annotated in the pomegranate reference genome and found that most members of groups I to III were annotated as the zinc finger protein CONSTANS-LIKE, which corresponds to their conserved domains distribution. Members of groups IV to V were annotated as B-box zinc finger proteins, which lacked the CCT domain near the C-terminus (Table 1). Notably, PgBBX6 and PgBBX22, though not annotated as BBX members in NCBI, contain the conserved BBX domain near the N-terminus. Therefore, some PgBBX genes were newly identified in this study and were not previously annotated in the reference genome.

Table 1.
Molecular characteristics, prediction of subcellular localization, and their NCBI annotations of PgBBX gene family members.
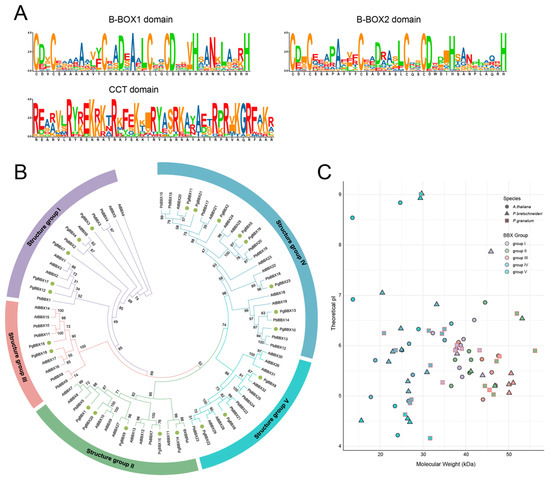
Figure 1.
Phylogenetic analysis and physicochemical properties of BBX proteins in pomegranate, Arabidopsis, and Chinese white pear. (A) Conserved amino acid sequence of B-box and CCT domain in pomegranate BBX proteins. (B) Phylogenetic relationship of BBX members of pomegranate (PgBBX), Arabidopsis (AtBBX), and Chinese white pear (PbBBX). (C) Distribution patterns of theoretical pI and molecular weight values for PgBBX, AtBBX, and PbBBX members.
Consistent with the physicochemical properties of AtBBX and PbBBX members in the corresponding groups, PgBBX genes in group II and III exhibited higher average molecular weights (MW) of 48.51 KDa, which were 1.26-, 1.68-, and 1.67-fold greater than those of group I (38.48 KDa), IV (28.94 KDa), and V (29.03 KDa) members, respectively (Table 1; Figure 1C). The majority of BBX members from Arabidopsis, Chinese white pear, and pomegranate were acidic proteins, with pI values typically below 7. Notably, group III members displayed lower theoretical pI values, averaging 5.36, which contrasted with other groups. Group V members exhibited more variability in theoretical pI values (Table 1; Figure 1C). Subcellular localization predictions showed that, with the exception of PgBBX10 and 13, most PgBBX members were predicted to be located in the nucleus (Table 1).
3.2. Phylogenetic Classification, Gene Structure, and Conserved Motif Composition of BBX Genes in Pomegranate, Arabidopsis, and Chinese White Pear
The evolutionary conservation can also be observed through the gene structure and the composition and distribution of motif elements. We examined the gene structures of a total of 80 BBX members from Arabidopsis, Chinese white pear, and pomegranate, revealing distinct patterns among the groups (Figure 2B). Typically, genes in group I (86.7%) and III (90%) contained two CDS. In group II, 13 genes (76.5%) possessed four to five CDS. The number of CDS varied in group IV, with 17 genes (68%) consisting of two to three CDS, and approximately 28% of members featuring five to seven CDS. In group V, members exhibited a relatively minimal number of CDS; specifically, eight genes (57.1%) contained a single CDS, while the remaining six members had two CDS.
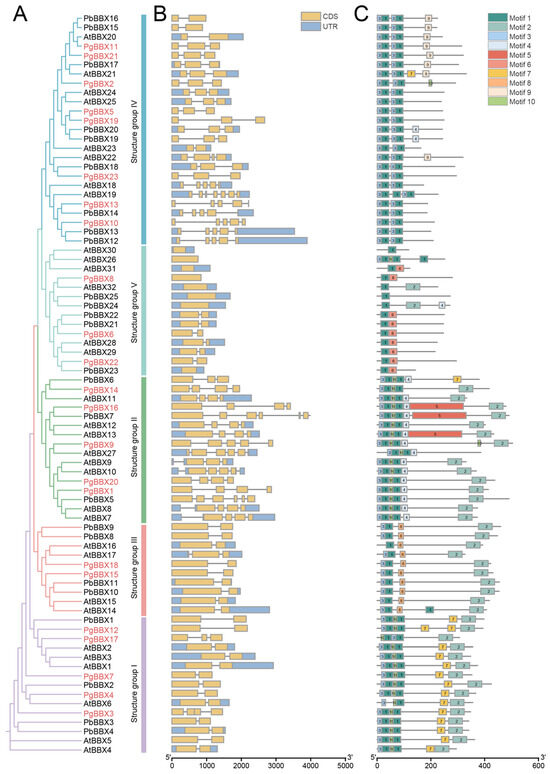
Figure 2.
Gene structure and conserved motif distribution among BBX members from pomegranate, Arabidopsis, and Chinese white pear. (A) Members were classified into five groups based on their phylogenetic relationship, with PgBBX members indicated by red highlighting. (B) Gene structures of the 80 BBX genes across the three species. (C) Composition and distribution of conserved motifs among BBX members. A total of ten distinct motifs, each with a different color, were identified using the MEME web server.
The composition and arrangement of motif elements correlated with the distribution of conserved domains (Figure 2C). Motif 1, exhibiting high sequence similarity with the B-BOX domain, was found extensively in the N-terminus of the 80 BBX members (Figure 1A). Notably, members of group I, II, and IV, which were characterized by the presence of two B-BOX domains, also consistently contained two instances of motif 1. Motif 2, associated with the CCT domain, was typically located in the C-terminus of members from groups I, II, and III. Additionally, certain motif elements were observed in specific groups, such as motif 4 in group II, motif 6 in group V, motif 7 in group I, and motif 8 in group III, which may contribute to the functional divergence among these groups.
3.3. Collinearity Relationship of BBX Genes
With the exception of the chromosome NC_045134, PgBBX genes were unevenly distributed across the other seven chromosomes (Figure 3A). Both the segmental and tandem duplications of the PgBBX gene family were analyzed. However, no tandem duplications were observed. Eight segmentally duplicated PgBBX gene pairs were identified within the pomegranate genome, including one pair from group I, one from group III, five from group IV, and one from group V. Chromosome NC_045133 contained the maximal number of PgBBX members and exhibited six collinearity relationships with other PgBBX genes (Figure 3A).
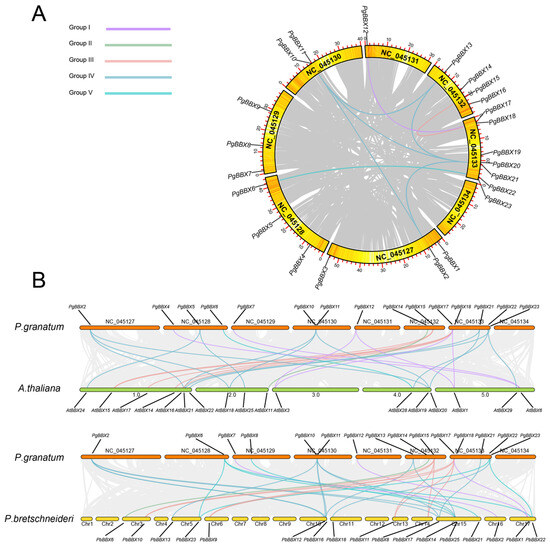
Figure 3.
