Abstract
The solvent-producing bacterium Clostridium acetobutylicum is able to grow on a variety of carbohydrates. The main hexose transport system is the phosphoenolpyruvate-dependent phosphotransferase system (PTS). When the gene glcG that encodes the glucose transporter was inactivated, the resulting mutant glcG::int(1224) grew as well as the wild type, yet its glucose consumption was reduced by 17% in a batch fermentation. Transcriptomics analysis of the phosphate-limited continuous cultures showed that the cellobiose transporter GlcCE was highly up-regulated in the mutant glcG::int(1224). The glcCE mutation did not affect growth and even consumed slightly more glucose during solventogenesis growth compared to wild type, indicating that GlcG is the primary glucose-specific PTS. Poor growth of the double mutant glcG::int(1224)-glcCE::int(193) further revealed that GlcCE was the secondary glucose PTS and that there must be other PTSs capable of glucose uptake. The observations obtained in this study provided a promising foundation to understand glucose transport in C. acetobutylicum.
1. Introduction
Clostridium acetobutylicum is able to grow on a variety of carbohydrates and thus encodes a diverse set of metabolic enzymes and transporters for hexoses and pentoses uptake. Transcriptional analysis of differential carbohydrate utilization by C. acetobutylicum unveiled that the hexoses were primarily taken up by the phosphoenolpyruvate phosphotransferase systems (PTS) [1,2]. A typical PTS contains two soluble components, enzyme I (EI), the histidine-containing protein (HPr) and a membrane-bound protein enzyme II (EII).
The EII complex incorporates three domains, termed IIA, IIB and IIC, in which the IIC is integral membrane protein to translocate carbohydrate into cytoplasm [3]. They are mainly responsible for the absorption of a large number of carbohydrates (such as hexose, 6- deoxyhexose, amino sugar, N-acetylamino sugar, gluconic acid, pentitol, ascorbic acid and disaccharides) in the process of energy transportation and catalyze their conversion into their respective phosphate esters.
Nevertheless, the diffusion of other carbohydrates, mainly pentose, is driven by ATP hydrolysis or ion gradients [4,5]. Non-PTS could also be used to transport a small amount of glucose if all PTS were deficient [6]. PTS is not only a sugar transport system (which mediates the intake and phosphorylation of carbohydrates) but also has a very strong regulatory capacity. PTS participates in the regulation of the central carbon and nitrogen metabolism [7,8,9,10,11,12], the regulation of metal ion homeostasis [13,14,15] and the regulation of bacterial virulence [16,17,18]. Increasingly, studies show that PTS is closely related to the stress response [19,20,21,22].
Bioinformatic analysis of the genome sequence of C. acetobutylicum ATCC 824, which was published in 2001, identified 13 putative PTS EII enzymes in this strain [1,23]. Although the family of these 13 putative PTS EII domains were assigned by phylogenetic analysis and the probable substrates were inferred, a considerable amount of functional characters were still required. For glucose uptake, it was predicted that the glcG gene (CAC0570) coded for a glucose transporter in C. acetobutylicum. However, after glcG was inactivated, the mutant fermented glucose as efficiently as the parent strain in a batch culture [6,24]. This result suggested that an additional glucose transporter exists in this organism.
Despite its importance, the lack of basic information of glucose uptake systems in C. acetobutylicum has limited the understanding of glucose assimilation in this strain. In contrast, glucose uptake in Escherichia coli had been studied extensively. There is a main glucose transporter that is very efficient for glucose uptake, and the maltose, mannose and galactose transport systems are also able to transport glucose into the cytoplasm [25,26,27]. Therefore, it of interest to investigate whether a similar glucose transport mechanism exists in C. acetobutylicum.
Moreover, a more detailed physiological analysis will further increase our appreciation of the importance of the PTS in this organism [28]. The complex metabolism of C. acetobutylicum is characterized by two sequential phases in a batch culture [29,30]. This culture grows fast during acidogenesis, and the main products are acetate and butyrate. Then, solventogenesis occurs as the growth rate slows down, the acids are re-assimilated, and the solvents (butanol, acetone and ethanol) become main products. The physiological process and metabolic change have been investigated extensively in batch fermentations [29,31,32,33].
However, some details cannot be observed easily due to the unstable growth rate, pH and glucose consumption of a batch culture. Phosphate-limited continuous fermentation (chemostat cultivation) of C. acetobutylicum makes it possible to keep the cells in a steady acidogenesis or solventogenesis simply by controlling the pH [34,35,36]. Since all the cells are identical in continuous culture, this is preferred to be used for ‘omics’ studies.
DNA microarray is a powerful tool to monitor the expression of all the genes of bacteria. It is commonly used to investigate bacterial metabolism and gene function [37]. In the present work, the ClosTron technology was used to inactivate the genes of glucose transporter candidates. The mutants and wild type strain were cultivated in batch and continuous cultures. Then, the growth phenotype and transcriptome analysis of the mutants were compared to the wild type, revealing a series of PTSs and their preference order in C. acetobutylicum.
2. Materials and Methods
2.1. Bacterial Strains and Generation of ClosTron Mutants
All bacterial strains and plasmids in this study are listed in Table S1. C. acetobutylicum ATCC 824 was stored as spore suspensions at 4 °C. Cultures were inoculated with spores in clostridial growth medium (CGM) [38] and heat-shocked at 80 °C for 10 min, and then the strain was cultured at 37 °C overnight as a pre-culture. Procedures requiring anaerobic conditions were performed in an anaerobic chamber (COY system, Grass Lake, MI, USA). Hungate tubes (Ochs GmbH, Bovenden, Germany), and serum bottles (Müller and Krempel AG, Bülach, Switzerland) were used for liquid cultivation in CGM.
