Abstract
Infant feces and breast milk are sources of lactic acid bacteria with interesting functional and technological properties. Therefore, we focused on the isolation and characterization of new potential probiotic strains from infant feces and breast milk. Nine strains were identified based on 16S rRNA sequences of Lacticaseibacillus rhamnosus, Limosilactobacillus reuteri, and Lactobacillus gasseri. Subsequently, these isolates were evaluated for their safety (antibiotic resistance, hemolytic, and enzymatic activity) and functional properties (gastrointestinal condition tolerance, hydrophobicity, auto-aggregation, adhesion to Caco-2/HT-29 cell lines, antimicrobial and antioxidant properties, and the production of exopolysaccharides). All the isolates were non-hemolytic and resistant to streptomycin, kanamycin, and vancomycin. They also demonstrated antimicrobial activity against five selected pathogens (Staphylococcus aureus, Bacillus cereus, Escherichia coli, Salmonella sp., and Enterococcus faecalis) and the production of exopolysaccharides. Isolates Lacticaseibacillus rhamnosus S1F and S6C together with Lactobacillus gasseri M8C showed the highest ability to survive 3.0 g/L of bile salts and a good adhesion property, which is important for gastrointestinal tract (GIT) colonization or the inhibition of pathogens. Following in vivo characterization, three strains S1F and S6C from infant feces, and M6C from breast milk, showed probiotic properties with potential applications in the food industry.
1. Introduction
Evidence of the relationship between microbial composition and an individual’s physical and psychological state has recently been growing [1,2,3]. Thus, the consumption of foods that bring health benefits is becoming increasingly common among the public. International market competition and increasing public interest in healthy lifestyles challenge innovations in the probiotic bacteria research sector. In addition to the commonly used microorganisms, efforts are being made to identify new strains of probiotic bacteria that positively affect human health. As our understanding of the intricate relationship between the microbiota and human health continues to evolve, harnessing the probiotic potential of the family Lactobacillaceae is a significant step toward personalized medicine and improved health outcomes for infants and beyond [4].
In recent years, potential probiotic microorganisms, such as lactic acid bacteria, bifidobacteria, and yeasts, have been isolated from different sources, including dairy and non-dairy fermented foods or the gastrointestinal tract of humans, animals, and insects [5,6,7,8]. However, the selection of new probiotics requires a systematic approach to characterize these candidates. According to the FAO/WHO [9], probiotics must be assessed as safe, must survive the inhospitable conditions of the gastrointestinal tract (low pH and bile salts), and have good functional and technological properties, such as antimicrobial activity against pathogens or adherence to the epithelial surface.
The most common sources of potentially beneficial microorganisms are fermented dairy products and other fermented foods, such as cabbage and other vegetables. However, many studies have addressed the isolation and characterization of novel potential probiotic strains from breast milk [10,11,12] or from the stools of adults and infants [13,14]. Breast milk provides a suitable environment for health-promoting microbial species because of its prebiotic milk oligosaccharide content [15]. Therefore, it is a source of lactic acid bacteria with remarkable properties [2]. Bacteria isolated from stool have been attributed to properties such as high resistance to bile salts, low pH, and lysozyme tolerance, as well as their ability to adhere to the intestinal epithelium [16]. Therefore, this study focused on the isolation of new strains from the genera Lactobacillus, Lacticaseibacillus, and Limosilactobacillus from infant feces and breast milk and investigated their probiotic potential. First, the isolates were evaluated for safety parameters (antibiotic resistance, hemolytic activity, and enzyme activity). Subsequently, functional attributes, such as resistance to gastrointestinal conditions, adhesion capabilities, antimicrobial and antioxidant activities, and exopolysaccharide production, were evaluated using in vitro methods.
2. Materials and Methods
2.1. Isolation and Identification
Samples of infant feces and breast milk were obtained from six volunteer donors (mothers and their children; Prague, Czech Republic). Voluntary donors signed informed consent forms. The fecal samples were sourced from healthy six to ten weeks old, breast-fed infants. Breast milk samples were collected in sterile tubes, and fecal samples were collected in tubes containing Wilkins Chalgren broth (Merck, Darmstadt, Germany). Strains from family Lactobacillaceae were isolated using two different media: Wilkins-Chalgren agar (Merck, Darmstadt, Germany) enriched with glacial acetic acid 0.1% (v/v; Sigma Aldrich, Prague, Czech Republic); and MRS agar, pH 5.7 (De Man, Rogosa and Sharp) (Merck, Germany). The cultivation was carried out anaerobically (37 °C, 72 h). First, the newly isolated strains were tested to determine bacterial cell morphology, and Gram staining was performed using an Eclipse E200 microscope (Nikon Europe B.V., Prague, Czech Republic). Subsequently, the molecular identification of the isolated bacteria was performed based on the alignment of the 16S SSU rRNA gene sequences using the BLAST alignment tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi; accessed on 25 August 2021). Bacterial DNA was extracted using a DNeasy® UltraClean® Microbial Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The 16S SSU rRNA gene was amplified using the fD1 (5′-CCG AAT TCG TCG ACA ACA GAG TTT GAT CCT GGC TCA G-3′) and rP2 (5′-CCC GGG ATC CAA GCT TAC GGC TAC CTT GTT ACG ACT T-3′) primers [17]. PCR products were separated on a 1% agarose gel (SeaKem® LE; Lonza, Rockland, ME, USA), stained with GelRed® (Biotium, Fremont, QC, Canada) at 70 V for 60 min, and visualized using the GENE GENIUS Bio Imaging System (Syngene, Frederick, MD, USA). All PCR products were treated with 2 µL of ExoSAP-ITTM according to the manufacturer’s instructions (ThermoFisher Scientific, Baltic UAB, Vilnius, Lithuania), and sequenced by Eurofins Genomics Germany GmbH (Ebersberg, Germany) using fD1. The pure cultures of identified isolates were preserved in MRS broth (pH 5.7) with the addition of 20% glycerol (Sigma Aldrich, Czech Republic) at −20 °C.
2.2. Hemolytic Activity
The hemolytic activity of the isolates was determined using the method described by Sui et al. [18], with minor modifications. Bacterial cultures in the exponential growth phase were streaked onto Columbia blood agar plates (Merck, Darmstadt, Germany) enriched with 5% sheep blood. The plates were incubated anaerobically for 48 h at 37 °C. Thereafter, the hemolysis zones were observed.
2.3. Antibiotic Susceptibility
The antibiotic resistance of the nine potential probiotic strains was assessed using the disc diffusion method [19]. Tested antibiotics were selected according to the European Food Safety Authority guidelines (EFSA; [20]): gentamicin (30 µg), erythromycin (15 µg), clindamycin (2 µg), streptomycin (25 µg), quinupristin/dalfopristin (15 µg), kanamycin (30 µg), chloramphenicol (30 µg), vancomycin (5 µg), ampicillin (10 µg), and tetracycline (30 µg; Oxoid, Hampshire, UK). The MRS agar plates with tested antibiotics, and strains were cultivated anaerobically at 37 °C for 24 h. Thereafter, the diameter of the inhibition zone was measured, and the resistance or sensitivity of strains to the antibiotics was evaluated such as resistant (R) when the inhibition zone had a diameter of <14 mm, intermediate (I) =15–19 mm, and sensitive (S) > 20 mm.
2.4. Antimicrobial Activity
The antimicrobial activity of the tested isolates against five pathogenic bacteria (Bacillus cereus CCM 869, Escherichia coli CCM 3954, Enterococcus faecalis CCM 4224, Staphylococcus aureus CCM 3958, and Salmonella enterica serotype Typhimurium CCM 7205) obtained from the Czech Collection of Microorganisms (Brno, Czech Republic) was determined using a microtiter plate assay following the methods described by Kostelac et al. [21], with slight modifications. Briefly, the acid supernatants were separated by centrifugation (5000× g, 5 min) from the medium cultivated at 37 °C for 24 h. Subsequently, 20 µL of the pathogen suspension at a density of 3 McF, 20 µL BHI (brain heart infusion, Merck), and 160 µL supernatant was placed into the microtiter plate (As,0) and incubated for 48 h at 37 °C. After 48 h, the absorbance was measured at 620 nm using a Tecan Infinite M200 spectrometer (Tecan Group, Männedorf, Switzerland). A mixture of 20 µL pathogen suspension and 180 µL BHI (Ac) was used as the control. The measurements were made in triplicates. The inhibition level was determined using the following equation:
2.5. Hydrophobicity
The hydrophobicity was determined using the partially modified MATH method described by Krausová et al. [22]. The cells were harvested by centrifugation (5 min, 5000× g), washed twice with phosphate saline buffer (pH 7.0) and resuspended to an optical density of 0.50–0.60 at 620 nm (H0) (Onda-V10 PLUS, Giorgio Bormac, Capri, Italy). Thereafter, 5 mL of bacterial suspension was mixed with 150 μL hexane (Merck, Darmstadt, Germany) and vortexed for 1 min. After 30 min at room temperature, the hydrophobicity of the aqueous phase was measured spectrophotometrically at 620 nm (Ht). Hydrophobicity was calculated using the following formula:
Hydrophobicity (%) = ((H0 − Ht)/H0) × 100.