Syntenic relationships of BBX genes. (A) Chromosomal localization and syntenic relationships of PgBBX genes in pomegranate genome. (B) Comparative synteny analysis of PgBBX genes with BBX members from Arabidopsis and Chinese white pear. The gene pairs, represented by colored lines, correspond to their group classification.
Moreover, we explored the synteny relationships between BBX genes in pomegranate and those in Arabidopsis and Chinese white pear (Figure 3B). A total of 18 PgBBX genes showed synteny, forming 27 orthologous gene pairs with AtBBX genes and 31 with PbBBX genes. Notably, thirteen PgBBX genes showed collinearity with both AtBBX and PbBBX genes, and an additional four genes exhibited synteny with either AtBBX or PbBBX genes. Among the different groups, group IV PgBBX genes showed twenty-five gene pairs with AtBBX or PbBBX genes, followed by group III with thirteen gene pairs and group I and V with nine gene pairs. In contrast, group II had the fewest, with only two gene pairs.
3.4. Cis-Acting Elements in the Promoter Regions of PgBBX Genes
Cis-acting elements, located within the promoter regions of genes, serve as switches that interact with TFs to initiate or suppress transcription, thereby determining the characteristics of gene expression. A 1500 nt sequence upstream of each PgBBX gene was extracted for identification of cis-acting elements (Figure 4). In total, 723 elements across 41 categories were identified and classified into five main types: light-responsive elements (LREs), phytohormone-responsive elements (PHREs), stress-responsive elements (SREs), development-related elements (DREs), and transcription factor binding elements (TFBEs) (Figure 4A). Notably, LREs were the most prevalent, with 234 elements across 15 categories. Each PgBBX gene promoter contained at least five LREs. Abundant PHREs were present in the promoter region of all PgBBX genes, ranging from 3 to 18, potentially responsible for the induction by various phytohormones, including methyl jasmonate (AAGAA-motif, CGTCA-motif, and TGACG-motif), abscisic acid (ABRE), auxin (AuxRR-core and TGA-element), ethylene (ERE), gibberellin (GARE-motif, P-box, and TATC-box), zein (O2-site), and salicylic acid (TCA-element). Moreover, the promoter regions of PgBBX genes contained 4 to 14 SREs and were responsive to various stresses such as anaerobic induction (ARE and GC-motif), oxidative stress (as-1), dehydration (DRE core), low temperature (LTR), drought inducibility (MBS), defense and stress (TC-rich repeats), and wounding (WRE3 and WUN-motif). In contrast, only 2% of the elements were related to development (Figure 4B). Additionally, 12% of elements were TFBEs, including 68 MYB and 16 W-box binding elements (Figure 4A,B). The proportion of the five types of cis-acting elements was similar across different groups (Figure 4C), suggesting that PgBBX genes may share redundant expression-regulated pathways.
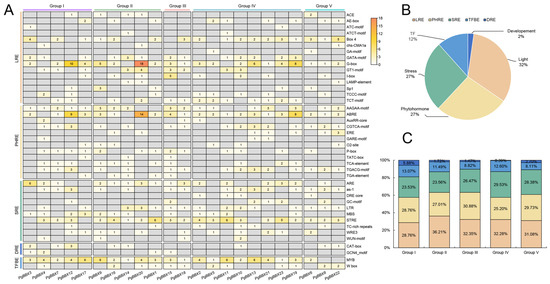
Figure 4.
Analysis of cis-acting elements of PgBBX genes. (A) Abundance of five types of cis-acting elements in the promoter of PgBBX genes across different groups, including light-responsive elements (LREs), phytohormone-responsive elements (PHREs), stress-responsive elements (SREs), development-related elements (DREs), and transcription factor binding elements (TFBEs). (B) Proportion of the five types of cis-acting elements within the promoters of PgBBX genes. (C) Distribution of the five types of cis-acting elements among different groups of PgBBX gene promoters.
3.5. GO Term Enrichment of PgBBX Proteins
To clarify the potential functions of PgBBX proteins, we performed GO annotation and enrichment analysis (Figure 5). The enriched GO terms associated with PgBBX were categorized into molecular function, cellular component, and biological process. All PgBBX proteins were characterized as zinc finger TFs, with several GO terms of molecular function, such as double-stranded DNA binding (GO:0003690), transcription regulatory region nucleic acid binding (GO:0001067), transcription regulator activity (GO:0140110), zinc ion binding (GO:0008270), and other functions. Importantly, the cellular component GO term for the nucleus (GO:0005634) was exclusively enriched, aligning with their role in DNA binding. Moreover, a range of biological process GO terms responding to light signaling were enriched, suggesting that PgBBX proteins function as TFs in regulating multiple aspects of plant growth and development, including photomorphogenesis, shade avoidance, and photoprotection.
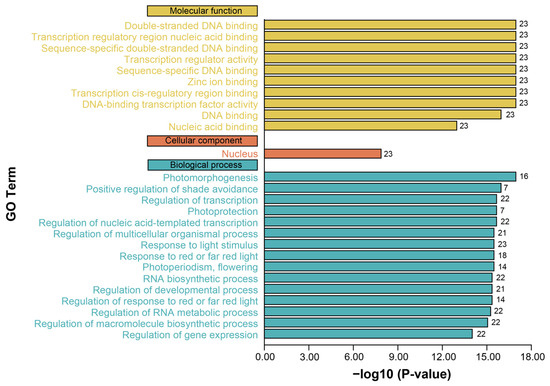
Figure 5.
GO function annotation of PgBBX proteins. The number on the right of the column indicates the candidate PgBBX members associated with each corresponding GO term.
3.6. Expression Characteristic of PgBBX Genes in Diverse Organs and Fruit Tissues
The expression patterns of PgBBX genes were examined in three organs (root, leaf, and flower) and three fruit tissues (inner seed coat, outer seed coat, and pericarp) at 50, 95, and 140 days after pollination (DAPs) [69]. Compared to the expression levels in the root, more PgBBX genes were expressed in the leaf (Figure 6A). Among these genes, a subset was also highly expressed in the flower, including PgBBX3, PgBBX7, PgBBX14, PgBBX1, PgBBX15, and PgBBX5. Compared to the other organs and tissues, PgBBX20 was primarily expressed in the flower. During fruit ripening, the expression patterns of most PgBBX genes did not correlate with the ripening process of the three fruit tissues. However, PgBBX5 showed a continuous increase in expression levels throughout fruit ripening. At 95 DAPs, the expression of PgBBX5 was upregulated to 2.03-, 2.61-, and 1.75-fold in the inner seed coat, outer seed coat, and pericarp, respectively, compared to 50 DAPs. At 140 DAPs, the expression of PgBBX5 further increased to 4.21-, 3.87-, and 2.14-fold in the three fruit tissues, respectively, compared to 50 DAPs. Moreover, similar expression characteristics of PgBBX5 were observed in the inner and outer seed coat across two pomegranate cultivars at 50, 95, and 140 DAPs (Figure 6B) [70]. Furthermore, the expression of PgBBX5 was highly consistent with the dynamic changes in key genes in anthocyanin biosynthesis and transport during fruit development, including PgCHS, PgF3’H, PgDFR, PgANS, PgUFGT, and PgGSTF6 (Figure 6C). These results suggest that PgBBX5 may play a positive role in the regulation of anthocyanin accumulation, thereby affecting color alteration during fruit development.