Chromosomal genes of C. acetobutylicum were inactivated using ClosTron technology. The targeted genes in present study were glcG (encoding PTS EII and CAC0570) and glcCE (encoding PTS cellobios-specific component IIC and CAC0386). The glcG gene encodes three typical domains, IIA, IIB and IIC. The mutant C. acetobutylicum glcG::int(1224) was obtained using the ClosTron technology, and the insertion site was targeted to domain C (IIC). The gene glcCE is located in a putative cellobiose transporter operon (CAC0383-CAC0386), and CAC0386 is annotated as domain C (IIC) in this operon. As the integral membrane protein, IIC takes charge of translocating the substrate to cytoplasm, which is the targeted insertion to make the mutant.
The procedure of construction and electroporation of the retargeted plasmid pMTL007C-E2 was as described previously [39,40,41]. For the double mutant (glcG::int(1224)-glcCE::int(193)), the mutant glcG was used as the parent strain. The retargeted plasmid pMTL007C-E2 of glcCE and the plasmid pAN2 were transformed to the mutant C. acetobutylicum glcG::int(1224), and the transformants were selected using the procedure described for ClosTron technology. The ClosTron mutants were verified by gene check primers listed in Table S1, and the correct insertion sites of intron were validated by Southern hybridization.
Southern hybridization was performed by digesting chromosomal DNA overnight with the HindIII and running on a 0.8% agarose gel. Transfer and hybridization was performed according to standard protocols as described previously with an intron-specific probe, which was generated from the PCR product using the primers intron II-F and intron II-R listed in Table S1 with pMTL007C-E2 plasmid DNA as the template [42]. In addition, the ClosTron mutants were stored at −80 °C in CGM containing 30% glycerol.
2.2. Batch and Continuous Fermentation Experiments
Batch fermentations were performed in minimal medium at 37 °C using 50 g/L glucose or cellobiose as the carbon source. Two independent cultivations were performed for each strain (wild type or mutants), and the average value of the OD600 was used for the growth curve. The minimal medium was prepared as described previously in 1 L distilled water containing the following ingredients: glucose, 50 g; (NH4)2SO4, 2 g; KH2PO4, 1 g; K2HPO4, 1 g; MgSO4·7H2O, 0.1 g; NaCl, 0.01 g; NaMoO4·2H2O, 0.01 g; CaCl2·2H2O, 0.01 g; MnSO4·H2O, 0.015 g; FeSO4·7H2O, 0.015 g; biotin, 0.1 mg; thiamine hydrochloride, 2 mg; and p-aminobenzoic acid, 2 mg [35].
The phosphate-limited minimal medium is a modified minimal medium used in phosphate-limited continuous fermentation. Phosphate is the limiting factor; thus, there is no K2HPO4 and only 0.07 g/L KH2PO4 (0.5 mM) in the phosphate-limited minimal medium. The pH of the medium was adjusted to 2 using H2SO4, and the glucose concentration in this medium was 40 g/L [43]. The continuous fermentation experiment was performed in a one-liter Biostat B plus fermenter system (Sartorius BBI Systems, Milsungen, Germany) with 0.75 liter working volume.
For the continuous fermentation, the stains (wild type and mutants) were cultured in minimal medium for about 12 h in the fermenter, and then the fermenter was connected to the phosphate-limited minimal medium to start the continuous fermentation. The dilution rate was 0.075 h−1, and the pH was adjusted automatically to 5.7 for acidogenic growth and 4.5 for solventogenic growth by adding 2 M KOH [36,44]. Cell pellets, for RNA isolation, were collected by centrifugation at −20 °C and 9000 rpm (Hettich Universal 320 R, Tuttlingen, Germany) for 15 min. Then, the pellets without supernatant were quickly frozen in liquid nitrogen and stored at −80 °C until RNA extraction.
2.3. Analytical Methods
Cell growth was determined by measuring the optical density at 600 nm in an Ultraspec 3300 pro UV/vis spectrophotometer (GE-Healthcare, Munich, Germany). The glucose in culture was determined by D-Glucose (GOPOD Format) Kit (Megazyme, Wicklow, Ireland).
2.4. DNA Microarray Experiments
The array contained 3840 oligonucleotides representing 99.8% of all annotated open reading frames (ORFs) in C. acetobutylicum, including all ORFs located on the megaplasmid (pSOL). Moreover, 13 putative PTS EII enzymes are encoded by 26 genes, which are all included in the array [1]. RNA isolation and DNAse treatment procedures were performed as previously described in detail [36]. For a microarray experiment in the present study, RNA was isolated from wild type and the mutant cultivated at the same fermentation condition and labeled with different fluorescent dyes by reverse transcription.
Therefore, one RNA (DNA-free) sample of wild type was normally labeled with Cy-3 (GE Healthcare, Munich, Germany), and the other RNA sample of the mutant was labeled with Cy-5 (GE Healthcare, Munich, Germany). The labeled samples were then mixed and hybridized with the array. The cDNA labeling, hybridization, DNA chips preparation, data normalization (GenePix Pro 6.0 software, Axon Instruments, Seattle, WA, USA) and data analysis were described in detail previously [36,37]. The transcriptional data of expression ratios of all the 26 genes in PTS were reproduced in two independent biological experiments. The average of two expression ratios of one gene was the final ratio of this gene in the present study.
3. Results and Discussion
3.1. Generation and Verification of the Mutants C. acetobutylicum glcG::int(1224), glcCE::int(193) and glcG::int(1224)-glcCE::int(193)
To investigate the glucose transporters in C. acetobutylicum, glcG and glcCE were targeted for insertion inactivation using ClosTron mutagenesis as described previously [39,40]. ClosTron technology is a proven system to construct single gene knockouts in clostridia, especially in C. acetobutylicum [41,45]. To allow the construction of multiple ClosTron mutants in C. acetobutylicum, the ermB gene is used as a selection marker. It can be deliberately excised by FLP-recombinase, and thus the mutants lose the erythromycin resistance [39].