2.6. Auto-Aggregation
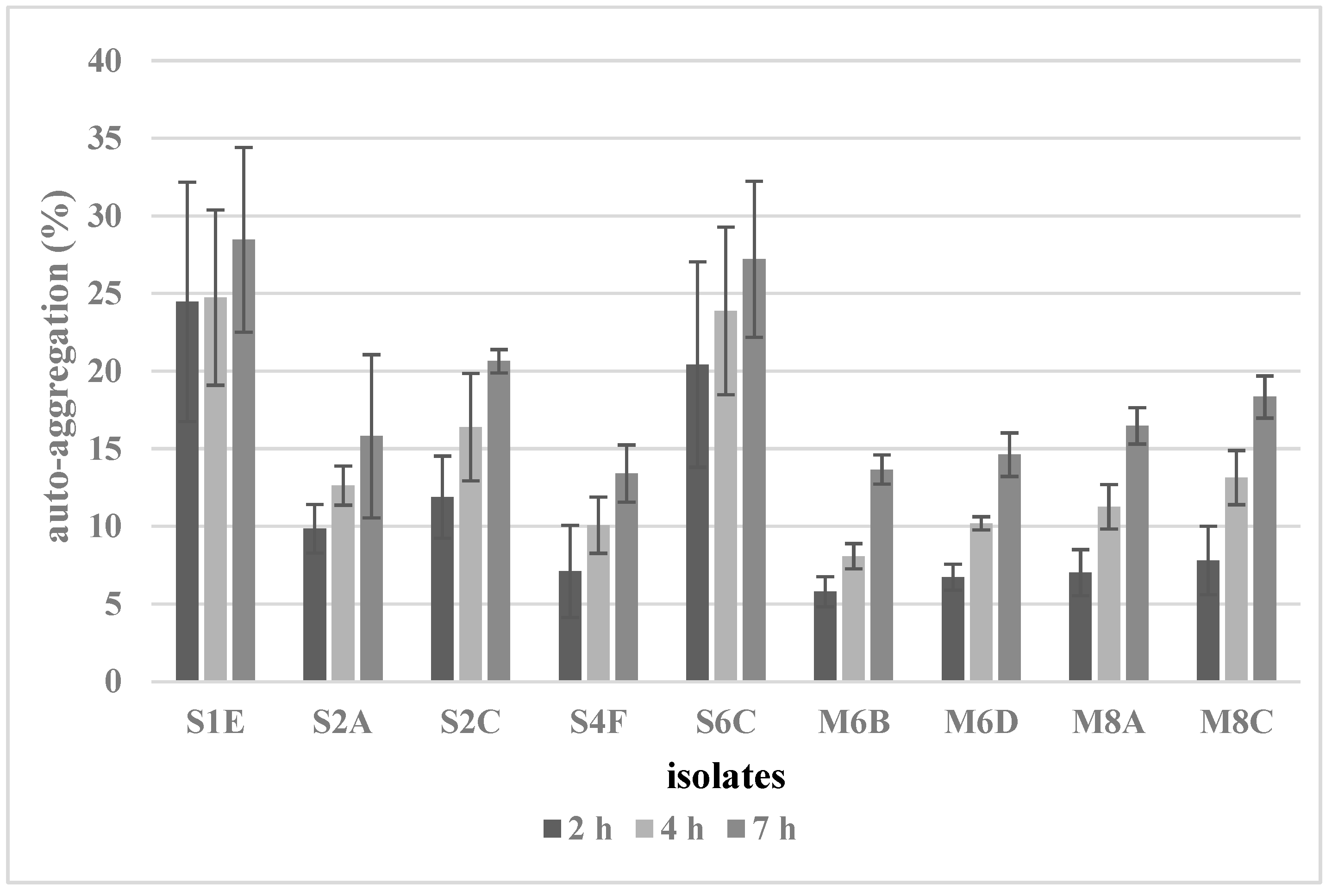
The auto-aggregation of potential probiotic strains was evaluated using the method described by Reuben et al. [5], with some modifications. In brief, overnight-growth cultures of each strain (18 h, 37 °C) were centrifuged (7 min, 5000× g), washed twice, and resuspended in PBS to an optical density of 0.50–0.60 at 600 nm (A0). The samples were incubated at 37 °C for 7 h. The absorbance of the suspension was measured after 2, 4, and 7 h of incubation and auto-aggregation was calculated as follows:
auto-aggregation (%) = (1 − (Ah/A0)) × 100.
2.7. Adherence Assay
The adherence of isolates from the genera Limosilactobacillus, Lactobacillus, and Lacticaseibacillus to mixed human colon adenocarcinoma cell lines Caco-2 (ATCC HTB-37) and HT-29 (ATCC HTB 38; American Type Culture Collection, Rockville, MD, USA) was tested according to the method described by Musilova et al. [23]. A mixture of Caco-2 and HT-29 cells (9:1) prepared as described above was seeded in 24-well plates at a final concentration of 4 × 104 cells/mL. The isolates were cultivated overnight, washed thrice with PBS, and added to the seeded well plates at a density of 107 CFU/mL. Subsequent incubation was performed for 90 min at 37 °C. After incubation, the cells were detached from the wells. The remaining suspensions containing viable adherent bacterial strains were diluted and cultivated on MRS agar (Merck, Darmstadt, Germany) at 37 °C for 3 d under anaerobic conditions. The adherence of the isolates to HT-29/Caco-2 cells was evaluated as the percentage of adhered cells compared to the total cell count.
2.8. Scavenging of DPPH Radical
The DPPH (1,1-diphenyl-2-(2,4,6-trinitrophenyl) hydrazyl) radical scavenging assay was performed according to the method described by Hyrslova et al. [24].
2.9. Resistance to Gastrointestinal Tract (GIT) Condition
The simulation of human gastric juice was performed according to the method described by Kostelac et al. [21], with slight modifications. Briefly, bacterial cells from the tested strains were harvested by centrifugation (7 min, 5500× g), washed twice with PBS (pH 7.0), and resuspended in the simulated gastric juice (SFG). SFG (pH 8.0) was prepared using pancreatin (1 g/L) (Sigma-Aldrich, Prague, Czech Republic) and bile salts (1.5 and 3.0 g/L) (Sigma-Aldrich, Czech Republic). The ability to survive under GIT conditions was evaluated by cell viability (%) using the ratio of cell counts (MRS agar, 37 °C, 72 h) at the beginning and after 4 h. The measurements were made in triplicates.
2.10. Production of Exopolysachaccharides
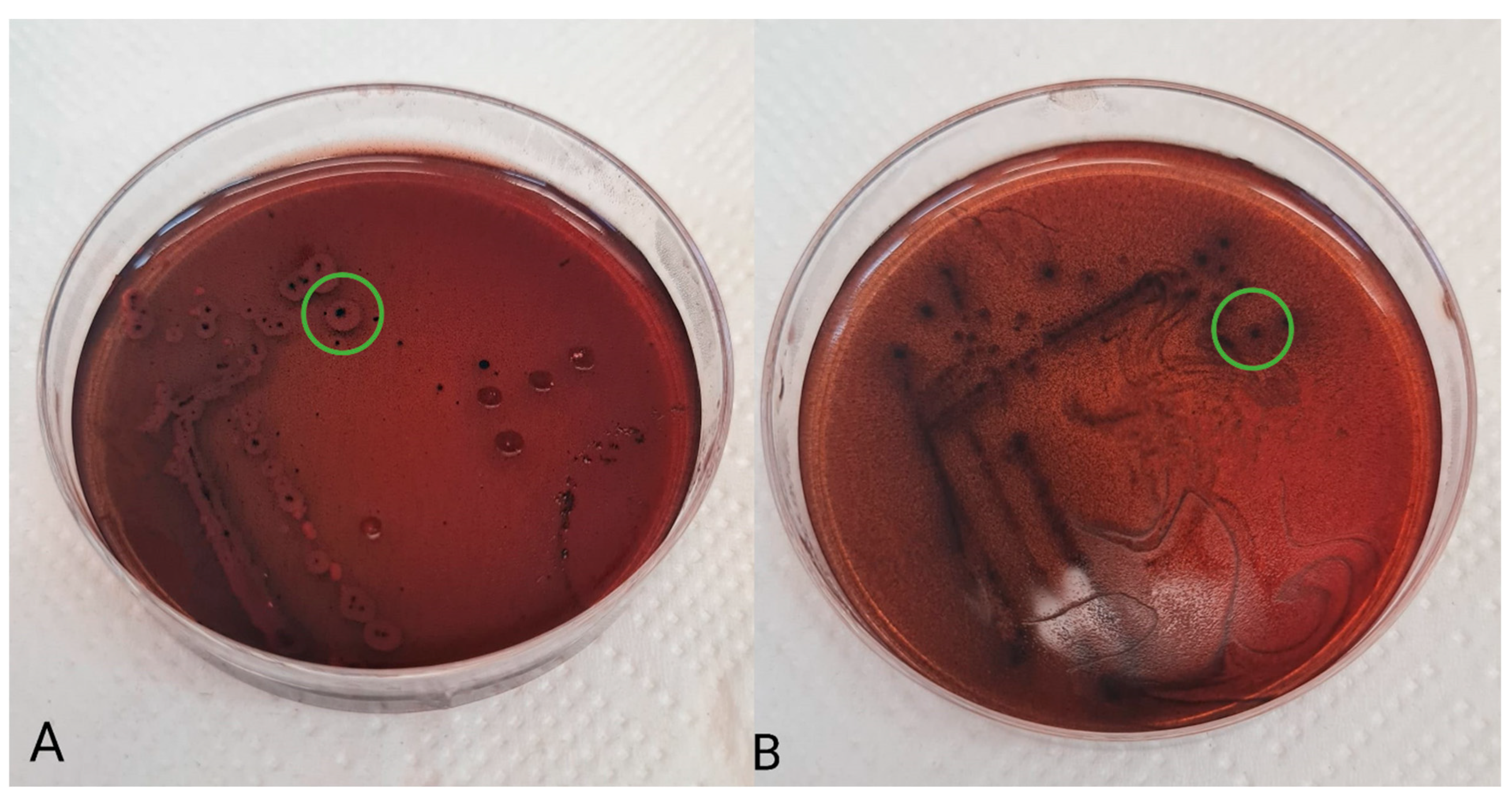
To assess the production of exopolysaccharides, the following basal medium was used: MRS-0 agar (10 g/L tryptone, 8 g/L meat extract, 4 g/L yeast extract, 2 g/L K2HPO4, 1 mL/L Tween 80, 2 g/L ammonium citrate, 5 g/L sodium acetate, 0.2 g/L MgSO4·7 H2O, 0.04 g/L MnSO4·4 H2O, and 12 g/L agar) supplemented with 40 g/l carbohydrates. Glucose (G), lactose (L), and sucrose (S) were used as carbohydrate sources in this study. After sterilization (121 °C, 15 min), Kongo red solution (0.8 g/L) was added to the agar medium. Separated and washed bacterial suspensions were streaked onto plates with the prepared agar media and overlaid with agar. Incubation was performed at 37 °C for 72 h. The ability of bacterial isolants to produce exopolysaccharides was demonstrated by the black coloring of the center of the colonies (Figure 1).