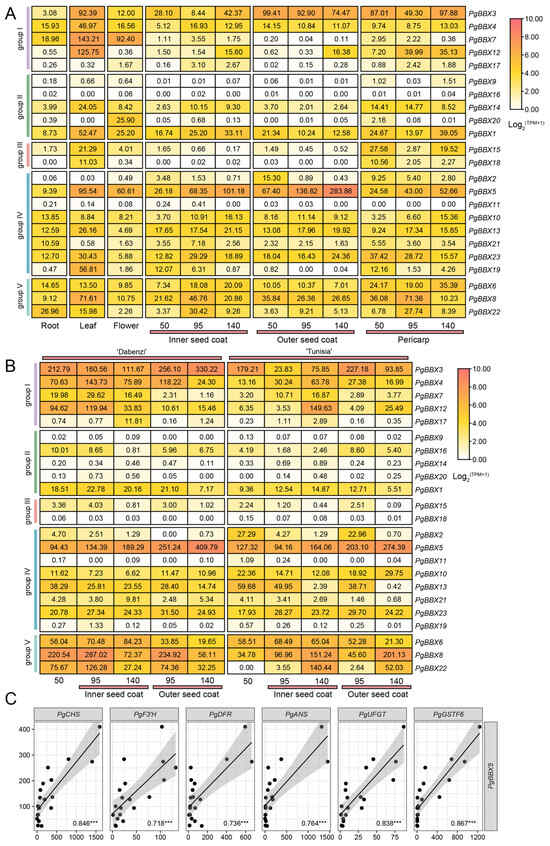
Figure 6.
Expression profiles of PgBBX genes in different organs and their dynamic changes during fruit ripening. (A) Expression levels of PgBBX genes in root, leaf, flower, and various fruit tissues at 50, 95, and 140 DAPs. (B) Comparative expression of PgBBX genes in inner and outer seed coats of ‘Dabenzi’ and ‘Tunisia’ cultivars at 50, 95, and 140 DAPs. (C) Correlation between PgBBX5 and key genes in anthocyanin biosynthesis and transport (PgCHS, PgF3’H, PgDFR, PgANS, PgUFGT, PgGSTF6) based on TPM values during fruit ripening. *** indicates p < 0.001.
3.7. PgBBX5 May Play a Role in Anthocyanin Accumulation During Fruit Ripening
To clarify the function of PgBBX5, the sequence characteristics of PgBBX5 were examined in comparison with those of its homologs, including AtBBX24 [13], PhBBX24 [74], FaBBX24 [27], MdCOL4 [17], and PyBBX24 [24], which are involved in the regulation of anthocyanin content. Compared to other BBX members of group IV, PgBBX5 showed closer phylogenetic relationships with the anthocyanin-related BBX homologs (Figure 7A). Multiple protein sequence alignment revealed that PgBBX5 shared similar protein sequences with the anthocyanin-related BBX homologs (Figure 7B), which exhibited high sequence similarities, with an average identity of 70.1% with its BBX homologs (Figure 7C). Although these members presented more than 70% random coil on average, the N-terminus was conserved in the predicted secondary and tertiary structures (Figure 7D,E), which was consistent with the conserved domain distribution (Figure 7B).
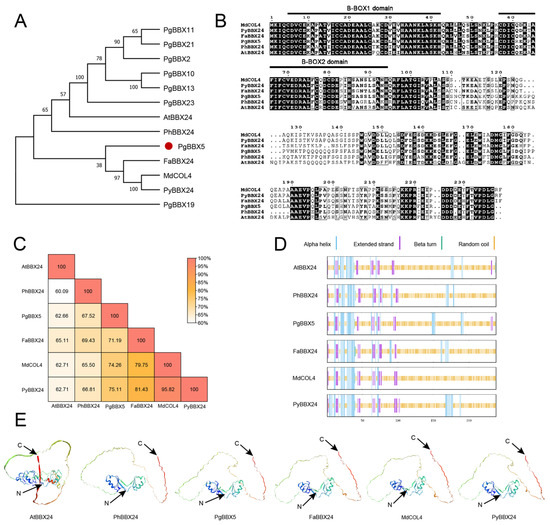
Figure 7.
Comparative analysis of sequence similarity among PgBBX5 homologs. (A) Phylogenetic analysis of PgBBX5 homologs, including AtBBX24 (AT1G06040), PhBBX24 (Peinf101Scf01164g02015), FaBBX24 (maker-Fvb4-4-snap-gene-165.32-mRNA-1), MdCOL4 (HM122534), PyBBX24 (EVM0042308), and group IV PgBBX proteins. (B) Multiple sequence alignment and conserved domain identification for PgBBX5 homologs. (C) The sequence similarity among PgBBX5 homologs. The predicted secondary (D) and tertiary (E) structures of PgBBX5 homologs. The C and N in the tertiary structures denote the C-terminal and N-terminal of the protein, respectively.
To evaluate the regulatory role of the PgBBX5 gene in anthocyanin metabolism, we monitored the dynamic changes in anthocyanin content and the expression patterns of PgBBX5, along with several key structural genes involved in anthocyanin accumulation during the development of ‘Tunisia’ fruits. The pericarp color transitioned from green to deep red, coincident with increased anthocyanin accumulation (Figure 8A). Specifically, at 135 DAPs, the anthocyanin content in the pericarp was 12.3 times higher than that at 15 DAPs (Figure 8B). The RT-qPCR results revealed a significant upregulation of key structural genes involved in anthocyanin biosynthesis (PgCHS, PgF3’H, PgDFR, PgANS, and PgUFGT) and transport (PgGSTF6), which primarily contributed to the observed increase in anthocyanin content (Figure 8C). PgBBX5 expression also increased during fruit ripening (Figure 8C), positively correlating with the changes in anthocyanin content and the relative expression of key structural genes in anthocyanin biosynthesis and transport (Figure 8D). These findings suggest that PgBBX5, a member of the BBX family, potentially plays a regulatory role in anthocyanin accumulation during pomegranate fruit ripening.
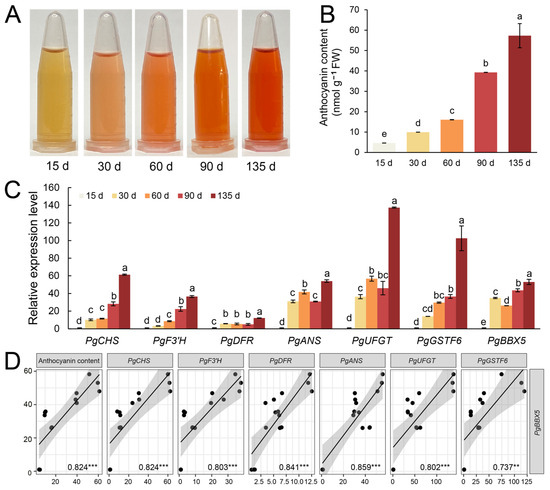
Figure 8.