The length of glcG was 1998 bp, and the target site was designed to be located at 1224/1225 bp. The length of glcCE was 1353 bp, and the target site was designed to be located at 193/194 bp. The strains, plasmids and primers used are listed in Table S1. Putative mutants (glcG::int(1224) and glcCE::int(193)) were obtained by screening the CGM plates. Erythromycin-resistant clones (putative mutants) were verified by gene check primers (CAC0570F, CAC0570R and CAC0386F, CAC0386R) as listed in Table S1. The results showed that wild type genome DNA gave a band of ~300 bp, while the correct mutants exhibited products of ~2.1 kb, which were 1.8 kb longer than the wild type (Figure S1).
For the double mutant (glcG::int(1224)-glcCE::int(193)), the correct retargeted plasmids, pMTL007C-E2-glcCE (Table S1) was transformed into the mutant C. acetobutylicum glcG::int(1224). Since the parent strain, C. acetobutylicum glcG::int(1224) already contained the erythromycin marker, the concentration of erythromycin used for selection increased from 5 to 15 μg/μL. The colonies obtained on the selection plates that grew faster were inoculated into fresh CGM medium and cultured overnight at 37 °C. Then, these colonies were verified by PCR using gene check primers (CAC0570F, CAC0570R and CAC0386F, CAC0386R) as listed in Table S1. The correct mutants exhibited products of ~2.1 kb, which were 1.8 kb longer than those of the wild type (Figure S1).
To further investigate the insertion sites of the intron, the two 2.1 kb PCR products from genomic DNA of clones containing the insertion in glcG and glcCE, respectively, were sequenced. Analysis of the sequences proved that the intron inserted the glcG at 1224/1225 bp and the glcCE at 193/194 bp, respectively.
These results verified that glcG and glcCE were inactivated by insertion of the ClosTron-derived group II intron into the desired positions and the correct glcG and glcCE insertion mutants were obtained. To further confirm that only one copy of the intron existed in the chromosomal DNA of the mutants C. acetobutylicum glcG::int(1224) and glcCE::int(193), Southern hybridization was performed with an intron-specific probe (Table S1). For the double mutant (glcG::int(1224)-glcCE::int(193)), two copies of the intron were inserted into the chromosomal DNA, showing two bands on the nylon membrane (Figure S2).
No hybridization signal was observed for wild type genomic DNA, and genomic DNA of the glcG::int(1224) and glcCE::int(193) mutants both exhibited a single band, demonstrating that strains glcG::int(1224) and glcCE::int(193) possessed only one copy of the intron on the chromosome. Two bands appeared in the lane of the double mutant, with one the same as that of the mutant glcG::int(1224) and the other identical to that of the mutant glcCE::int(193) (Figure S2), which verified that two copies of intron were found in the chromosomal DNA of the mutant glcG::int(1224)-glcCE::int(193) and that the two introns were inserted into glcG and glcCE genes, respectively.
3.2. The Phenotypes of Wild Type and the Mutant C. acetobutylicum glcG::int(1224) in Batch and Phosphate-Limited Continuous Fermentations
The mutant C. acetobutylicum glcG::int(1224) cultured in minimal medium with 60 g/L glucose exhibited similar growth phenotypes; however, the glucose consumption was reduced by ~17% compared to wild type (Figure 1), which led to an ~25% reduction in butanol production (Figure S3A,B). Under the same conditions, wild type and the mutant glcG::int(1224) achieved steady-state growth during acidogenic and solventogenic growth in the continuous fermentation (Figure 2). The optical density of the wild type was almost identical to that observed for the mutant glcG::int(1224) during acidogenic and solventogenic growth.
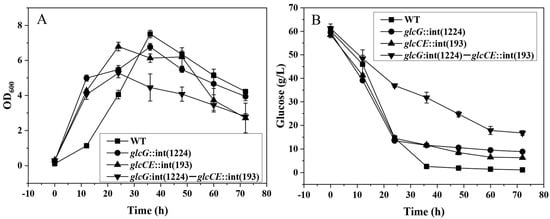
Figure 1.
Growth curves (A) and residual glucose concentrations (B) of C. acetobutylicum wild type, mutants glcG::int(1224), glcCE::int(193) and glcG::int(1224)-glcCE::int(193) in minimal medium with glucose as the sole carbon source. (■), Wild type; (●), glcG::int(1224); (▲), glcCE::int(193); and (▼), glcG::int(1224)-glcCE::int(193).
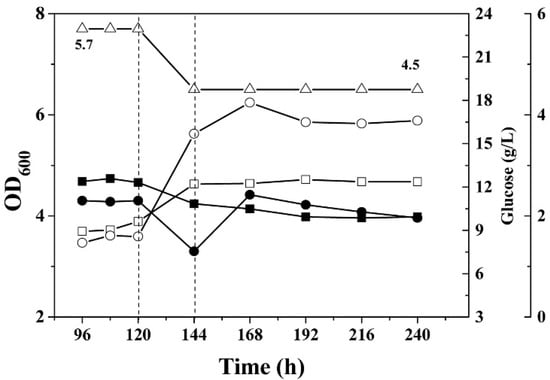
Figure 2.
Growth curves and residual glucose concentration in phosphate-limited continuous culture of C. acetobutylicum wild type and the mutant glcG::int(1224). (△), pH; (■), OD600 of wild type; (●) OD600 of glcG::int(1224); (□), residual glucose during wild type cultivation; and (○), residue glucose during cultivation of glcG::int(1224). The dashed line indicates the initiation of dynamic pH shift from 5.7 to 4.5 in continuous fermentation.
There was 40 g/L glucose in the phosphate-limited continuous medium. Assays of the residual glucose concentration in the continuous culture indicated approximately 9 g/L glucose during acidogenic growth for the wild type and the mutant glcG::int(1224), while approximately 12 g/L glucose was determined for the wild type and 17.5 g/L glucose for the mutant glcG::int(1224) in the continuous culture during solventogenic growth.
Due to inactivation of the glcG gene, the rate of glucose uptake by the mutant glcG::int(1224) was lower than that of wild type; additionally, some previous studies showed the key role of GlcG in glucose transport [6,24,46], which suggested that GlcG (encoded by glcG, CAC0570) indeed acted as EII of PTS for glucose transport, that an alternative PTS can compensate the loss of glcG in C. acetobutylicum, and that some of them may be bifunctional, such as those in E. coli, S. cerevisiae and other microorganisms [26,27,47].