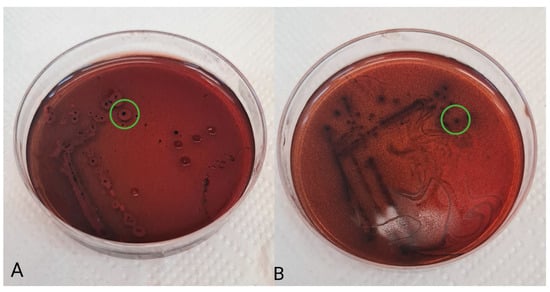
Figure 1.
Production of exopolysaccharides (green circle) by Lactobacillus reuteri 2SA (A) and Lacticaseibacillus rhamnosus S6C (B).
2.11. Statistical Analysis
ANOVA with a post hoc Scheffé-test for multiple comparisons was used to evaluate the results, with statistical significance set at p < 0.05. For the comparison of only two datasets, a two-sample t-test (p < 0.05) was used instead of ANOVA. A Real Statistics Resource Pack software (Release 7.2; Charles Zaiontz) was used to perform the tests.
3. Results
3.1. Identification and Safety Assessment
Isolates from infant feces and breast milk, as shown in Table 1, belong to the new nomenclature of the genus Lactobacillus to three genera: Lactobacillus, Limosilactobacillus, and Lacticaseibacillus [25]. Specifically, Lactobacillus gasseri, Lacticaseibacillus rhamnosus, and Limosilactobacillus reuteri strains were identified using 16S rRNA gene sequencing. Potential probiotics must possess non-hemolytic properties to be safe for use. We confirmed that all the isolates tested in this study complied with this requirement. The resistance of these strains to commonly used antibiotics (n = 10) was evaluated. All the strains were shown to be resistant to streptomycin, kanamycin, and vancomycin. However, the results showed that the tested isolates were intermediately sensitive or sensitive to the other tested antibiotics, with the exception of gentamycin (Table 2). Three strains of Lacticasebacillus rhamnosus (S4F, S6C, and M8A) were resistant to all four abovementioned antibiotics.

Table 1.
List of isolated strains from genera Lactobacillus, Limosilactobacillus, and Lacticaseibacillus.

Table 2.
Antibiotic resistance profile of tested strains.
The extracellular enzyme activity was monitored using a commercial API ZYM kit (Table 3). All tested isolates were positive for esterase (C4), lipase-esterase (C8), leucinarylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α- and β-galactosidase, and β-glucosidase. Eight of the nine strains exhibited valine arylamidase activity. Seven strains tested positive for the enzyme α-glucosidase. Alkaline phosphatase, lipase (C14), trypsin, α-chymotrypsin, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase were not detected in any of the isolates.

Table 3.
Enzymatic activity.
3.2. Hydrophobicity of Cell Surface and Aggregation Potential
The MATH method was used to assess the cell surface hydrophobicity of potential probiotic isolates. The tested strains displayed significantly different degrees of affinity for hexane (Table 4). Hydrophobicity was the highest in the Lactobacillus gasseri M6C strain (57.1%), followed by Lacticaseibacillus rhamnosus M8A (50.8%), and S1E (46.1%). The mean values obtained were also significantly different between the origins and genera.

Table 4.
Probiotic properties of tested isolates.
Auto-aggregation is an important probiotic ability that helps reduce pathogen colonization. In this study, the auto-aggregation of the tested isolates was observed at 2, 4, and 7 h (Figure 2). The auto-aggregation abilities of isolates had a variable range between 5.8 and 28.5%, depending on incubation time and strain. Lacticaseibacillus rhamnosus S1E and S6C showed the strongest percentages of auto-aggregation (28.5% and 27.2%) after 7 h.
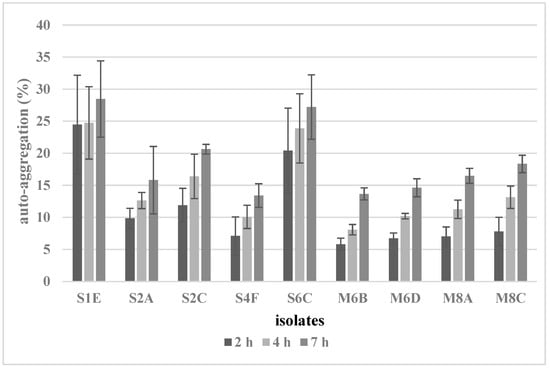
Figure 2.
Auto-aggregation of selected isolates.
3.3. Adherence Assay
Caco-2 and HT-29 cell lines at a 9:1 ratio were used as models of the intestinal epithelium for in vitro determination of the adherence of isolates from the genera Lactobacillus, Lacticaseibacillus, and Limosilactobacillus. Lactobacillus gasseri M8C showed the strongest adherence capacity (6.0%) compared to the other tested isolates. Both Limosilactobacillus reuteri S2A and S2C strains isolated from infant feces also proved good adhesion capacities of >3.5%.
3.4. Antioxidant Activity of the Tested Isolates
The antioxidant capacity of the potential probiotic isolates was evaluated using DPPH radical scavenging activity. The DPPH radical scavenging activity of these strains ranged from 18.4 to 69.3%, as shown in Table 4. Mainly, two strains of Lacticaseibacillus rhamnosus, S4F and S6C, isolated from infant feces exhibited considerably higher DPPH activity (69.3%) than the other isolates.
3.5. Bile Salt Tolerance
The effects SFG with pancreatin and 1.5 or 3.0 g/L bile salts are presented in Table 4. Two strains of Lacticaseibacillus rhamnosus S1E and S4F, significantly showed the highest viability (>85%) after 4 h incubation in simulated gastric juice containing both concentrations of bile salts. Remaining strains proved a good bile salt tolerance for the lower concentration (>67%) but the higher concentration led to a decrease in cell viability of approximately 10–30%.
3.6. Production of Exopolysaccharides
To determine the production of exopolysaccharides by the tested strains, MRS agar enriched with Congo red, glucose, sucrose, or lactose was used. All isolates produced exopolysaccharides on all three agar types (Table 5). Nevertheless, the highest production was observed on MRS agar supplemented with glucose.

Table 5.
Production of exopolysaccharides by isolates from breast milk and infant feces.