PgBBX5 plays a potential role in anthocyanin accumulation during ‘Tunisia’ fruit ripening. Anthocyanin extraction of the fruit pericarp samples collected at 15, 30, 60, 90, and 135 DAPs (A) and the content (B). Relative expression (C) and correlation (D) between PgBBX5 and key anthocyanin biosynthetic and transport genes (PgCHS, PgF3’H, PgDFR, PgANS, PgUFGT, PgGSTF6) throughout fruit ripening. Different letters in (B,C) indicate statistically significant differences between the samples (one-way ANOVA followed by a post hoc Tukey test, p < 0.05). The asterisk in (D) indicates p < 0.05, ** indicates p < 0.01, and *** indicates p < 0.001.
3.8. Subcellular Localization of PgBBX5
The subcellular localization of PgBBX5 was predicted to be nuclear (Table 1). To confirm this prediction, 35S:PgBBX5-GFP was constructed and transiently expressed in tobacco leaves. The 35S:GFP of the empty vector served as a negative control. Fluorescence was determined using a confocal laser scanning microscope, and the PgBBX5-GFP fusion protein was found exclusively in the nucleus, corresponding with the localization of the nuclear marker protein (mCherry) (Figure 9). In contrast, the 35S:GFP control was distributed in the nucleus, cytosol, and plasma membrane of the tobacco leaf (Figure 9). These results indicate that PgBBX5 is a nuclear-localized protein.
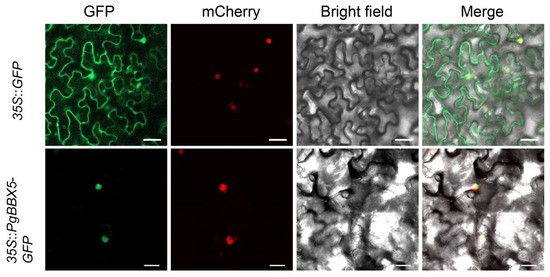
Figure 9.
The subcellular localization of PgBBX5 in tobacco leaves. GFP: green fluorescent protein. mCherry: the nuclear marker. Bar = 20 μm.
4. Discussion
BBX genes are crucial for plant growth and secondary metabolites. Pomegranates, rich in phenolic compounds with anthocyanins in the seed coat and pericarp, causing their color variation, have known core structural genes and some TFs involved in anthocyanin biosynthesis, but the role of BBX genes remains unclear [59,60,61,62,63,64]. This study explores the potential contributions of BBX genes to pomegranate anthocyanin metabolism. We identified a total of 23 BBX genes at the genome-wide scale in pomegranate (Table 1). This number is similar to that found in pepper [41] and blueberry [42], and exceeds the numbers found in sugar beet [37], Chinese chestnut [38], pineapple [39], and woodland strawberry [40]; however, it is lower than in Arabidopsis [2], Chinese white pear [44], and other species, suggesting that the BBX gene family exhibits a diverse number of genes across species. Based on the presence of B-Box domains and the CCT domain, BBX genes can be distinctly classified into five groups across various plants [75]. Accordingly, the BBX genes from pomegranate, Arabidopsis, and Chinese white pear were also categorized into these five groups (Figure 1B). Members within the same group shared similar characteristics, whereas distinct differences were observed across groups, including MW and pI values (Figure 1C), gene structure, and conserved motifs composition (Figure 2), suggesting that the members of the same group have been relatively intact across species during evolution. These findings indicate that BBX genes were present before the divergence of Arabidopsis, Chinese white pear, and pomegranate. Consequently, our results support the hypothesis that the different structural groups of BBX genes originated at an early stage of green plant evolution [75].
Gene duplication is a pivotal force driving the expansion, diversification, and functional innovation within gene families. Segmental and tandem duplications are recognized as the predominant patterns of gene duplication [76]. Specifically, in the BBX gene family, segmental duplications appear to be the predominant mechanism for driving the expansion of the gene family. In rice, segmental duplications were found in 18 OsBBX genes, whereas no tandem duplications were identified [49]. Similarly, 13 segmental duplication gene pairs but no tandem duplications were observed in the pear genome [44]. Universal segmental duplication events were also found in the BBX gene family in Chinese chestnut [38], pineapple [39], pepper [41], grapevine [43], cucumber [45], Moso bamboo [46], Tartary buckwheat [48], tomato [50], I. trifida [51], maize [52], soybean [55], and cultivated peanut [57]. Consequently, in the present study, collinearity analysis showed that eight PgBBX gene pairs formed by segmental duplication types (Figure 3A), which contributed to the expansion of PgBBX gene family members rather than tandem duplication in pomegranate. Moreover, abundant homologous gene pairs were identified between pomegranate and both Arabidopsis and Chinese white pear, suggesting a common ancestry for BBX genes among these species (Figure 3B) [44]. Corresponding to their closer phylogenetic relationship, similar gene structure, and motif distributions (Figure 1 and Figure 2), PgBBX5 exhibited collinearity with AtBBX24 (Figure 3B), implying that it may also be involved in anthocyanin biosynthesis regulation [13].
Recent research has highlighted the critical functions of specific BBX proteins in mediating light-responsive plant growth and developmental processes, such as seed germination, photomorphogenesis, shade avoidance, and secondary metabolite pathways [77]. Accordingly, numerous light-responsive elements have been identified in the promoter regions of BBX genes across various plant species, such as rice [49], pepper [41], R. chingii [28], grapevine [43], cucumber [45], Moso bamboo [46], tomato [50], cultivated peanut [57], and pomegranate (Figure 4). Moreover, several light-controlled GO terms were significantly enriched in biological processes, including photomorphogenesis (GO:0009640), positive regulation of shade avoidance (GO:1902448), photoprotection (GO:0010117), and response to light stimulus (GO:0009416) (Figure 5). For instance, light is essential for anthocyanin biosynthesis in fruit peels. Several BBX genes were highly induced by light irradiation and formed protein complexes with ELONGATED HYPOCOTYL 5 (HY5) to regulate the expression of anthocyanin structural genes and transcription factors in a light-dependent manner, such as MdBBX22 in apple [19], PpBBX16 and PpBBX18 in red pear [21,22], and CsBBX22 in citrus [78]. Correspondingly, at least eight LREs were identified in the promoter region of PgBBX5, a key candidate TF responsible for anthocyanin accumulation in pomegranate, suggesting that PgBBX5 may also respond to light induction during pomegranate fruit ripening (Figure 4). Furthermore, various PHREs and SREs were found across species, including in pomegranate [28,41,43,45,46,49,50,57] (Figure 4). These findings indicate that PgBBX genes may have diverse functions in response to light, multiple phytohormones, and stress.