Moreover, the fermentation of C. acetobutylicum can be divided into two distinctive growth phases [30,33]. In most studies, the characterization of mutants is done in batch cultures [41,45]. However, this does not allow characterization of differences between wild type and mutants specific for acidogenic or solventogenic growth. Phosphate-limited continuous fermentations make it possible to compare the phenotypes of wild type and the mutants during two-phase growth [34,35,36].
In present work, the results showed that there was a great advantage in studying the glucose transporters in C. acetobutylicum using phosphate-limited continuous fermentation. The glucose consumption of wild type and the mutant glcG::int(1224) were identical in batch fermentations (Figure 1A), whereas they were clearly different during the solventogenic growth in phosphate-limited continuous fermentation (Figure 2). Due to the same growth rate of all the cells in the continuous culture, it was also important to analyze the expression profile differences caused by the mutation between the wild type and the mutant.
3.3. Transcription Analysis of the Mutant C. acetobutylicum glcG::int(1224) as Compared to Wild Type
To test whether another PTS can compensate for the inactivation of GlcG, DNA microarray experiments were conducted to detect changes in the transcription level of all the 13 PTS EII protein genes caused by the glcG mutation compared to wild type. Cells of steady-state acidogenic growth (t = 120 h) and solventogenic growth (t = 240 h) of the chemostat culture of the mutant glcG::int(1224) and wild type were used for microarray experiments (Figure 2). The microarray data of all the 13 PTS EII protein genes are listed in Table 1.

Table 1.
Transcriptional profiles of all the PTS enzyme II protein genes of mutants C. acetobutylicum glcG::int(1224) and glcCE::int(193) compared to the wild type.
Assays of the transcription profiles of all the PTS EII protein genes of the mutant glcG::int(1224) compared to the wild type indicated that the gene expression of five PTSs were up-regulated (Table 1). Among these five PTSs, a cluster of genes (CAC0383-CAC0386), encoding a putative cellobiose-specific transporter exhibited the most pronounced elevated transcription in the mutant glcG::int(1224) during acidogenic and solventogenic growth in continuous culture.
In addition, the transcription of genes encoding a fructose-specific transporter (CAC0233-CAC0234) also increased compared to the wild type. Furthermore, slight changes in the expression of the other three PTSs occurred, including a putative mannitol-specific transporter (CAC0154-CAC0156), a putative N-acetylglucosamine-specific transporter (CAC1353-CAC1354) and another putative glucose-specific transporter (CAC3425-CAC3427). These results suggested that the putative cellobiose transporter is able to transport glucose in the cell in the case of a GlcG inactivation.
3.4. The Phenotypes and Transcription Analysis of Wild Type and the Mutant C. acetobutylicum glcCE::int(193) in Batch and Phosphate-Limited Continuous Fermentations
Sequence analysis suggested that the operon (CAC0383-CAC0386) encodes a cellobiose transporter in C. acetobutylicum [1]. However, there was no experimental evidence to prove this in vivo until now. The gene glcCE (CAC0386) encoding a PTS EIIC in this operon was inactivated by ClosTron insertion and the resulting mutant glcCE::int(193) showed poor growth in the batch fermentation with cellobiose as the sole carbon source (Figure 3). The highest optical density the mutant glcCE::int(193) achieved was approximately 4, which was significantly lower than that of wild type with the highest optical density of 9 in the batch culture with cellobiose as the sole carbon source.
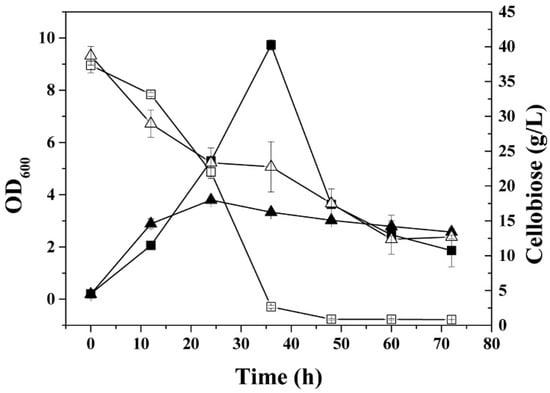
Figure 3.
Growth curves and residual cellobiose concentrations of the C. acetobutylicum wild type and the mutant glcCE::int(193) cultured in minimal medium with cellobiose as the sole carbon source. (■), OD600 of wild type; (▲), OD600 of glcCE::int(193); (□), residual cellobiose during cultivation of wild type; and (△), residual cellobiose during cultivation of glcCE::int(193).
There was still 30% of cellobiose left in the glcCE::int(193) culture, which suggested that the transporter encoded by the operon (CAC0383-CAC0386) was indeed a cellobiose-specific transporter (Figure 3). Whereas the weak growth of glcCE::int(193) on cellobiose and the 70% cellobiose consumption suggested there is a secondary cellobiose transporter in C. acetobutylicum, which needs to be identified in a future study.
To test whether this cellobiose-specific transporter is able to transport glucose, the mutant glcCE::int(193) was cultured in minimal medium in the presence of 60 g/L glucose and in a phosphate-limited continuous culture. The results illustrated that the mutant glcCE::int(193) and wild type exhibited similar growth phenotypes in both batch and continuous fermentation (Figure 1 and Figure 4) and that the concentration of butanol was nearly identical (Figure S3C), suggesting that the glcCE mutation did not affect the growth and glucose uptake of the strain. The mutant glcCE::int(193) even consumed slightly more glucose and produced more butanol during solventogenesis growth compared to that of wild type in the continuous fermentation (Figure 4 and Table S2), indicating that energy consumption of each glucose transporter was different.