3.7. Antimicrobial Activity
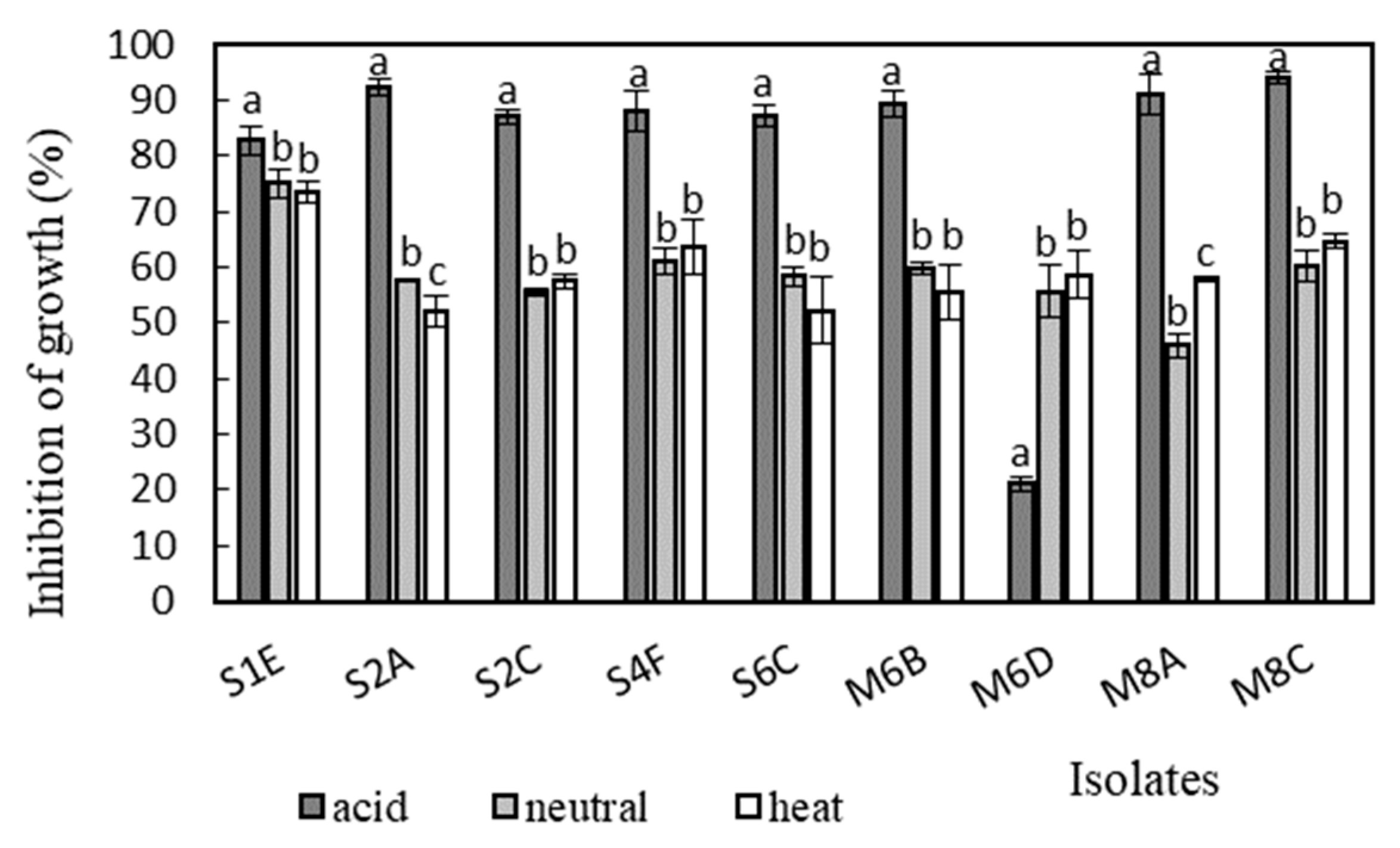
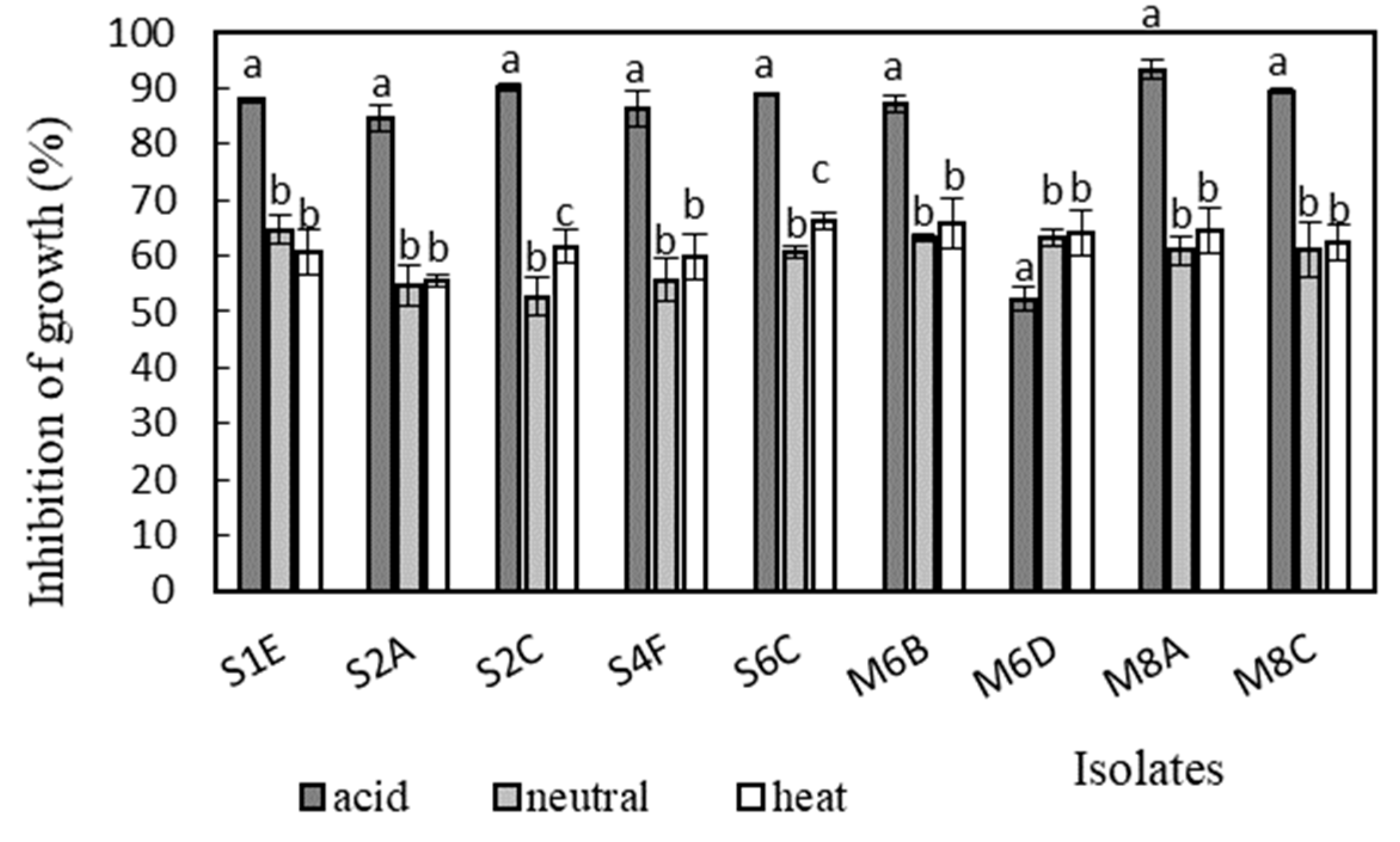
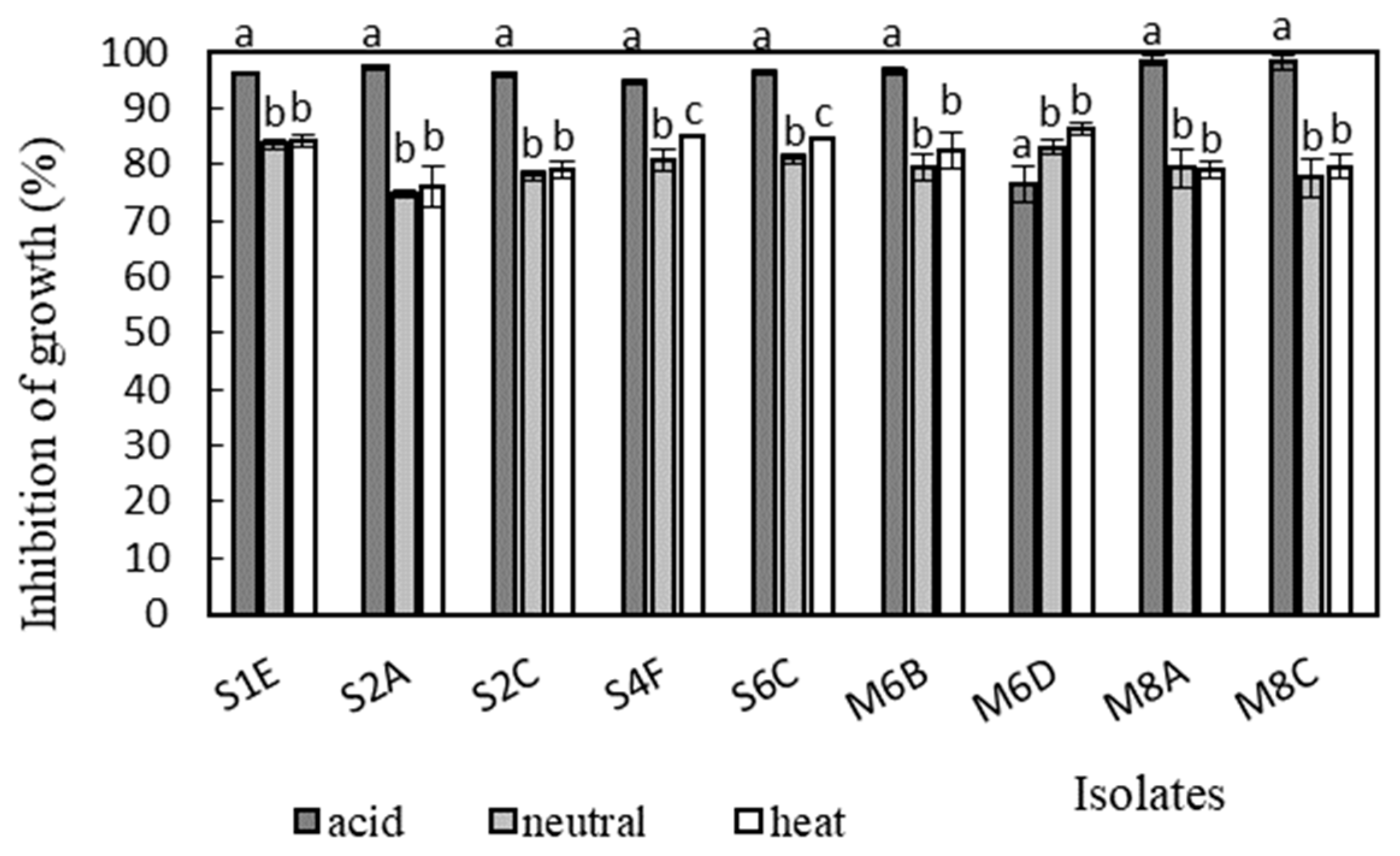
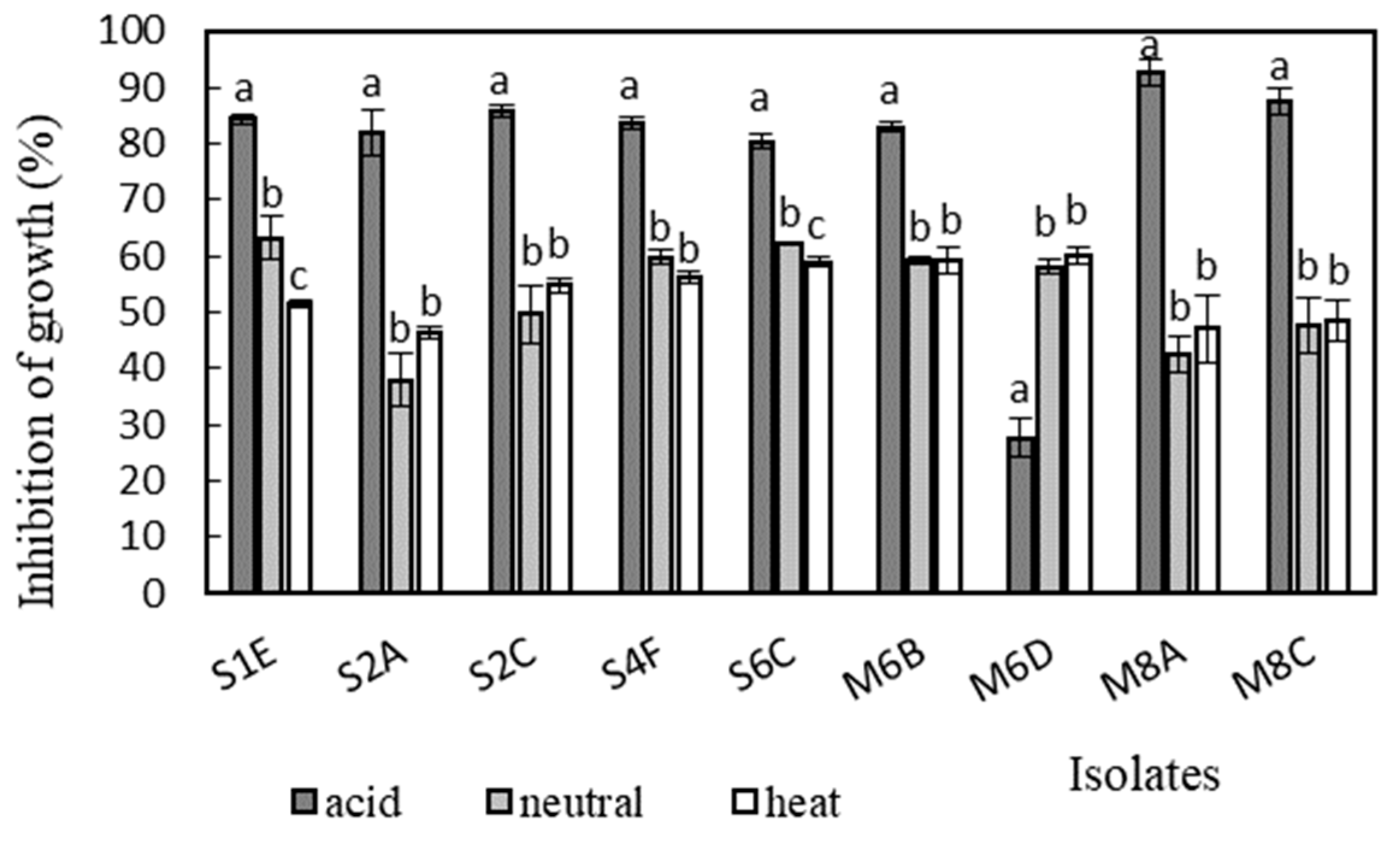
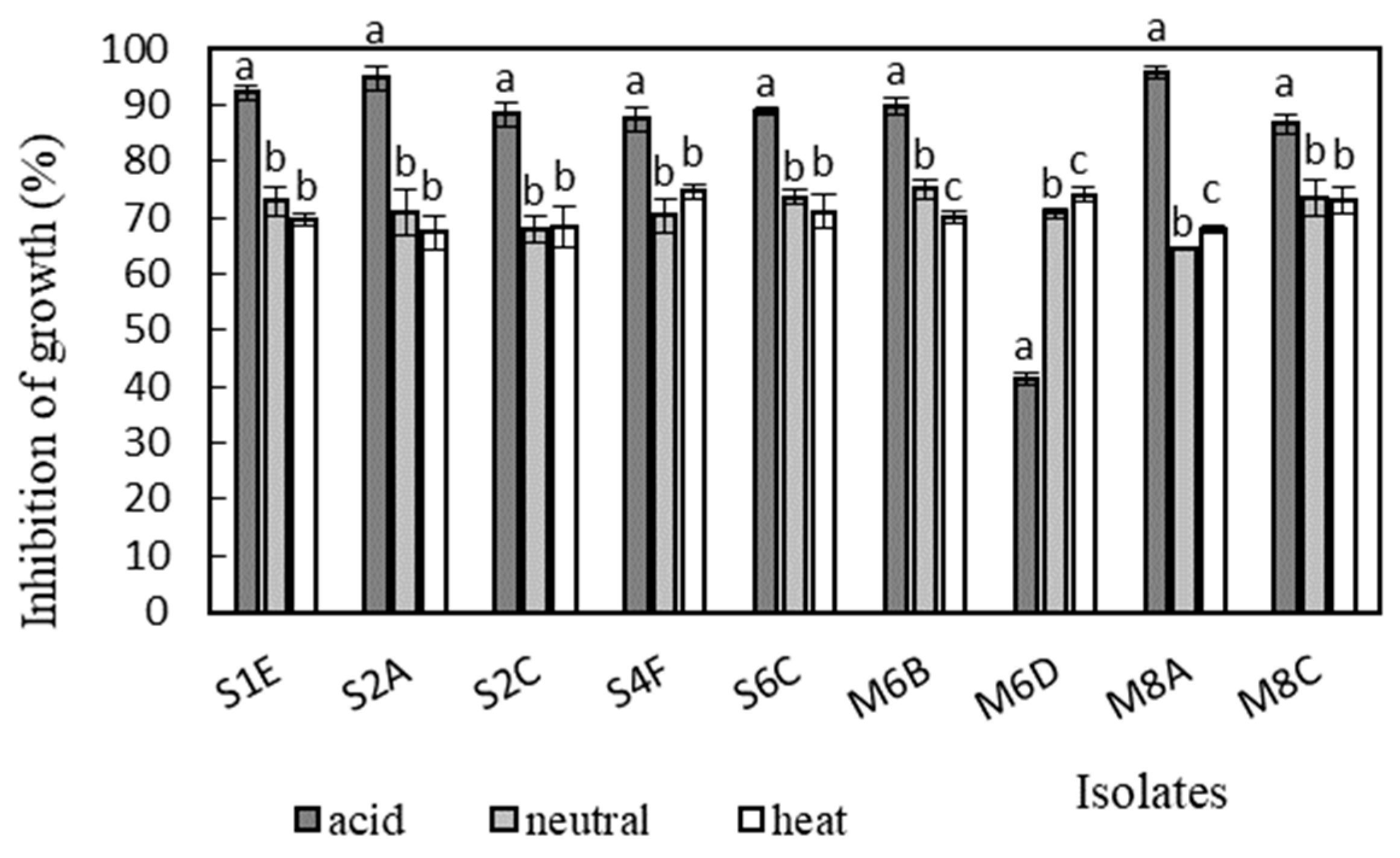
Antimicrobial activity was determined in the supernatants of the isolates against Bacillus cereus CCM 869, Escherichia coli CCM 3954, Enterococcus faecalis CCM 4224, Staphylococcus aureus CCM 3958, and Salmonella enterica serotype Typhimurium CCM 7205, which are known to cause various infectious diseases. Acidic supernatants inhibited the growth of most of the tested pathogens (>80%), as is shown in Figure 3, Figure 4, Figure 5, Figure 6 and Figure 7. One exception was Lactobacillus gasseri M6D, for which the acidic supernatant showed the lowest