During evolution, homologous genes with close phylogenetic relationships typically maintain conserved functions across various species. For instance, R2R3-MYB genes within subgroup 4 (SG4), SG5, SG6, and SG7 serve as upstream regulatory factors in flavonoid synthesis [79]. SG6 members primarily activate anthocyanin biosynthesis, while SG4 members function as repressors. SG5 genes contribute to proanthocyanidin (PA) biosynthesis, and SG7 genes generally promote the flavonol pathway [79]. Similarly, homologs of AtBBX21 and AtBBX22 have been identified as activators in the anthocyanin pathway [80], such as MdCOL11 [15], MdBBX22 [19], PpBBX16, PpBBX18 [21,22], PavBBX6, PavBBX9 [25], FaBBX22 [26], FtBBX22 [34], CsBBX22 [78], and OsBBX14 [33]. However, in our study, the expression patterns of PgBBX17 and PgBBX23, which are homologs of AtBBX21 and AtBBX22, respectively, were analyzed using two transcriptome datasets (Figure 1 and Figure 6). The results indicated that these two genes were not upregulated in different pomegranate fruit tissues during ripening (Figure 1 and Figure 6). This suggests that PgBBX17 and PgBBX23 may not play a significant role in anthocyanin accumulation in pomegranate, unlike the characterized BBX22 homologs in apple [19], pear [21,22], sweet cherry [25], strawberry [26], Tartary buckwheat [34], citrus [78], and rice [33]. Moreover, our study has revealed that PgBBX5 exhibited increased expression during fruit ripening and displayed a significant positive correlation with the dynamic changes in anthocyanin content and the expression of anthocyanin biosynthetic and transport genes (Figure 6 and Figure 8). This suggests that PgBBX5 may act as a positive regulator of anthocyanin accumulation in pomegranate. To better understand the role of PgBBX5, we compared it with other homologs of AtBBX24. AtBBX24 and its homologs have been reported to have diverse functions in anthocyanin biosynthesis [81]. For instance, in pears, PpBBX21 restricted the formation of the PpBBX18-PpHY5 complex and repressed the expression of MYB and other anthocyanin-related genes, resulting in decreased anthocyanin content in both Arabidopsis and pear [22]. Similarly, in apple, MdCOL4 interacted with MdHY5 to inhibit the expression of MdMYB1 and directly bound to the promoters of MdANS and MdUFGT, suppressing their expression and thereby inhibiting anthocyanin biosynthesis [17]. Grape hyacinth MaBBX51 also negatively regulated anthocyanin biosynthesis by physically interacting with MaBBX20, MaBBX20-MaHY5 complex, or MaMybA to inhibit the transcription of anthocyanin-related genes [32]. However, recent studies demonstrated that some homologs of AtBBX24 play positive roles in the anthocyanin pathway. In petunia flower petals, PhBBX24 (Peinf101Scf01164g02015) was markedly increased in response to white light and could interact with PhHY5 to upregulate anthocyanin synthesis [74]. In strawberry fruits, FaBBX24 synergistically regulated anthocyanin accumulation with FaMYB5 [27]. In pear, PyBBX24 ΔN14, with a 14 bp deletion in the third exon, promoted anthocyanin accumulation in pear, strawberry, and tobacco [24]. Additionally, LvBBX24 in lilies positively regulated anthocyanin accumulation in lily petals by binding to the LvMYB5 promoter [35]. PgBBX5, containing two B-BOX domains, was classified into group IV (Figure 1A). It clustered with AtBBX24, PhBBX24, FaBBX24, MdCOL4, and PyBBX24 (Figure 1A,B), shared similar protein sequences with them (Figure 7), and exhibited nuclear localization indicative of its role as a transcriptional factor (Figure 9). In contrast to the negative regulatory roles of some AtBBX24 homologs, PgBBX5 showed a positive correlation with anthocyanin accumulation and related gene expression in our study. This indicated that homologs of AtBBX24 have functionally diverged. Although our findings imply that PgBBX5 may function as a positive regulator of anthocyanin accumulation, further validation of its specific role is needed. This could have potential applications in genetic transformation and molecular breeding for pomegranate fruit color regulation.
5. Conclusions
In this study, we conducted a systematic investigation of the pomegranate BBX gene family, focusing particularly on their roles in anthocyanin accumulation during fruit ripening. The PgBBX genes were classified into five groups, with members within each group being more conserved based on their phylogenetic relationship, physicochemical properties, gene structure, and conserved motif composition. We identified that segmental duplications have driven the expansion of the pomegranate BBX gene family. Light-responsive elements were the most prevalent in the PgBBX promoters. Transcriptome sequencing, RT-qPCR analysis, and multiple sequence alignment of homologs revealed that PgBBX5 potentially serves as a positive regulator in promoting anthocyanin accumulation, although further verification is required.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae11050507/s1, Table S1: The primers used in this study.
Author Contributions
Conceptualization, L.L. and J.Z.; formal analysis, L.L.; data curation, L.L. and J.Z.; writing—original draft preparation, L.L.; writing—review and editing, L.L. and J.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Huaibei Normal University 2024 Annual Surplus Fund Support Project (No. 2024ZK24 and 2024ZK25).
Data Availability Statement
The original contributions presented in the study are included in the article; further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.H. The Arabidopsis B-Box zinc finger family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef]
- Robson, F.; Costa, M.M.R.; Hepworth, S.R.; Vizir, I.; Piñeiro, M.; Reeves, P.H.; Putterill, J.; Coupland, G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001, 28, 619–631. [Google Scholar] [CrossRef]
- Xiong, C.; Luo, D.; Lin, A.; Zhang, C.; Shan, L.; He, P.; Li, B.; Zhang, Q.; Hua, B.; Yuan, Z.; et al. A tomato B-Box protein SlBBX20 modulates carotenoid biosynthesis by directly activating Phytoene Synthase 1, and is targeted for 26S proteasome-mediated degradation. New Phytol. 2018, 221, 279–294. [Google Scholar] [CrossRef]