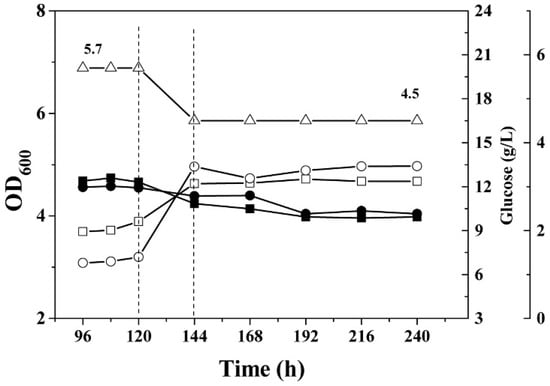
Figure 4.
Growth curves and residual glucose concentration in phosphate-limited continuous culture of the C. acetobutylicum wild type and the mutant glcCE::int(193). (△), pH; (■), OD600 of wild type; (●) OD600 of glcCE::int(193); (□), residual glucose during cultivation of wild type; and (○), residual glucose during cultivation of glcCE::int(193). The dashed line indicates the initiation of dynamic pH shift from 5.7 to 4.5 in the continuous fermentation.
On the other side, the growth and sugar consumption of the mutant glcCE::int(193) both decreased in the batch culture with cellobiose as the sole carbon source (Figure 3) and the concentration of acetone and butanol were significantly less as compared to those of wild type (Figure S4), indicating that one of the PTSs of C. acetobutylicum, GlcCE, played a key role in cellobiose transport. To further investigate the effects the of glcCE mutation, DNA microarray experiments were conducted to detect transcription level changes of all 13 PTS EII protein genes compared to the wild type.
During acidogenic and solventogenic stead-state growth of the mutant glcCE::int(193) on glucose, the transcriptional levels of genes for the glucose transporter (glcG, CAC0570) and a putative mannitol-specific transporter (CAC0154-CAC0156) were identical to that of wild type (Table 1). The genes coding for a putative fructose-specific transporter (CAC0233-CAC0234), a putative N-acetylglucosamine-specific transporter (CAC1353-CAC1354) and a putative glucose-specific transporter (CAC3425-CAC3427) were even slightly repressed compared to the wild type.
These results confirmed that GlcG (CAC0570) is the predominate glucose transporter in C. acetobutylicum. When it worked normally in the culture with glucose as carbon source, all of the other PTSs retired to the second line. To prove that the cellobiose transporter can also transport glucose, it was necessary to construct a double mutant in which both of the glcG and glcCE genes were inactivated. In addition, the genes for GlcCE (CAC0383-CAC0386) in wild type grown on cellobiose were up-regulated compared to those grown on glucose, indicating that GlcCE is the primary cellobiose transporter of C. acetobutylicum. The operon (CAC2951-2956) in mutant glcCE::int(193) grown on cellobiose were up-regulated compared to the wild type, implying that CAC2956 may encode the secondary cellobiose transporter of C. acetobutylicum (Table S3).
3.5. The Phenotype of the Mutant C. acetobutylicum glcG::int(1224)-glcCE::int(193) in Batch and Phosphate-Limited Continuous Fermentations
The mutant C. acetobutylicum glcG::int(1224)-glcCE::int(193) was grown in a fermenter with 0.75 L minimal medium as the working volume, and then the pH was set to 5.7. After overnight cultivation, the OD600 of the culture achieved 4.5, and then continuous fermentation started with phosphate-limited minimal medium at the dilution rate of 0.075 h−1. The growth result demonstrated that the mutant glcG::int(1224)-glcCE::int(193) was unable to achieve steady-state growth during continuous fermentation (Figure 5).
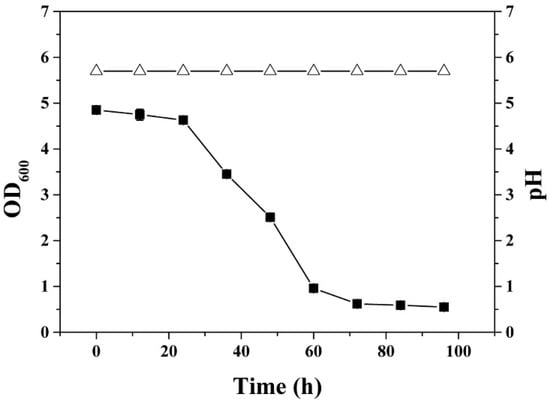
Figure 5.
Growth curve of the mutant C. acetobutylicum glcG::int(1224)-glcCE::int(193) in phosphate-limited continuous culture. (△), pH; and (■), OD600 of glcG::int(1224)-glcCE::int(193).
Under the same fermentation condition, wild type, mutants glcG::int(1224) and glcCE::int(193) consumed approximately 32 g/L glucose (the total glucose concentration was 40 g/L) and exhibited comparable steady-state growth during acidogenic grow in continuous fermentation (Figure 2 and Figure 4). We speculated that the double mutant is not able to assimilate enough glucose to support steady-state growth in continuous fermentation due to the inactivation of glcG and glcCE. Therefore, the cells were washed out of the fermenter in continuous culture.
Despite that the double mutant was not able to grow in the continuous fermentation, it was able to grow in batch culture with 60 g/L glucose as the sole carbon source. The highest optical density that the double mutant reached was approximately an OD600 of 5, which was lower than those of the wild type, mutants glcG::int(1224) and glcCE::int(193) with a highest OD600 of 7. At the end of batch fermentation, the glucose consumption and butanol production were 30% and 44% lower than the wild type, respectively, and the output of other products also decreased to different degrees (Figure 1 and Figure S3D).
These results also showed that the simultaneous inactivation of the primary glucose transporter and the cellobiose transporter impaired the growth of the strain on glucose. Importantly, the mutant glcG::int(1224)-glcCE::int(193) was still able to grow in the batch fermentation, implying that yet another PTS could transport glucose into cytoplasm except the glucose- and cellobiose-specific transporters.