antimicrobial activity. The neutralization and heat treatment of the supernatants decreased their antimicrobial activities, mainly against B. cereus, E. coli, and Salmonella enterica serotype Typhimurium.
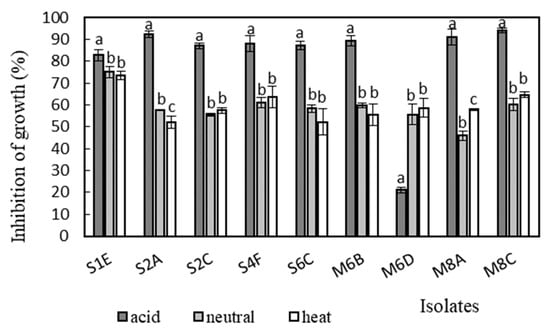
Figure 3.
Antimicrobial activity of isolates against B. cereus CCM 869. Data with different superscripts (a–c) are significantly different (p < 0.05).
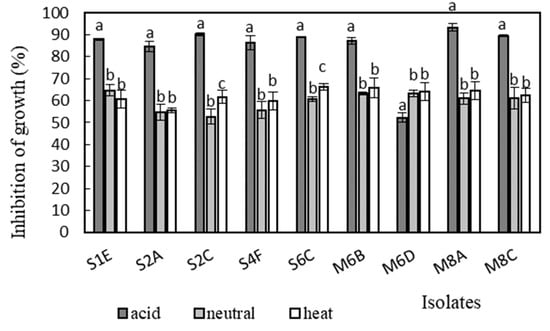
Figure 4.
Antimicrobial activity of isolates against E. coli CCM 3954. Data with different superscripts (a–c) are significantly different (p < 0.05).
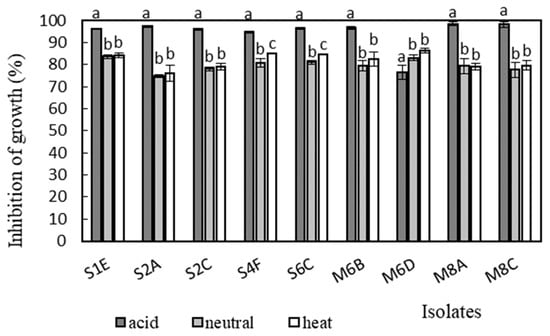
Figure 5.
Antimicrobial activity of isolates against E. faecalis CCM 4224. Data with different superscripts (a–c) are significantly different (p < 0.05).