- Yin, Y.; Shi, H.; Mi, J.; Qin, X.; Zhao, J.; Zhang, D.; Guo, C.; He, X.; An, W.; Cao, Y.; et al. Genome-Wide identification and analysis of the BBX gene family and its role in carotenoid biosynthesis in wolfberry (Lycium barbarum L.). Int. J. Mol. Sci. 2022, 23, 8440. [Google Scholar] [CrossRef]
- Ma, J.; Dai, J.; Liu, X.; Lin, D. The transcription factor CaBBX20 regulates capsanthin accumulation in pepper (Capsicum annuum L.). Sci. Hortic. 2023, 314, 111907. [Google Scholar] [CrossRef]
- Gao, X.R.; Zhang, H.; Li, X.; Bai, Y.W.; Peng, K.; Wang, Z.; Dai, Z.R.; Bian, X.F.; Zhang, Q.; Jia, L.C.; et al. The B-Box transcription factor IbBBX29 regulates leaf development and flavonoid biosynthesis in sweet potato. Plant Physiol. 2022, 191, 496–514. [Google Scholar] [CrossRef]
- Song, Z.; Zhao, F.; Chu, L.; Lin, H.; Xiao, Y.; Fang, Z.; Wang, X.; Dong, J.; Lyu, X.; Yu, D.; et al. The GmSTF1/2-GmBBX4 negative feedback loop acts downstream of blue-light photoreceptors to regulate isoflavonoid biosynthesis in soybean. Plant Commun. 2024, 5, 100730. [Google Scholar] [CrossRef]
- He, W.; Liu, H.; Li, Y.; Wu, Z.; Xie, Y.; Yan, X.; Wang, X.; Miao, Q.; Chen, T.; Rahman, S.; et al. Genome-wide characterization of B-box gene family in Artemisia annua L. and its potential role in the regulation of artemisinin biosynthesis. Ind. Crop Prod. 2023, 199, 116736. [Google Scholar] [CrossRef]
- He, W.; Liu, H.; Wu, Z.; Miao, Q.; Hu, X.; Yan, X.; Wen, H.; Zhang, Y.; Fu, X.; Ren, L.; et al. The AaBBX21–AaHY5 module mediates light-regulated artemisinin biosynthesis in Artemisia annua L. J. Integr. Plant Biol. 2024, 66, 1735–1751. [Google Scholar] [CrossRef]
- Xu, D.; Jiang, Y.; Li, J.; Lin, F.; Holm, M.; Deng, X.W. BBX21, an Arabidopsis B-Box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. USA 2016, 113, 7655–7660. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.S.J.; Maloof, J.N.; Wu, S.H. COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol. 2011, 156, 228–239. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Holm, M.; Botto, J.F. Molecular Interactions of BBX24 and BBX25 with HYH, HY5 HOMOLOG, to modulate Arabidopsis seedling development. Plant Signal. Behav. 2013, 8, e25208. [Google Scholar] [CrossRef]
- Plunkett, B.J.; Henry-Kirk, R.; Friend, A.; Diack, R.; Helbig, S.; Mouhu, K.; Tomes, S.; Dare, A.P.; Espley, R.V.; Putterill, J.; et al. Apple B-box factors regulate light-responsive anthocyanin biosynthesis genes. Sci. Rep. 2019, 9, 17762. [Google Scholar] [CrossRef]
- Bai, S.; Saito, T.; Honda, C.; Hatsuyama, Y.; Ito, A.; Moriguchi, T. An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 2014, 240, 1051–1062. [Google Scholar] [CrossRef]
- Fang, H.; Dong, Y.; Yue, X.; Hu, J.; Jiang, S.; Xu, H.; Wang, Y.; Su, M.; Zhang, J.; Zhang, Z.; et al. The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 2019, 42, 2090–2104. [Google Scholar] [CrossRef]
- Fang, H.; Dong, Y.; Yue, X.; Chen, X.; He, N.; Hu, J.; Jiang, S.; Xu, H.; Wang, Y.; Su, M.; et al. MdCOL4 interaction mediates crosstalk between UV-B and high temperature to control Fruit coloration in apple. Plant Cell Physiol. 2019, 60, 1055–1066. [Google Scholar] [CrossRef]
- Zhang, B.; Zhu, Z.Z.; Qu, D.; Wang, B.C.; Hao, N.N.; Yang, Y.Z.; Yang, H.J.; Zhao, Z.Y. MdBBX21, a B-Box protein, positively regulates light-induced anthocyanin accumulation in apple peel. Front. Plant Sci. 2021, 12, 774446. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Hao, Y.J. MdBBX22 regulates UV-B-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome-mediated degradation. Plant Biotechnol. J. 2019, 17, 2231–2233. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Espley, R.V.; Kui, L.W.; Bi, S.Q.; You, C.X.; Hao, Y.J. An apple B-Box protein MdBBX37 modulates anthocyanin biosynthesis and hypocotyl elongation synergistically with MdMYBs and MdHY5. Plant Cell Physiol. 2019, 61, 130–143. [Google Scholar] [CrossRef]
- Bai, S.; Tao, R.; Tang, Y.; Yin, L.; Ma, Y.; Ni, J.; Yan, X.; Yang, Q.; Wu, Z.; Zeng, Y.; et al. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 2019, 17, 1985–1997. [Google Scholar] [CrossRef]
- Bai, S.; Tao, R.; Yin, L.; Ni, J.; Yang, Q.; Yan, X.; Yang, F.; Guo, X.; Li, H.; Teng, Y. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 2019, 100, 1208–1223. [Google Scholar] [CrossRef]
- Ou, C.; Zhang, X.; Wang, F.; Zhang, L.; Zhang, Y.; Fang, M.; Wang, J.; Wang, J.; Jiang, S.; Zhang, Z. A 14 nucleotide deletion mutation in the coding region of the PpBBX24 gene is associated with the red skin of "Zaosu Red" pear (Pyrus pyrifolia White Pear Group): A deletion in the PpBBX24 gene is associated with the red skin of pear. Hortic. Res. 2020, 7, 39. [Google Scholar] [CrossRef]
- Yang, G.; Sun, M.; Brewer, L.; Tang, Z.; Nieuwenhuizen, N.; Cooney, J.; Xu, S.; Sheng, J.; Andre, C.; Xue, C.; et al. Allelic variation of BBX24 is a dominant determinant controlling red coloration and dwarfism in pear. Plant Biotech. J. 2024, 22, 1468–1490. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, Y.; Sun, Y.; Zhang, X.; Du, B.; Turupu, M.; Yao, Q.; Gai, S.; Tong, S.; Huang, J.; et al. Two B-Box proteins, PavBBX6/9, positively regulate light-induced anthocyanin accumulation in sweet cherry. Plant Physiol. 2023, 192, 2030–2048. [Google Scholar] [CrossRef]
- Liu, Y.; Ye, Y.; Wang, Y.; Jiang, L.; Yue, M.; Tang, L.; Jin, M.; Zhang, Y.; Lin, Y.; Tang, H. B-Box transcription factor FaBBX22 promotes light-induced anthocyanin accumulation in strawberry (Fragaria × ananassa). Int. J. Mol. Sci. 2022, 23, 7757. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Yue, M.; Jiang, L.; Zhang, N.; Luo, Y.; Chen, Q.; Zhang, Y.; Wang, Y.; Li, M.; et al. FaMYB5 interacts with FaBBX24 to regulate anthocyanin and proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa). Int. J. Mol. Sci. 2023, 24, 12185. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, G.; Chen, J.; Ying, Y.; Yao, L.; Li, X.; Teixeira, J.A.; Yu, Z. Role of Rubus chingii BBX gene family in anthocyanin accumulation during fruit ripening. Front. Plant Sci. 2024, 15, 1427359. [Google Scholar] [CrossRef]
- Liu, W.; Mu, H.; Yuan, L.; Li, Y.; Li, Y.; Li, S.; Ren, C.; Duan, W.; Fan, P.; Dai, Z.; et al. VvBBX44 and VvMYBA1 form a regulatory feedback loop to balance anthocyanin biosynthesis in grape. Hortic. Res. 2023, 10, uhad176. [Google Scholar] [CrossRef]