3.6. Transcription Analysis of the Phosphotransferase Systems of the Mutant C. acetobutylicum glcG::int(1224)-glcCE::int(193) Compared to Wild Type
The simultaneous inactivation of gene glcG and glcCE genes resulted in weak growth of the double mutant, suggesting that a third glucose transporter exists in C. acetobutylicum. To test which PTS was able to transport glucose except for GlcG and GlcCE, a DNA microarray experiment was conducted to detect changes of the transcription level of all 13 PTS EII protein genes compared to the wild type in batch culture. Cells of the mutant C. acetobutylicum glcG::int(1224)-glcCE::int(193) and wild type were harvested after 12 h of batch culture and were used for RNA isolation and subsequent microarray experiments (Figure 1) to identify any compensatory adjustment of gene expression of PTS caused by the mutations of the glucose transporter (encoded by CAC0570) and the cellobiose transporter (encoded by CAC0383-CAC0386) in the double mutant.
The microarray data of all the 13 PTS genes is shown in Table 2. The result indicated that only three PTSs showed changes in the expression patterns of the double mutant as compared to wild type. Among these three PTSs, the expression of genes encoding a mannitol-specific transporter (CAC0154-CAC0156) was up-regulated by 2.8-fold, while the expression of genes encoding a putative fructose-specific transporter (CAC0233-CAC0234) and a putative N-acetylglucosamine-specific transporter (CAC1353-CAC1354) were down-regulated compared to those of the wild type (Table 2). This result suggested that the mannitol-specific transporter is possibly able to transport glucose, which has to be further studied.

Table 2.
Transcriptional profiles of all the PTS enzyme II protein genes of mutant C. acetobutylicum glcG::int(1224)-glcCE::int(193) compared to the wild type in the batch culture.
4. Conclusions
GlcG is the primary glucose transporter of C. acetobutylicum, whereas the cellobiose-specific transporter GlcCE is capable of transporting glucose into cytoplasm when GlcG is inactivated, revealing that it is a secondary glucose transporter. Each PTS is responsible for transporting a certain carbohydrate but may be multifunctional. Transcription analysis suggested that the PTSs encoded by CAC0154 and CAC2956 may be a tertiary glucose and secondary cellobiose transporter in C. acetobutylicum, respectively. On the basis of the data, we propose a model for glucose uptake in C. acetobutylicum (Figure 6), which provides a foundation to study the PTSs in this microorganism.
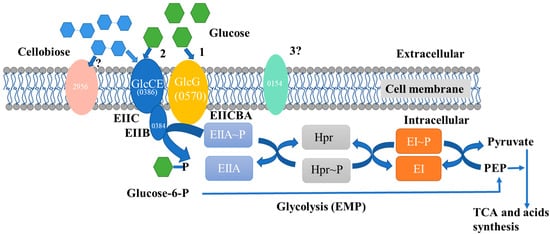
Figure 6.
Glucose uptake model for C. acetobutylicum. 1, 2 and 3: work order of the glucose transporters; PEP: phosphoenolpyruvate; EI: enzyme I; EII: enzyme II; Hpr: histidine-containing protein; and 0154, 0384, 0386, 0570 and 2956: the gene ID of C. acetobutylicum.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation9010064/s1, Figure S1:PCR verification of C. acetobutylicum glcG::int(1224), glcCE::int(193) and glcG::int(1224)-glcCE::int(193).; Figure S2: Southern hybridization to demonstrate the presence of intron in the constructed C. acetobutylicum mutants; Figure S3: Product concentrations of the C. acetobutylicum wild type (A) glcG::int(1224) (B), glcCE::int(193) (C), and glcG::int(1224)-glcCE::int(193) (D) in the batch culture with glucose as the sole carbon source; Figure S4: Product concentrations of the C. acetobutylicum wild type (A) and glcCE::int(193) (B) in the batch culture with cellobiose as the sole carbon source; Table S1: Strains, plasmids and primers used in this study; Table S2: Glucose consumption, cell density and product yields of the C. acetobutylicum wild type, mutants glcG::int(1224) and glcCE::int(193) in the phosphate-limited continuous fermentation; Table S3: Upregulated genes of C. acetobutylicum wild type and the mutant glcCE::int(193) grown on glucose and cellobiose via DNA microarray.
Author Contributions
Conceptualization, Z.L. and A.E.; Methodology, K.Z.; Validation, D.J.; Formal Analysis, M.W.; Investigation, L.G.; Resources, W.L.; Data Curation, Z.L.; Writing—Original Draft Preparation, K.Z.; Writing—Review and Editing, A.E.; Visualization, K.Z.; Supervision, Z.L.; Project Administration, A.E.; Funding Acquisition, W.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by the Shandong Energy Institute (Single-cell Center project), China Petrochemical Corporation (Sinopec, 20223702031404) and EU Marie Curie Seventh Framework Program under grant agreement no. 237942 (CLOSTNET).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The DNA microarray data had been submitted to EMBL-EBL with the Accession No. E-MTAB-12054.
Acknowledgments
The authors would like to thank Nigel P. Minton and John T. Heap from the University of Nottingham, UK, for kindly providing the ClosTron plasmids.