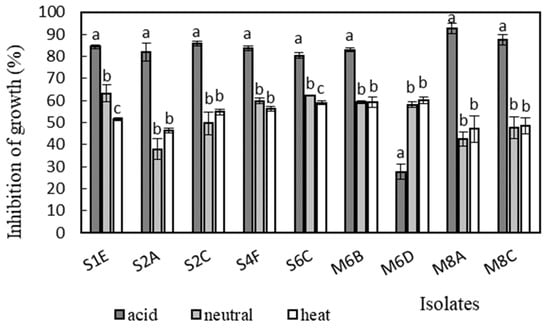
Figure 6.
Antimicrobial activity of isolates against Salmonella enterica serotype Typhimurium CCM 7205. Data with different superscripts (a–c) are significantly different (p < 0.05).
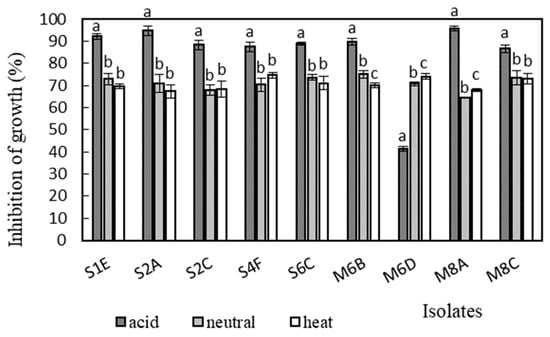
Figure 7.
Antimicrobial activity of isolates against S. aureus CCM 3958. Data with different superscripts (a–c) are significantly different (p < 0.05).
4. Discussion
In recent years, potential probiotic microorganisms, such as lactic acid bacteria, bifidobacteria, and yeasts, have been isolated from different sources, including dairy and non-dairy fermented foods, and the gastrointestinal tract of humans, animals, and insects [5,6,7,8]. Nevertheless, the selection of new probiotics requires a systematic approach to characterize these candidates. According to the FAO/WHO [9], probiotics must be assessed as safe, survive in the inhospitable conditions of the gastrointestinal tract (low pH and bile salts), and have good functional and technological properties, such as antimicrobial activity against pathogens or adherence to the epithelial surface. In this study, nine strains of the genera Lactobacillus, Lacticaseibacillus, and Limosilactobacillus were isolated from breast milk and infant feces, and identified by 16S rRNA sequencing using the NCBI database. Subsequently, these strains were characterized for safety (resistance to ATB and hemolytic enzymatic activity) and functional properties, such as resistance to the harsh conditions of the digestive tract, adhesion abilities (hydrophobicity, auto-aggregation, and adherence to the co-culture of Caco-2 and HT-29 cell lines), antimicrobial and antioxidant activities, and the production of exopolysaccharides.
Currently, antibiotics are used on a large scale, and their resistance is a serious problem. For this reason, both the EFSA and WHO/FAO issued regulations for testing the antibiotic profiles of probiotic and potentially probiotic microorganisms [9,20,26]. Evaluating the antibiotic resistance of potential probiotic microorganisms is important to limit the gene transfer for antibiotic resistance between common microbiota and pathogens [12]. All tested isolates were evaluated as resistant to the aminoglycoside antibiotics streptomycin and kanamycin, and the glycopeptide antibiotic vancomycin. Vancomycin is a glycopeptide antibiotic that inhibits cell wall synthesis [27]. However, vancomycin resistance genes are chromosomally coded; therefore, there is no risk of transfer as in the case of plasmids. Lactobacilli are generally vancomycin-resistant and naturally resistant to aminoglycoside antibiotics. Gentamicin is also an aminoglycoside antibiotic; however, only three strains in this study were resistant to gentamicin. Similar results have been reported in previous studies [28,29,30]. Sensitivity to gentamicin can be explained by its ability to cross the bacterial membrane more efficiently than the other aminoglycoside antibiotics (such as streptomycin or kanamycin) owing to differences in the chemical structures of antibiotics and their different interactions with bacterial membranes [31]. However, the tested isolates were sensitive to the remaining therapeutically important antibiotics. In the present study, no hemolytic activity was observed in any of the tested isolates. These results are in agreement with studies that observed hemolytic activity in lactobacilli isolated from the stool of infants [12], children [32], and adults [33], or from breast milk [2]. Probiotic microorganisms must also be examined for the production of appropriate enzymes to prevent the synthesis of toxic compounds and ensure host safety [34]. None of the isolates showed the activity of β-glucuronidase, which is a bacterial carcinogenic enzyme [34]. From a safety perspective, all tested isolates showed favorable properties.
One of the crucial criteria for selecting a potential probiotic strain is its ability to survive the inhospitable conditions of the human GIT (low pH and high levels of bile salts). Tested strains were exposed to simulated gastric juice containing pancreatin and two different concentrations of bile salts (1.5 and 3.0%) for 4 h. The concentration of bile salts in the human intestine can vary depending on the type of food intake and the secretion of pancreatic enzymes [35]. However, its average concentration ranges from 0.1 to 0.5% [36]. Lacticaseibacillus rhamnosus S1E and S4F, isolated from infant feces, showed the highest viability (>85%) after exposure to both concentrations of bile salts. Resistance to bile salts can influence a wide range of factors, such as bacterial cell wall structure, transcription, and the expression of proteins or BSH enzymes. Ali et al. [37] evaluated the probiotic properties of Limosilactobacillus reuteri PSC102 isolated from pig feces. This strain exhibited tolerance toward low pH and bile salts and did not produce harmful enzymes or possess hemolytic activity, similar to the strains used in this study. The strain L. rhamnosus WEI 30 and Lcb. rhamnosus WEI 41, isolated from piglets were also characterized by high resistance (>80%) to bile salts (3.0%) [38].
Adherence to the intestinal epithelium is important for the interaction between probiotic microorganisms and the host and influences the time of the potential beneficial effect of probiotics [39]. The adhesion properties of the newly isolated strains were evaluated by testing the hydrophobicity of the cell surface, auto-aggregation, and adherence to a co-culture of Caco-2 and HT-29 cells. Our results revealed significant differences in the hydrophobicity of the tested isolates. These differences were also observed within species and genera. This is also confirmed by the results described in the study of de Souza et al. [40], in which the hydrophobicity of different strains of Lacticaseibacillus casei ranged from 9.66 to 69.36%, similar to this work, and for L. fermentum strains, the hydrophobicity values ranged from 0.30 to 68.81%. Similar differences in the hydrophobicity of newly isolated probiotic strains have been reported by Krausová et al. [22]. Generally, these results indicate that the hydrophobicity of the cell surface is a unique property of each strain. De Souza et al. [40] reported that bacteria with higher hydrophobicity can adhere better to epithelial cells and significantly influence the microbial composition in the intestine. This claim is in contradiction to another study by Todorov/Dicks [41], which determined a high percentage of hydrophobicity (75–80%) of Lacticaseibacillus rhamnosus ST461BZ and ST462BZ; however, these strains proved a lower ability to adhere to HT-29 cell lines. In the present study, the strain Lactobacillus gasseri M6C, isolated from breast milk, showed the highest ability to adhere to Caco-2/HT-29 cells and the highest cell surface hydrophobicity to hexane. Nevertheless, other strains (Limosilactobacillus reuteri S2A and S2C) with higher adherence abilities had the lowest hydrophobicity. These results indicate that hydrophobic interactions may facilitate the adhesion process through initial interactions with the intestinal surface; however, they are not a precondition for high bacterial adherence. In addition to hydrophobicity, auto-aggregation may also be correlated with the ability to adhere. A correlation between these properties has been described in studies by MacKenzie et al. [42] and Krausová et al. [22], in which Limosilactobacillus reuteri strains showed high auto-aggregation and good adherence. In contrast, the correlation between auto-aggregation and adhesion capacity was refuted by Chaffanel et al. [43]. No association between aggregation and adhesion capacity was observed for any of the 14 Streptococcus salivarius strains tested. The results of this study suggest that the processes of auto-aggregation and adherence are probably mediated by distinct mechanisms involving different surface molecules. In this study, the auto-aggregation ability of isolates had a variable range between 5.8 and 28.5%, depending on time and strain. Lacticaseibacillus rhamnosus S1E and S6C showed the strongest percentages of auto-aggregation (28.5% and 27.2%) after 7 h.
Lactic acid bacteria produce a wide range of antimicrobial compounds including organic acids, bacteriocins, and hydrogen peroxide [44]. The mentioned production of organic acids (acetic acid, lactic acid, 3-phenyl lactic acid, 4-hydroxyphenyl lactic acid, and benzoic acid) leads to a decrease in pH, which can inhibit the growth of pathogens [21,45]. In this study, acidic supernatants inhibited the growth of the five tested pathogens (B. cereus, E. coli, E. faecalis, S. aureus, and Salmonella enterica serotype Typhimurium) probably because of the presence of organic acids and a low pH. The neutralization of these supernatants caused a significant decrease in their antimicrobial activity, similar to the results of other studies [21,46,47]. The heat treatment of neutralized supernatants did not affect their antimicrobial activity compared to neutralized supernatants. This indicates the presence of thermostable compounds with antimicrobial activity, such as thermostable bacteriocins. The thermostable bacteriocins (gassericin and plantaricin) produced by Lactobacillus gasseri and Lactiplatibacillus plantarum have been described previously [48,49].
Antioxidant activity is an important property by which cells protect themselves from oxidative stress [50]. Recently, LABs have been studied for their ability to actively participate in the reduction in reactive oxygen species (ROS) levels [51,52]. The DPPH radical scavenging method was used to determine the antioxidant capacity in our study. The two strains of Lacticaseibacillus rhamnosus S4F and S6C, isolated from infant feces, showed the highest antioxidant activity (69.3%). Kim et al. [8] also determined the antioxidant activities of strains of the genera Lactobacillus and Lacticaseibacillus that only reached values of 21.6 and 27.5%, respectively [8]. In another study by Oh et al. [53], the values of antioxidant activities of Lacticaseibacillus rhamnosus and Lactobacillus gasseri isolated from infant feces did not exceed 40% [53]. On the contrary, another study described antioxidant activity in the range of 76–88% for Lacticaseibacillus rhamnosus obtained from breast milk [16]. These results suggest that antioxidant activity is highly strain-specific and does not depend on the origin of the strain. In the future, it will be important to test the antioxidant activity of the isolates using other methods, such as scavenging of hydroxyl radicals or ABTS radical methods.