- Li, C.; Pei, J.; Yan, X.; Cui, X.; Tsuruta, M.; Liu, Y.; Lian, C. A poplar B-box protein PtrBBX23 modulates the accumulation of anthocyanins and proanthocyanidins in response to high light. Plant Cell Environ. 2021, 44, 3015–3033. [Google Scholar] [CrossRef]
- Luo, D.; Xiong, C.; Lin, A.; Zhang, C.; Sun, W.; Zhang, J.; Yang, C.; Lu, Y.; Li, H.; Ye, Z.; et al. SlBBX20 interacts with the COP9 signalosome subunit SlCSN5-2 to regulate anthocyanin biosynthesis by activating SlDFR expression in tomato. Hortic. Res. 2021, 8, 163. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, J.; Tian, S.; Hao, W.; Du, L. Two B-Box proteins, MaBBX20 and MaBBX51, coordinate light-induced anthocyanin biosynthesis in grape hyacinth. Int. J. Mol. Sci. 2022, 23, 5678. [Google Scholar] [CrossRef]
- Kim, D.H.; Park, S.; Lee, J.Y.; Ha, S.H.; Lee, J.G.; Lim, S.H. A rice B-Box protein, OsBBX14, finely regulates anthocyanin biosynthesis in rice. Int. J. Mol. Sci. 2018, 19, 2190. [Google Scholar] [CrossRef]
- Deng, J.; Zhang, L.; Wang, L.; Zhao, J.; Yang, C.; Li, H.; Huang, J.; Shi, T.; Zhu, L.; Damaris, R.N.; et al. The complex FtBBX22 and FtHY5 positively regulates light-induced anthocyanin accumulation by activating FtMYB42 in Tartary buckwheat sprouts. Int. J. Mol. Sci. 2024, 25, 8376. [Google Scholar] [CrossRef]
- Gao, Z.; Sun, Y.; Zhu, Z.; Ni, N.; Sun, S.; Nie, M.; Du, W.; Irfan, M.; Chen, L.; Zhang, L. Transcription factors LvBBX24 and LvbZIP44 coordinated anthocyanin accumulation in response to light in lily petals. Hortic. Res. 2024, 11, uhae211. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, H.; Qu, D.; Zhu, Z.; Yang, Y.; Zhao, Z. The MdBBX22–miR858–MdMYB9/11/12 module regulates proanthocyanidin biosynthesis in apple peel. Plant Biotechnol. J. 2022, 20, 1683–1700. [Google Scholar] [CrossRef]
- Song, H.; Ding, G.; Zhao, C.; Li, Y. Genome-wide identification of B-Box gene family and expression analysis suggest its roles in responses to cercospora leaf spot in sugar beet (Beta vulgaris L.). Genes 2023, 14, 1248. [Google Scholar] [CrossRef]
- Yu, L.; Wang, D.; Huang, R.; Cao, F.; Guo, C.; Zhang, J. Genome-wide identification, characterization and expression profile analysis of BBX gene family in Chinese chestnut (Castanea mollissima). Plant Biotechnol. Rep. 2023, 18, 129–142. [Google Scholar] [CrossRef]
- Ouyang, Y.; Pan, X.; Wei, Y.; Wang, J.; Xu, X.; He, Y.; Zhang, X.; Li, Z.; Zhang, H. Genome-wide identification and characterization of the BBX gene family in pineapple reveals that candidate genes are involved in floral induction and flowering. Genomics 2022, 114, 110397. [Google Scholar] [CrossRef]
- Xu, D.; Wang, H.; Feng, X.; Ma, Y.; Huang, Y.; Wang, Y.; Ding, J.; Chen, H.; Wu, H. Genome-wide identification, phylogenetic and expression analysis of the B-BOX gene family in the woodland strawberry (Fragaria vesca). Horticulturae 2023, 9, 842. [Google Scholar] [CrossRef]
- Wang, J.; Yang, G.; Chen, Y.; Dai, Y.; Yuan, Q.; Shan, Q.; Pan, L.; Dai, L.; Zou, X.; Liu, F.; et al. Genome-wide characterization and anthocyanin-related expression analysis of the B-BOX gene family in Capsicum annuum L. Front. Genet. 2022, 13, 847328. [Google Scholar] [CrossRef]
- Liu, X.; Sun, W.; Ma, B.; Song, Y.; Guo, Q.; Zhou, L.; Wu, K.; Zhang, X.; Zhang, C. Genome-wide analysis of blueberry B-box family genes and identification of members activated by abiotic stress. BMC Genom. 2023, 24, 584. [Google Scholar] [CrossRef]
- Wei, H.; Wang, P.; Chen, J.; Li, C.; Wang, Y.; Yuan, Y.; Fang, J.; Leng, X. Genome-wide identification and analysis of B-BOX gene family in grapevine reveal its potential functions in berry development. BMC Plant Biol. 2020, 20, 72. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jiao, C.; Jin, Q.; Lin, Y.; Cai, Y. B-BOX genes: Genome-wide identification, evolution and their contribution to pollen growth in pear (Pyrus bretschneideri Rehd.). BMC Plant Biol. 2017, 17, 156. [Google Scholar] [CrossRef]
- Obel, H.O.; Cheng, C.; Li, Y.; Tian, Z.; Njogu, M.K.; Li, J.; Lou, Q.; Yu, X.; Yang, Z.; Ogweno, J.O.; et al. Genome-wide identification of the B-Box gene family and expression analysis suggests their potential role in photoperiod-mediated β-carotene accumulation in the endocarp of cucumber (Cucumis sativus L.) fruit. Genesis 2022, 13, 658. [Google Scholar] [CrossRef]
- Ma, R.; Chen, J.; Huang, B.; Huang, Z.; Zhang, Z. The BBX gene family in Moso bamboo (Phyllostachys edulis): Identification, characterization and expression profiles. BMC Genom. 2021, 22, 535. [Google Scholar] [CrossRef]
- Li, Y.; Tong, Y.; Ye, J.; Zhang, C.; Li, B.; Hu, S.; Xue, X.; Tian, Q.; Wang, Y.; Li, L.; et al. Genome-wide characterization of B-Box gene family in Salvia miltiorrhiza. Int. J. Mol. Sci. 2023, 24, 2146. [Google Scholar] [CrossRef]
- Zhao, J.; Li, H.; Huang, J.; Shi, T.; Meng, Z.; Chen, Q.; Deng, J. Genome-wide analysis of BBX gene family in Tartary buckwheat (Fagopyrum tataricum). PeerJ 2021, 9, e1193. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Weng, X.; Wang, L.; Xie, W. The rice B-Box zinc finger gene family: Genomic identification, characterization, expression profiling and diurnal analysis. PLoS ONE 2012, 7, e48242. [Google Scholar] [CrossRef]
- Bu, X.; Wang, X.; Yan, J.; Zhang, Y.; Zhou, S.; Sun, X.; Yang, Y.; Ahammed, G.J.; Liu, Y.; Qi, M.; et al. Genome-wide characterization of B-Box gene family and its roles in responses to light quality and cold stress in tomato. Front. Plant Sci. 2021, 12, 698525. [Google Scholar] [CrossRef]
- Hou, W.; Ren, L.; Zhang, Y.; Sun, H.; Shi, T.; Gu, Y.; Wang, A.; Ma, D.; Li, Z.; Zhang, L. characterization of BBX family genes and their expression profiles under various stresses in the sweet potato wild ancestor Ipomoea trifida. Sci. Hortic. 2021, 288, 110374. [Google Scholar] [CrossRef]
- Xu, X.; Li, W.; Yang, S.; Zhu, X.; Sun, H.; Li, F.; Lu, X.; Cui, J. Identification, evolution, expression and protein interaction analysis of genes encoding B-box zinc-finger proteins in maize. J. Integr. Agr. 2023, 22, 371–388. [Google Scholar] [CrossRef]