Conflicts of Interest
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
References
- Mitchell, W.J.; Tangney, M. Carbohydrate Uptake by the Phosphotransferase System and Other Mechanisums; Durre, P., Ed.; CRC Press: Boca Raton, FL, USA, 2005; pp. 155–175. [Google Scholar]
- Servinsky, M.D.; Kiel, J.T.; Dupuy, N.F.; Sund, C.J. Transcriptional analysis of differential carbohydrate utilization by Clostridium acetobutylicum. Microbiology 2010, 156, 3478–3491. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H., Jr.; Reizer, J. Proposed uniform nomenclature for the proteins and protein domains of the bacterial phosphoenolpyruvate: Sugar phosphotransferase system. J. Bacteriol. 1992, 174, 1433–1438. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Mitchell, W.J.; Tangney, M.; Blaschek, H.P. Evidence for the presence of an alternative glucose transport system in Clostridium beijerinckii NCIMB 8052 and the solvent-hyperproducing mutant BA101. Appl. Environ. Microbiol. 2005, 71, 3384–3387. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, W.J. The Phosphotransferase System in Solventogenic Clostridia. J. Mol. Microbiol. Biotechnol. 2015, 25, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Gu, Y.; Ning, Y.; Yang, Y.; Mitchell, W.J.; Jiang, W.; Yang, S. Confirmation and elimination of xylose metabolism bottlenecks in glucose phosphoenolpyruvate-dependent phosphotransferase system-deficient Clostridium acetobutylicum for simultaneous utilization of glucose, xylose, and arabinose. Appl. Environ. Microbiol. 2011, 77, 7886–7895. [Google Scholar] [CrossRef]
- Deutscher, J. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2008, 11, 87–93. [Google Scholar] [CrossRef]
- Wang, X.; Xia, K.; Yang, X.; Tang, C. Growth strategy of microbes on mixed carbon sources. Nat. Commun. 2019, 10, 1279. [Google Scholar] [CrossRef]
- Viana, R.; Monedero, V.; Dossonnet, V.; Vadeboncoeur, C.; Perez-Martinez, G.; Deutscher, J. Enzyme I and HPr from Lactobacillus casei: Their role in sugar transport, carbon catabolite repression and inducer exclusion. Mol. Microbiol. 2000, 36, 570–584. [Google Scholar] [CrossRef]
- Doucette, C.D.; Schwab, D.J.; Wingreen, N.S.; Rabinowitz, J.D. Alpha-ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat. Chem. Biol. 2011, 7, 894–901. [Google Scholar] [CrossRef]
- Magasanik, B. The Regulation of Nitrogen-Utilization in Enteric Bacteria. J. Cell Biochem. 1993, 51, 34–40. [Google Scholar] [CrossRef]
- Ninfa, A.J.; Jiang, P. PII signal transduction proteins: Sensors of alpha-ketoglutarate that regulate nitrogen metabolism. Curr. Opin. Microbiol. 2005, 8, 168–173. [Google Scholar] [CrossRef]
- Lee, C.R.; Cho, S.H.; Yoon, M.J.; Peterkofsky, A.; Seok, Y.J. Escherichia coli enzyme IIANtr regulates the K+ transporter TrkA. Proc. Natl. Acad. Sci. USA 2007, 104, 4124–4129. [Google Scholar] [CrossRef]
- Choi, J.; Ryu, S. Regulation of Iron Uptake by Fine-Tuning the Iron Responsiveness of the Iron Sensor Fur. Appl. Environ. Microbiol. 2019, 85, e03026-18. [Google Scholar] [CrossRef]
- Luttmann, D.; Heermann, R.; Zimmer, B.; Hillmann, A.; Rampp, I.S.; Jung, K.; Gorke, B. Stimulation of the potassium sensor KdpD kinase activity by interaction with the phosphotransferase protein IIA(Ntr) in Escherichia coli. Mol. Microbiol. 2009, 72, 978–994. [Google Scholar] [CrossRef]
- Yoo, W.; Kim, D.; Yoon, H.; Ryu, S. Enzyme IIA(Ntr) Regulates Salmonella Invasion Via 1,2-Propanediol And Propionate Catabolism. Sci. Rep. 2017, 7, 44827. [Google Scholar] [CrossRef]
- Lim, S.; Seo, H.S.; Jeong, J.; Yoon, H. Understanding the multifaceted roles of the phosphoenolpyruvate: Phosphotransferase system in regulation of Salmonella virulence using a mutant defective in ptsI and crr expression. Microbiol. Res. 2019, 223–225, 63–71. [Google Scholar] [CrossRef]
- Zhi, Y.; Lin, S.M.; Jang, A.Y.; Ahn, K.B.; Ji, H.J.; Guo, H.C.; Lim, S.; Seo, H.S. Effective mucosal live attenuated Salmonella vaccine by deleting phosphotransferase system component genes ptsI and crr. J. Microbiol. 2019, 57, 64–73. [Google Scholar] [CrossRef]
- Dalebroux, Z.D.; Swanson, M.S. ppGpp: Magic beyond RNA polymerase. Nat. Rev. Microbiol. 2012, 10, 203–212. [Google Scholar] [CrossRef]
- Karstens, K.; Zschiedrich, C.P.; Bowien, B.; Stulke, J.; Gorke, B. Phosphotransferase protein EIIA(Ntr) interacts with SpoT, a key enzyme of the stringent response, in Ralstonia eutropha H16. Microbiology 2014, 160, 711–722. [Google Scholar] [CrossRef]
- Lee, J.; Park, Y.H.; Kim, Y.R.; Seok, Y.J.; Lee, C.R. Dephosphorylated NPr is involved in an envelope stress response of Escherichia coli. Microbiology 2015, 161, 1113–1123. [Google Scholar] [CrossRef]
- Wu, S.Y.; Yu, P.L.; Wheeler, D.; Flint, S. Transcriptomic study on persistence and survival of Listeria monocytogenes following lethal treatment with nisin. J. Glob. Antimicrob. Resist. 2018, 15, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Nolling, J.; Breton, G.; Omelchenko, M.V.; Makarova, K.S.; Zeng, Q.; Gibson, R.; Lee, H.M.; Dubois, J.; Qiu, D.; Hitti, J.; et al. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 2001, 183, 4823–4838. [Google Scholar] [CrossRef] [PubMed]