EPSs produced by lactic acid bacteria are generally considered GRASs and may represent a new generation of natural, healthy, and prebiotic food compounds. Owing to their unique physicochemical properties, they can replace many substances used to improve the sensory properties. Because of their ability to retain large amounts of water and form gel-like and viscous structures, EPS play an important role in the food, pharmaceutical, and cosmetic industries. However, their disadvantages include small quantities and difficult isolation and purification [54,55]. Some studies have described the positive health effects of EPS, such as effective cholesterol-lowering, antioxidant, anticancer, immunomodulatory, and prebiotic activities [56]. In the present study, EPS production was observed in all nine isolates in a medium containing all the tested substrates (glucose, lactose, and sucrose). The production of EPS from different carbohydrate sources has been observed in previous studies by Badel et al. [54], Oleksy-Sobczak et al. [57], and Rendueles et al. [58].
5. Conclusions
In conclusion, this study focused on the isolation and investigation of potential probiotic microorganisms from breast milk and infant feces. Nine strains were identified by 16S rRNA sequencing Lactobacillus gasseri, Lacticaseibacillus rhamnosus, and Limosilactobacillus reuteri. All tested isolates were non-hemolytic and sensitive to therapeutically important antibiotics. The acidic supernatants of the isolates inhibited selected pathogenic microorganisms (B. cereus, E. coli, S. aureus, Salmonella, and E. faecalis). Additionally, partial inhibition was observed after treatment with the neutralized and heated supernatants. However, a more complex characterization of the mechanisms of antimicrobial activities of the tested strains, such as the production of organic acids or bacteriocins, remains an area that needs further investigation and may be considered a limitation of this study. The evaluation of the results showed that the two strains, Lacticaseibacillus rhamnosus S1F and S6C, isolated from infant feces, together with Lactobacillus gasseri M8C, isolated from breast milk, showed the highest ability to survive in the presence of high concentrations of bile salts and a good adhesion property, which is important for the colonization of the GIT or the inhibition of pathogens. The three isolates mentioned here could be considered potential probiotics with future applications in the food industry. Nevertheless, other in vitro property tests need to be performed to confirm their probiotic status.
Author Contributions
Conceptualization, I.H.; Methodology, I.H., V.D. and J.C.; Formal analysis, I.H., V.D., J.C. and I.M.; Investigation, V.D.; Resources, G.K.; Data curation, A.K.; Writing—original draft, I.H. and J.C.; Writing—review & editing, G.K., A.K., J.S. and S.M.; Visualization, A.K.; Supervision, G.K. and S.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Agriculture of the Czech Republic Institutional Support No. MZE-RO1423 and no. QK1910024 and QK22010186.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are presented in the paper.
Conflicts of Interest
The authors of the manuscript “Functional and probiotic characterization of newly isolated strains from infant feces and breast milk” declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper of interest.
References
- Asan-Ozusaglam, M.; Gunyakti, A. A new probiotic candidate bacterium from human milk: Limosilactobacillus vaginalis MA-10. Acta Aliment. 2021, 50, 13–21. [Google Scholar] [CrossRef]
- Damaceno, Q.S.; Gallotti, B.; Reis, I.M.; Totte, Y.C.; Assis, G.B.; Figueiredo, H.C.; Silva, T.F.; Azevedo, V.; Nicoli, J.R.; Martins, F.S. Isolation and identification of potential probiotic bacteria from human milk. Probiotics Antimicrob. Proteins 2021, 15, 491–501. [Google Scholar] [CrossRef]
- de Melo Pereira, G.V.; de Oliveira Coelho, B.; Magalhães Júnior, A.I.; Thomaz-Soccol, V.; Soccol, C.R. How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv. 2018, 36, 2060–2076. [Google Scholar] [CrossRef]
- Singh, T.P.; Natraj, B.H. Next-generation probiotics: A promising approach towards designing personalized medicine. Crit. Rev. Microbiol. 2021, 47, 479–498. [Google Scholar] [CrossRef] [PubMed]
- Reuben, R.C.; Roy, P.C.; Sarkar, S.L.; Rubayet Ul Alam, A.S.M.; Jahid, I.K. Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties. J. Dairy Sci. 2020, 103, 1223–1237. [Google Scholar] [CrossRef] [PubMed]
- Sionek, B.; Szydłowska, A.; Zielińska, D.; Neffe-Skocińska, K.; Kołożyn-Krajewska, D. Beneficial bacteria isolated from food in relation to the next generation of probiotics. Microorganisms 2023, 11, 1714. [Google Scholar] [CrossRef]
- Shin, M.; Ban, O.H.; Jung, Y.H.; Yang, J.; Kim, Y. Genomic characterization and probiotic potential of Lactobacillus casei IDCC 3451 isolated from infant faeces. Lett. Appl. Microbiol. 2021, 72, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, J.S.; Kim, Y.; Jeong, Y.; Kim, J.E.; Paek, N.S.; Kang, C.H. Antioxidant and probiotic properties of Lactobacilli and bifidobacteria of human origins. Biotechnol. Bioprocess Eng. 2020, 25, 421–430. [Google Scholar] [CrossRef]
- FAO/WHO. Guidelines for the Evaluation of Probiotics in Food; Food and Agriculture Organization of the United Nations/World Health Organization: London, UK; London, ON, Canada, 2002. [Google Scholar]
- Asan-Ozusaglam, M.; Gunyakti, A. Lactobacillus fermentum strains from human breast milk with probiotic properties and cholesterol-lowering effects. Food Sci. Biotechnol. 2019, 28, 501–509. [Google Scholar] [CrossRef]
- Gunyakti, A.; Asan-Ozusaglam, M. Investigation of the potential use of Lactobacillus gasseri originated from human breast milk as food additive. LWT 2018, 93, 613–619. [Google Scholar] [CrossRef]
- Rodrigues da Cunha, L.; Ferreira, C.L.F.; Durmaz, E.; Goh, Y.J.; Sanozky-Dawes, R.; Klaenhammer, T. Characterization of Lactobacillus gasseri isolates from a breastfed infant. Gut Microbes 2012, 3, 15–24. [Google Scholar] [CrossRef]
- Rastogi, S.; Mittal, V.; Singh, A. In vitro assessment of antioxidant and antimicrobial potential of Lactobacillus gasseri strains isolated from human milk and infant faeces. J. Pure Appl. Microbiol. 2020, 14, 1305–1315. [Google Scholar] [CrossRef]
- Verdenelli, M.C.; Ghelfi, F.; Silvi, S.; Orpianesi, C.; Cecchini, C.; Cresci, A. Probiotic properties of Lactobacillus rhamnosus and Lactobacillus paracasei isolated from human faeces. Eur. J. Nutr. 2009, 48, 355–363. [Google Scholar] [CrossRef]
- Cheng, L.; Akkerman, R.; Kong, C.; Walvoort, M.T.C.; de Vos, P. More than sugar in the milk: Human milk oligosaccharides as essential bioactive molecules in breast milk and current insight in beneficial effects. Crit. Rev. Food Sci. Nutr. 2021, 61, 1184–1200. [Google Scholar] [CrossRef]
- Riaz Rajoka, M.S.; Mehwish, H.M.; Siddiq, M.; Haobin, Z.; Zhu, J.; Yan, L.; Shao, D.; Xu, X.; Shi, J. Identification, characterization, and probiotic potential of Lactobacillus rhamnosus isolated from human milk. Lebensm. Wiss. Technol. 2017, 84, 271–280. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Sui, Y.; Liu, J.; Liu, Y.; Wang, Y.; Xiao, Y.; Gao, B.; Zhu, D. In Vitro probiotic characterization of Lactobacillus strains from fermented tangerine vinegar and their cholesterol degradation activity. Food Biosci. 2021, 39, 100843. [Google Scholar] [CrossRef]
- Sharma, C.; Gulati, S.; Thakur, N.; Singh, B.P.; Gupta, S.; Kaur, S.; Mishra, S.K.; Puniya, A.K.; Gill, J.P.S.; Panwar, H. Antibiotic sensitivity pattern of indigenous lactobacilli isolated from curd and human milk samples. 3 Biotech 2017, 7, 53. [Google Scholar] [CrossRef] [PubMed]
- Kostelac, D.; Gerić, M.; Gajski, G.; Markov, K.; Domijan, A.M.; Čanak, I.; Jakopović, Ž.; Svetec, I.K.; Žunar, B.; Frece, J. Lactic acid bacteria isolated from equid milk and their extracellular metabolites show great probiotic properties and anti-inflammatory potential. Int. Dairy J. 2021, 112, 104828. [Google Scholar] [CrossRef]
- Krausová, G.; Hyršlová, I.; Hynštová, I. In Vitro evaluation of adhesion capacity, hydrophobicity, and auto-aggregation of newly isolated potential probiotic strains. Fermentation 2019, 5, 100. [Google Scholar] [CrossRef]
- Musilova, S.; Modrackova, N.; Doskocil, I.; Svejstil, R.; Rada, V. Influence of human milk oligosaccharides on adherence of bifidobacteria and clostridia to cell lines. Acta Microbiol. Immunol. Hung. 2017, 64, 415–422. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hyrslova, I.; Kana, A.; Kantorova, V.; Krausova, G.; Mrvikova, I.; Doskocil, I. Selenium accumulation and biotransformation in Streptococcus, Lactococcus, and Enterococcus strains. J. Funct. Foods 2022, 92, 105056. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- EFSA. Technical guidance—Update of the criteria used in the assessment of bacterial resistance to antibiotics of human or veterinary importance. Eur. Food Saf. Authority J. 2008, 6, 732. [Google Scholar]
- Morandi, S.; Silvetti, T.; Brasca, M. Biotechnological and safety characterization of Enterococcus lactis, a recently described species of dairy origin. Antonie Leeuwenhoek 2013, 103, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Liasi, S.; Azmi, T.; Hassan, M.; Shuhaimi, M.; Rosfarizan, M.; Ariff, A. Antimicrobial activity and antibiotic sensitivity of three isolates of lactic acid bacteria from fermented fish product, Budu. Malays. J. Microbiol. 2009, 5, 33–37. [Google Scholar]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2019, 85, e01738-18. [Google Scholar] [CrossRef]
- Georgieva, R.; Yocheva, L.; Tserovska, L.; Zhelezova, G.; Stefanova, N.; Atanasova, A.; Danguleva, A.; Ivanova, G.; Karapetkov, N.; Rumyan, N.; et al. Antimicrobial activity and antibiotic susceptibility of Lactobacillus and Bifidobacterium spp. intended for use as starter and probiotic cultures. Biotechnol. Biotechnol. Equip. 2015, 29, 84–91. [Google Scholar] [CrossRef]
- Suhonen, A. Antibiotic Susceptibility of Lactic Acid Bacteria. 2019. Available online: http://urn.fi/URN:NBN:fi:hulib-201905081914 (accessed on 20 September 2023).