- Liu, S.; Lyu, S.; Yang, Z.; Xu, G.; Zhang, Y.; Liu, Y.; Jin, J.; Deng, S. Genome-wide characterization of tobacco B-BOX gene family identified two close members play contrast roles under cold stress. Environ. Exp. Bot. 2023, 216, 105533. [Google Scholar] [CrossRef]
- Ye, Y.; Liu, Y.; Li, X.; Wang, G.; Zhou, Q.; Chen, Q.; Li, J.; Wang, X.; Tang, H. An evolutionary analysis of B-Box transcription factors in strawberry reveals the role of FaBBx28c1 in the regulation of flowering time. Int. J. Mol. Sci. 2021, 22, 11766. [Google Scholar] [CrossRef]
- Sui, C.; Cheng, S.; Wang, D.; Lv, L.; Meng, H.; Du, M.; Li, J.; Su, P.; Guo, S. Systematic identification and characterization of the soybean (Glycine max) B-Box transcription factor family. Biotechnol. Biotec. Eq. 2023, 37, 104–116. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Dai, Y.; Chen, X.; Wang, X. Genome-wide identification and expression analysis of the B-box gene family in the apple (Malus domestica Borkh.) genome. Mol. Genet. Genom. 2017, 293, 303–315. [Google Scholar] [CrossRef]
- Tang, H.; Yuan, C.; Shi, H.; Liu, F.; Shan, S.; Wang, Z.; Sun, Q.; Sun, J. Genome-wide identification of peanut B-BOXs and functional characterization of AhBBX6 in salt and drought stresses. Plants 2024, 13, 955. [Google Scholar] [CrossRef]
- Faria, G.M.L.; Silva, E.K. Pulsed electric field, ultrasound and microwave heating based extraction techniques for valorization of pomegranate peel byproducts: A review. J. Environ. Chem. Eng. 2024, 12, 113078. [Google Scholar] [CrossRef]
- Zhao, X.; Feng, Y.; Ke, D.; Teng, Y.; Yuan, Z. Comparative transcriptomic and metabolomic profiles reveal fruit peel color variation in two red pomegranate cultivars. Plant Mol. Biol. 2024, 114, 51. [Google Scholar] [CrossRef]
- Zhao, X.; Yuan, Z.; Feng, L.; Fang, Y. Cloning and expression of anthocyanin biosynthetic genes in red and white pomegranate. J. Plant Res. 2015, 128, 687–696. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, X.; Wang, C.; Feng, L.; Yin, Y.; Li, J. PgMYB1 positively regulates anthocyanin accumulation by activating PgGSTF6 in pomegranate. Int. J. Mol. Sci. 2023, 24, 6366. [Google Scholar] [CrossRef]
- Arlotta, C.; Puglia, G.D.; Genovese, C.; Toscano, V.; Karlova, R.; Beekwilder, J.; De, C.H.; Raccuia, S.A. MYB5-like and bHLH influence flavonoid composition in pomegranate. Plant Sci. 2020, 298, 110563. [Google Scholar] [CrossRef]
- Ben-Simhon, Z.; Judeinstein, S.; Nadler-Hassar, T.; Trainin, T.; Bar-Ya’akov, I.; Borochov-Neori, H.; Holland, D. A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 2011, 234, 865–881. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, X.; Li, B.; Zhao, X.; Shen, Y.; Yuan, Z. genome-wide identification and characterization of BZIP gene family and cloning of candidate genes for anthocyanin biosynthesis in pomegranate (Punica granatum). BMC Plant Biol. 2022, 22, 170. [Google Scholar] [CrossRef]
- Luo, X.; Li, H.X.; Wu, Z.K.; Yao, W.; Zhao, P.; Cao, D.; Yu, H.Y.; Li, K.D.; Poudel, K.; Zhao, D.G.; et al. The pomegranate (Punica granatum L.) draft genome dissects genetic divergence between soft- and hard-seeded cultivars. Plant Biotechnol. J. 2020, 18, 955–968. [Google Scholar] [CrossRef]
- Hu, J.; Huang, B.; Yin, H.; Qi, K.; Jia, Y.; Xie, Z.; Gao, Y.; Li, H.; Li, Q.; Wang, Z.; et al. PearMODB: A multiomics database for pear (Pyrus) genomics, genetics and breeding study. Database 2023, 2023, baad050. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A "one for all, all for one" bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-mapper V2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Qin, G.H.; Xu, C.Y.; Ming, R.; Tang, H.B.; Guyot, R.; Kramer, E.M.; Hu, Y.D.; Yi, X.K.; Qi, Y.J.; Xu, X.Y.; et al. The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J. 2017, 91, 1108–1128. [Google Scholar] [CrossRef]
- Li, J.; Liu, C.; Yu, Q.; Cao, Z.; Yang, Y.; Jia, B.; Su, Y.; Li, G.; Qin, G. Identification of sugar transporter (SWEET) genes involved in pomegranate seed coat sugar accumulation. 3 Biotech 2022, 12, 181. [Google Scholar] [CrossRef]
- Liu, L.B.; Zheng, J. Identification and expression analysis of the sucrose synthase gene family in pomegranate (Punica granatum L.). PeerJ 2022, 10, e12814. [Google Scholar] [CrossRef]
- Zheng, J.; Liu, L.; Tao, H.; An, Y.; Wang, L. Transcriptomic profiling of apple calli with a focus on the key genes for ALA-induced anthocyanin accumulation. Front. Plant Sci. 2021, 12, 640606. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fu, Z.; Shang, H.; Jiang, H.; Gao, J.; Dong, X.; Wang, H.; Li, Y.; Wang, L.; Zhang, J.; Shu, Q.; et al. Systematic identification of the light-quality responding anthocyanin synthesis-related transcripts in Petunia petals. Hortic. Plant J. 2020, 6, 428–438. [Google Scholar] [CrossRef]
- Crocco, C.D.; Botto, J.F. BBX proteins in green plants: Insights into their evolution, structure, feature and functional diversification. Gene 2013, 531, 44–52. [Google Scholar] [CrossRef]
- Moore, R.C.; Purugganan, M.D. The Early Stages of Duplicate Gene Evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef]
- Yadav, A.; Ravindran, N.; Singh, D.; Rahul, P.V.; Datta, S. Role of Arabidopsis BBX proteins in light signaling. J. Plant Biochem. Biotechnol. 2020, 29, 623–635. [Google Scholar] [CrossRef]
- Fu, J.L.; Liao, L.; Jin, J.; Lu, Z.; Sun, J.; Song, L.; Huang, Y.; Liu, S.; Huang, D.; Xu, Y.; et al. A transcriptional cascade involving BBX22 and HY5 finely regulates both plant height and fruit pigmentation in citrus. J. Integr. Plant Biol. 2024, 66, 1752–1768. [Google Scholar] [CrossRef]
- Zhao, L.; Gao, L.; Wang, H.; Chen, X.; Wang, Y.; Yang, H.; Wei, C.; Wan, X.; Xia, T. The R2R3-MYB, BHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct. Integr. Genom. 2013, 13, 75–98. [Google Scholar] [CrossRef]
- Chang, C.S.J.; Li, Y.; Chen, L.T.; Chen, W.C.; Hsieh, W.P.; Shin, J.; Jane, W.N.; Chou, S.J.; Choi, G.; Hu, J.M.; et al. LZF1, a HY5-Regulated Transcriptional Factor, Functions in Arabidopsis De-Etiolation. Plant J. 2008, 54, 205–219. [Google Scholar] [CrossRef]
- Job, N.; Yadukrishnan, P.; Bursch, K.; Datta, S.; Johansson, H. Two B-Box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol. 2018, 176, 2963–2976. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).