- Tangney, M.; Mitchell, W.J. Characterisation of a glucose phosphotransferase system in Clostridium acetobutylicum ATCC 824. Appl. Microbiol. Biotechnol. 2007, 74, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Boos, W.; Shuman, H. Maltose/maltodextrin system of Escherichia coli: Transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 1998, 62, 204–229. [Google Scholar] [CrossRef] [PubMed]
- Steinsiek, S.; Bettenbrock, K. Glucose transport in Escherichia coli mutant strains with defects in sugar transport systems. J. Bacteriol. 2012, 194, 5897–5908. [Google Scholar] [CrossRef]
- Hunter, I.S.; Kornberg, H.L. Glucose transport of Escherichia coli growing in glucose-limited continuous culture. Biochem. J. 1979, 178, 97–101. [Google Scholar] [CrossRef]
- Deutscher, J.; Francke, C.; Postma, P.W. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria (vol 70, pg 939, 2006). Microbiol. Mol. Biol. Rev. 2008, 72, 555. [Google Scholar] [CrossRef]
- Amador-Noguez, D.; Brasg, I.A.; Feng, X.J.; Roquet, N.; Rabinowitz, J.D. Metabolome remodeling during the acidogenic-solventogenic transition in Clostridium acetobutylicum. Appl. Environ. Microbiol. 2011, 77, 7984–7997. [Google Scholar] [CrossRef]
- Monot, F.; Martin, J.R.; Petitdemange, H.; Gay, R. Acetone and Butanol Production by Clostridium acetobutylicum in a Synthetic Medium. Appl. Environ. Microbiol. 1982, 44, 1318–1324. [Google Scholar] [CrossRef]
- Alsaker, K.V.; Papoutsakis, E.T. Transcriptional program of early sporulation and stationary-phase events in Clostridium acetobutylicum. J. Bacteriol. 2005, 187, 7103–7118. [Google Scholar] [CrossRef]
- Durre, P. Biobutanol: An attractive biofuel. Biotechnol. J. 2007, 2, 1525–1534. [Google Scholar] [CrossRef]
- Jones, D.T.; Woods, D.R. Acetone-butanol fermentation revisited. Microbiol. Rev. 1986, 50, 484–524. [Google Scholar] [CrossRef]
- Janssen, H.; Doring, C.; Ehrenreich, A.; Voigt, B.; Hecker, M.; Bahl, H.; Fischer, R.J. A proteomic and transcriptional view of acidogenic and solventogenic steady-state cells of Clostridium acetobutylicum in a chemostat culture. Appl. Microbiol. Biotechnol. 2010, 87, 2209–2226. [Google Scholar] [CrossRef]
- Bahl, H.; Andersch, W.; Gottschalk, G. Continuous Production of Acetone and Butanol by Clostridium acetobutylicum in a 2-Stage Phosphate Limited Chemostat. Eur. J. Appl. Microbiol. 1982, 15, 201–205. [Google Scholar] [CrossRef]
- Grimmler, C.; Janssen, H.; Krausse, D.; Fischer, R.J.; Bahl, H.; Durre, P.; Liebl, W.; Ehrenreich, A. Genome-Wide Gene Expression Analysis of the Switch between Acidogenesis and Solventogenesis in Continuous Cultures of Clostridium acetobutylicum. J. Mol. Microbiol. Biotechnol. 2011, 20, 1–15. [Google Scholar] [CrossRef]
- Ehrenreich, A. DNA microarray technology for the microbiologist: An overview. Appl. Microbiol. Biotechnol. 2006, 73, 255–273. [Google Scholar] [CrossRef]
- Wiesenborn, D.P.; Rudolph, F.B.; Papoutsakis, E.T. Thiolase from Clostridium acetobutylicum ATCC 824 and Its Role in the Synthesis of Acids and Solvents. Appl. Environ. Microbiol. 1988, 54, 2717–2722. [Google Scholar] [CrossRef]
- Heap, J.T.; Kuehne, S.A.; Ehsaan, M.; Cartman, S.T.; Cooksley, C.M.; Scott, J.C.; Minton, N.P. The ClosTron: Mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods 2010, 80, 49–55. [Google Scholar] [CrossRef]
- Heap, J.T.; Pennington, O.J.; Cartman, S.T.; Carter, G.P.; Minton, N.P. The ClosTron: A universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 2007, 70, 452–464. [Google Scholar] [CrossRef]
- Cooksley, C.M.; Zhang, Y.; Wang, H.; Redl, S.; Winzer, K.; Minton, N.P. Targeted mutagenesis of the Clostridium acetobutylicum acetone-butanol-ethanol fermentation pathway. Metab. Eng. 2012, 14, 630–641. [Google Scholar] [CrossRef]
- Sambrook, J.; Russel, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory, Ed.; Cold Spring Harbor: Long Island, NY, USA, 2001. [Google Scholar]
- Fischer, R.J.; Oehmcke, S.; Meyer, U.; Mix, M.; Schwarz, K.; Fiedler, T.; Bahl, H. Transcription of the pst operon of Clostridium acetobutylicum is dependent on phosphate concentration and pH. J. Bacteriol. 2006, 188, 5469–5478. [Google Scholar] [CrossRef] [PubMed]
- Janssen, H.; Grimmler, C.; Ehrenreich, A.; Bahl, H.; Fischer, R.J. A transcriptional study of acidogenic chemostat cells of Clostridium acetobutylicum—Solvent stress caused by a transient n-butanol pulse. J. Biotechnol. 2012, 161, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, D.; Honicke, D.; Ehrenreich, A.; Schmidt, M.; Weuster-Botz, D.; Bahl, H.; Lutke-Eversloh, T. Modifying the product pattern of Clostridium acetobutylicum: Physiological effects of disrupting the acetate and acetone formation pathways. Appl. Microbiol. Biotechnol. 2012, 94, 743–754. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Z.; Xin, X.; Bai, F.; Xue, C. Synergetic Engineering of Central Carbon, Energy, and Redox Metabolisms for High Butanol Production and Productivity by Clostridium acetobutylicum. Ind. Eng. Chem. Res. 2020, 59, 17137–17146. [Google Scholar] [CrossRef]
- Maier, A.; Volker, B.; Boles, E.; Fuhrmann, G.F. Characterisation of glucose transport in Saccharomyces cerevisiae with plasma membrane vesicles (countertransport) and intact cells (initial uptake) with single Hxt1, Hxt2, Hxt3, Hxt4, Hxt6, Hxt7 or Gal2 transporters. FEMS Yeast Res. 2002, 2, 539–550. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).