- Elkins, C.A.; Mullis, L.B. Bile-mediated aminoglycoside sensitivity in Lactobacillus species likely results from increased membrane permeability attributable to cholic acid. Appl. Environ. Microbiol. 2004, 70, 7200–7209. [Google Scholar] [CrossRef]
- Kõll, P.; Mändar, R.; Smidt, I.; Hütt, P.; Truusalu, K.; Mikelsaar, R.H.; Shchepetova, J.; Krogh-Andersen, K.; Marcotte, H.; Hammarström, L.; et al. Screening and evaluation of human intestinal lactobacilli for the development of novel gastrointestinal probiotics. Curr. Microbiol. 2010, 61, 560–566. [Google Scholar] [CrossRef]
- Vesterlund, S.; Vankerckhoven, V.; Saxelin, M.; Goossens, H.; Salminen, S.; Ouwehand, A.C. Safety assessment of Lactobacillus strains: Presence of putative risk factors in faecal, blood and probiotic isolates. Int. J. Food Microbiol. 2007, 116, 325–331. [Google Scholar] [CrossRef]
- Jeong, Y.; Kim, H.; Lee, J.Y.; Won, G.; Choi, S.I.; Kim, G.H.; Kang, C.H. The antioxidant, anti-diabetic, and anti-adipogenesis potential and probiotic properties of lactic acid bacteria isolated from human and fermented foods. Fermentation 2021, 7, 123. [Google Scholar] [CrossRef]
- Shokryazdan, P.; Sieo, C.C.; Kalavathy, R.; Liang, J.B.; Alitheen, N.B.; Faseleh Jahromi, M.; Ho, Y.W. Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. BioMed. Res. Int. 2014, 2014, 927268. [Google Scholar] [CrossRef]
- Reis, N.A.; Saraiva, M.A.F.; Duarte, E.A.A.; de Carvalho, E.A.; Vieira, B.B.; Evangelista-Barreto, N.S. Probiotic properties of lactic acid bacteria isolated from human milk. J. Appl. Microbiol. 2016, 121, 811–820. [Google Scholar] [CrossRef]
- Ali, M.S.; Lee, E.B.; Lim, S.K.; Suk, K.; Park, S.C. Isolation and identification of Limosilactobacillus reuteri PSC102 and evaluation of its potential probiotic, antioxidant, and antibacterial properties. Antioxidants 2023, 12, 238. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Qiao, L.; Liu, R.; Yao, H.; Gao, C. Potential probiotic properties of lactic acid bacteria isolated from the intestinal mucosa of healthy piglets. Ann. Microbiol. 2017, 67, 239–253. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Rastall, R.A.; Gibson, G.R.; Charalampopoulos, D.; Chatzifragkou, A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 2019, 103, 6463–6472. [Google Scholar] [CrossRef]
- de Souza, B.M.S.; Borgonovi, T.F.; Casarotti, S.N.; Todorov, S.D.; Penna, A.L.B. Lactobacillus casei and Lactobacillus fermentum strains isolated from mozzarella cheese: Probiotic potential, safety, acidifying kinetic parameters and viability under gastrointestinal tract conditions. Probiotics Antimicrob. Proteins 2019, 11, 382–396. [Google Scholar] [CrossRef]
- Todorov, S.D.; Dicks, L.M.T. Evaluation of lactic acid bacteria from kefir, molasses and olive brine as possible probiotics based on physiological properties. Ann. Microbiol. 2008, 58, 661–670. [Google Scholar] [CrossRef]
- MacKenzie, D.A.; Jeffers, F.; Parker, M.L.; Vibert-Vallet, A.; Bongaerts, R.J.; Roos, S.; Walter, J.; Juge, N. Strain-specific diversity of mucus-binding proteins in the adhesion and aggregation properties of Lactobacillus reuteri. Microbiology 2010, 156, 3368–3378. [Google Scholar] [CrossRef] [PubMed]
- Chaffanel, F.; Charron-Bourgoin, F.; Soligot, C.; Kebouchi, M.; Bertin, S.; Payot, S.; Le Roux, Y.; Leblond-Bourget, N. Surface proteins involved in the adhesion of Streptococcus salivarius to human intestinal epithelial cells. Appl. Microbiol. Biotechnol. 2018, 102, 2851–2865. [Google Scholar] [CrossRef]
- González, L.; Sandoval, H.; Sacristán, N.; Castro, J.M.; Fresno, J.M.; Tornadijo, M.E. Identification of lactic acid bacteria isolated from Genestoso cheese throughout ripening and study of their antimicrobial activity. Food Control 2007, 18, 716–722. [Google Scholar] [CrossRef]
- Šušković, J.; Kos, B.; Beganović, J.; Leboš Pavunc, A.; Habjanič, K.; Matošić, S. Antimicrobial activity–the most important property of probiotic and starter lactic acid bacteria. Food Technol. Biotechnol. 2010, 48, 296–307. [Google Scholar]
- Davoodabadi, A.; Dallal, M.M.S.; Foroushani, A.R.; Douraghi, M.; Harati, F.A.; Amin Harati, F. Antibacterial activity of Lactobacillus spp. isolated from the feces of healthy infants against enteropathogenic bacteria. Anaerobe 2015, 34, 53–58. [Google Scholar] [CrossRef]
- Frece, J.; Markov, K.; Kovačević, D. Determination of indigenous microbial populations, mycotoxins and characterization of potential starter cultures in Slavonian kulen. Meso Prvi Hrvatski Cas. Mesu 2010, 12, 92–99. [Google Scholar]
- Zhang, Q. Lactic Acid Bacteria and Bacteriocins. Lactic Acid Bacteria; Springer: Berlin/Heidelberg, Germany, 2019; pp. 61–91. [Google Scholar]
- Ratsep, M.; Naaber, P.; Koljalg, S.; Smidt, I.; Shkut, E.; Sepp, E. Effect of Lactobacillus plantarum strains on clinical isolates of Clostridium difficile in vitro. J. Probiotics Health 2014, 2, 1–5. [Google Scholar] [CrossRef]
- Mahmoudi, I.; Telmoudi, A.; Hassouna, M. Beneficial effects of probiotic Lactobacillus plantarum isolated from cow, goat and sheep raw milks. Acta Sci. Microbiol. 2018, 1, 17–20. [Google Scholar] [CrossRef]
- Kim, H.S.; Chae, H.S.; Jeong, S.G.; Ham, J.S.; Im, S.K.; Ahn, C.N.; Lee, J.M. In Vitro antioxidative properties of lactobacilli. Asian-Australas. J. Anim. Sci. 2006, 19, 262–265. [Google Scholar] [CrossRef]
- Pieniz, S.; Andreazza, R.; Anghinoni, T.; Camargo, F.; Brandelli, A. Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 2014, 37, 251–256. [Google Scholar] [CrossRef]
- Oh, N.S.; Joung, J.Y.; Lee, J.Y.; Kim, Y. Probiotic and anti-inflammatory potential of Lactobacillus rhamnosus 4B15 and Lactobacillus gasseri 4M13 isolated from infant feces. PLoS ONE 2018, 13, e0192021. [Google Scholar] [CrossRef]
- Badel, S.; Bernardi, T.; Michaud, P. New perspectives for lactobacilli exopolysaccharides. Biotechnol. Adv. 2011, 29, 54–66. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; De Vin, F.; Vaningelgem, F.; Degeest, B. Recent developments in the biosynthesis and applications of heteropolysaccharides from lactic acid bacteria. Int. Dairy J. 2001, 11, 687–707. [Google Scholar] [CrossRef]
- Farag, M.M.S.; Moghannem, S.A.M.; Shehabeldine, A.M.; Azab, M.S. Antitumor effect of exopolysaccharide produced by Bacillus mycoides. Microb. Pathog. 2020, 140, 103947. [Google Scholar] [CrossRef] [PubMed]
- Oleksy-Sobczak, M.; Klewicka, E.; Piekarska-Radzik, L. Exopolysaccharides production by Lactobacillus rhamnosus strains—Optimization of synthesis and extraction conditions. Lebensm. Wiss. Technol. 2020, 122, 109055. [Google Scholar] [CrossRef]
- Rendueles, O.; Kaplan, J.B.; Ghigo, J.M. Antibiofilm polysaccharides. Environ. Microbiol. 2013, 15, 334–346. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).